Abstract
Ribosome biogenesis has been associated with regulation of cell growth and cell division, but the molecular mechanisms that integrate the effect of ribosome biogenesis on these processes in mammalian cells remain unknown. To study the effect of impaired ribosome functions in vivo, we conditionally deleted one or two alleles of the 40S ribosomal protein S6 gene in T cells in the mouse. While complete deletion of S6 abrogated T-cell development, hemizygous expression did not have any effect on T-cell maturation in the thymus, but inhibited the accumulation of T cells in the spleen and lymph nodes, as a result of their decreased survival in the peripheral lymphoid organs. Additionally, TCR-mediated stimulation of S6-heterozygous T cells induced a normal increase in their size, but cell cycle progression was impaired. Genetic inactivation of p53 tumor suppressor rescued development of S6-homozygous null thymocytes and proliferative defect of S6-heterozygous T cells. These results demonstrate the existence of a p53-dependent checkpoint mechanism that senses changes in the fidelity of the translational machinery to prevent aberrant cell division or eliminate defective T cells in vivo. Failure to activate this checkpoint response could potentially lead to a development of pathological processes such as tumors and autoimmune diseases.
Keywords: S6 ribosomal protein, ribosome biogenesis, cell growth, cell proliferation, checkpoint
Cell growth (increase in cell mass) and cell division (increase in cell number) are separable processes that are coordinated in most dividing cells, probably to ensure that there are sufficient cellular components for survival and normal functioning of daughter cells (Conlon and Raff 1999). Although earlier studies suggested a tight coordination between cell growth and division, recent analyses in higher organisms very often reveal their loose coordination (Hartwell 1971; Nurse et al. 1976; Zetterberg and Larsson 1991; Neufeld et al. 1998; Stocker and Hafen 2000; Weinkove and Leevers 2000; Conlon et al. 2001; Du and Stillman 2002; Jorgensen and Tyers 2004; Sveiczer et al. 2004; O'Farrell et al. 2004).
Over the last few decades, major advances have been made in understanding the machinery that controls progression through the cell division cycle, in particular identifying cell cycle regulatory genes, including cyclins, cyclin-dependent kinases, and their inhibitors, as well as genes that monitor the distinct steps of cell cycle progression, termed cell cycle checkpoint genes (Elledge 1996; Sherr and Roberts 1999; Nurse 2000). A cell cycle checkpoint is defined as a regulatory pathway that controls the order and timing of cell cycle transitions, ensuring that critical events such as DNA replication, chromosome segregation, and probably many other events are completed with high fidelity (Nurse 2000; Jones et al. 2005). Mechanistically, cell cycle checkpoints establish the relationship between two unrelated biochemical events. The first represents the event in which a lesion takes place, and the second prevents the cell from progressing through the cell cycle until the first event has been repaired. The establishment of this relationship requires the presence of three components: (1) a defect (the lesion); (2) a sensor, which detects the defect; and (3) a target, a cell cycle regulator. Failure to activate these checkpoints allows a cell to divide when DNA is damaged or when chromosomes are incorrectly partitioned, leading to genomic instability. This is essential for the generation of cancer, as evidenced by the high frequency of mutations of cell cycle checkpoint genes in human cancer (Nurse 2000).
On the other hand, it is poorly understood what cell growth really is and how it is regulated (Conlon and Raff 1999). Cell growth is regulated by growth-factor and nutrient activation of several signaling pathways that control anabolic processes (Schmelzle and Hall 2000; Volarevic and Thomas 2001; Hafen 2004; Holland et al. 2004). Genetic analyses in Drosophila melanogaster had discovered that Ras, Myc, and PI3K-TOR signaling cascades regulate cell growth (Johnston et al. 1999; Weinkove et al. 1999; Prober and Edgar 2000; Johnston and Gallant 2002). It has been proposed that protein synthesis may be a key determinant of cell growth (Pardee 1989). Consistent with this, the Myc, Ras, and PI3K-TOR signaling pathways increase the overall rate of protein synthesis by stimulating either the rates of translation initiation and elongation or ribosome biogenesis (Holland et al. 2004). It has been suggested that deregulation of the molecular mechanisms controlling cell growth results in cells of altered size and can contribute to a variety of pathological conditions, including cancer (Holland et al. 2004). More than 30 years ago, Harvey Lodish postulated that the spectrum of translated mRNA varies with the overall rate of protein synthesis (Lodish 1974). Messenger RNAs that have low affinity for translational machinery are outcompeted with messages with high affinity when the rate of translation is reduced, whereas low- and high-affinity mRNAs are translated when protein synthesis becomes up-regulated, as in the case of hyperactivation of growth stimulating pathways (Rajasekhar et al. 2003; Mamane et al. 2004). The fact that many mRNAs with low affinity for the translational machinery encode many oncoproteins, growth factors, survival factors, and cell cycle regulators, suggests that a non-physiological increase in the rate of translation, as the result of deregulation of growth signaling pathways, and could cause malignant transformation. This notion is further supported by the discovery that rapamycin, an effective anticancer drug, inhibits a key regulator of protein synthetic machinery and cell growth, mTORC1 (Holland et al. 2004).
As mentioned above, the synthesis of proteins during cell growth and proliferation requires ribosome biogenesis (Thomas 2000; Warner et al. 2001). Genes that control ribosome biogenesis and protein translation have been identified in yeast as critical regulators of cell growth and cell size (Jorgensen et al. 2002; Jorgensen and Tyers 2004; Zhang et al. 2002). Ribosome biogenesis is one of the major energy-consuming processes in proliferating cells (Hadjiolov 1985; Lewis and Tollervey 2000; Thomas 2000; Warner et al. 2001; Fatica and Tollervey 2002; Moss 2004).
Given the enormous energy investment in ribosome biogenesis, its importance to cell growth and proliferation, and the observation that some human diseases are caused by defects in ribosome biogenesis, we and others had hypothesized that the mechanisms must have evolved to sense the fidelity of this process (Draptchinskaia et al. 1999; Volarevic et al. 2000; Ruggero and Pandolfi 2003; Holland et al. 2004; Olson 2004).
To discover such molecular mechanisms in a genetically defined in vivo mouse model, we induced a defect in this process by deleting one or both alleles of S6 ribosomal protein gene in the thymus by using CD4-Cre transgenic mice (Sawada et al. 1994; Wolfer et al. 2001). T lymphocytes are ideal for studying the relationship between ribosome biogenesis, cell growth, and proliferation since the initial response of T cells to antigenic stimulation is transformation of small resting cells into large blasts, followed by cell division (Rathmell et al. 2000; Frauwirth et al. 2002). Additionally, a long-term consequence of a defect in ribosome biogenesis could be followed in several experimental paradigms in vivo. In this report we show that in T-cell receptor (TCR)-stimulated T cells, fidelity of ribosome biogenesis is monitored by a p53-dependent checkpoint regulatory pathway that inhibits cell division or induces apoptosis of potentially defective cells.
Results
Specific deletion of the S6 gene in the thymus
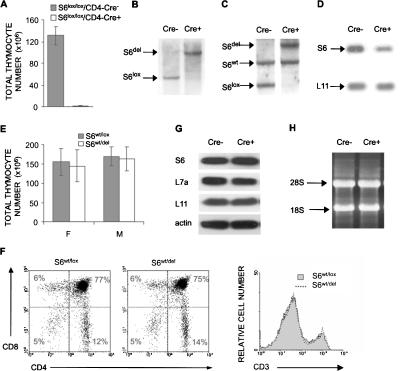
To generate a model for studying the effects of decreased S6 expression on development and function of T cells, we decided to delete both alleles of the S6 ribosomal gene in the thymus by using mice that express the Cre recombinase under the control of a CD4 promoter (Sawada et al. 1994; Wolfer et al. 2001). The CD4 promoter directs the expression of Cre recombinase in CD4+ CD8+ thymocytes, and, to a lesser extent, in CD4- CD8- thymocytes. The thymi from 57% of S6lox/lox/CD4-Cre+ mice could not be recovered (n = 23), and ones that were recovered manifested on average a 100-fold decreases in the cell number relative to S6lox/lox/CD4-Cre- control mice (Fig. 1A). Southern blot analysis of genomic DNA from pooled thymi showed that deletion of S6 is complete (Fig. 1B). These data demonstrate that S6 is absolutely required for T-cell development.
Figure 1.
Conditional deletion of the S6 gene in the thymus. (A) Total number of thymocytes in S6lox/lox/CD4-Cre- and S6lox/lox/CD4-Cre+ mice (n = 14 and 10, respectively). (B) Southern blot analysis of genomic DNA from pooled thymi of S6lox/lox/CD4-Cre+ and S6lox/lox/CD4-Cre- mice using S6 intron-specific probe. The positions of DNA fragments representing S6del and S6lox are shown. (C) Southern blot analysis of genomic DNA from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymi using S6 intron-specific probe. The positions of DNA fragments representing S6del, S6lox, and S6wt alleles are indicated. (D) Northern blot analysis of total RNA from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymi using S6 cDNA as a probe. The L11 cDNA probe was used as a control for loading the same quantity of total RNA on the gel. (E) Total number of thymocytes from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ mice. (F) S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ thymocytes were stained with CD4 and CD8 antibodies (left panel) or anti-CD3 antibodies (right panel), and analyzed by FACS. Cell surface markers are shown as coordinates. The numbers in the quadrant refer to the percentage of a particular T-cell population of live cells. (G) The protein expression of ribosomal proteins S6, L7a, and L11 in thymocytes from S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- mice was analyzed by Western blot. Reprobing with antibodies to actin served as a loading control. (H) Total RNA from 15 × 106 S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- thymocytes was separated in agarose gel containing formaldehyde, transferred to a nitrocellulose membrane, and stained with ethidium bromide. Positions of 28S and 18S rRNAs are shown.
Next, we decided to conditionally delete only one S6 allele in the thymus. The extent of S6 deletion in thymi from S6wt/lox/CD4-Cre+ mice was determined by Southern blot analysis. The blot revealed the expected 9.1-kb band, representing the S6 fragment after Cre-mediated recombination, and the absence of the 4.6-kb S6lox band, showing that deletion of the floxed S6 allele was complete (Fig. 1C). There was no effect on the wild-type S6 allele. To examine the effect of a single S6 allele deletion on S6 mRNA expression, total RNA from S6wt/lox/CD4-Cre+ thymi was analyzed by Northern blot using the S6-specific probe. S6wt/lox thymi contained easily detectable S6 mRNA. S6wt/del thymi showed ∼50% decrease in S6 mRNA expression relative to control thymi (Fig. 1D). Therefore, the remaining S6 allele cannot compensate for the loss of the other allele at the level of S6 mRNA in thymocytes. To examine the consequences of a conditional S6wt/del mutation on thymocyte development, we purified thymocytes from S6wt/lox/CD4-Cre+ mice, and determined their cellularity and subset representation. The total number of thymocytes in 4-wk-old S6wt/lox/CD4-Cre+ mice was similar to that of S6wt/lox/CD4-Cre- controls (Fig. 1E). FACS analysis of cells from S6wt/del thymi revealed that T-cell populations defined by CD4, CD8, and CD3 markers were also normal (Fig. 1F). Also, analysis of mutant CD4- CD8-, CD4+ CD8+, CD4+, and CD8+ populations of thymocytes with FACS confirmed that their size as measured by forward light scatter (FSC) was similar to the size of corresponding populations of control thymocytes (data not shown). In agreement with normal phenotype, the protein expression of S6 in S6wt/del thymi was indistinguishable from the control, suggesting that one S6 allele is sufficient to support normal S6 protein production during T-cell development (Fig. 1G). Consistent with these results is the observation that the amount of mature 18S and 28S rRNAs in mutant and control thymocytes was indistinguishable (Fig. 1H). These results suggest that T-cell development is unperturbed by conditional deletion of one S6 allele.
Effects of S6 heterozygous deletion in peripheral lymphoid organs
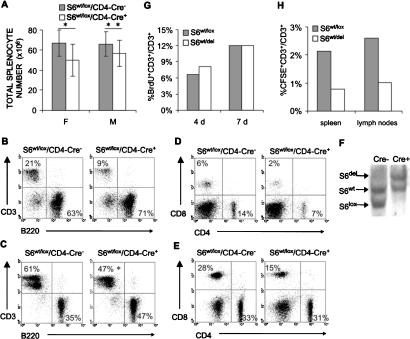
We next wanted to determine if deletion of one S6 allele has an effect on T-cell peripheral compartments in spleen and lymph nodes. The total number of splenocytes was decreased by 14% in male and 23% in female 4-wk-old S6wt/lox/Cre+ mice compared with S6wt/lox/Cre- controls (Fig. 2A). The percentage of T cells in the spleen was reduced from 21% in control mice to 9% in S6wt/lox/Cre+ mice (Fig. 2B; data not shown). When total splenic cell number was taken into account, S6wt/lox/CD4-Cre+ mice exhibited an even larger decrease of ∼60%-65%in the total number of T cells.
Figure 2.
Decreased number of S6wt/del T cells in the spleen and lymph nodes is a consequence of their decreased survival. (A) Total cell number in spleens from female (F) and male (M) S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ mice (n = 10, for each group). Error bars represent standard deviation. For females, (*) p < 0.05 versus S6wt/lox/CD4-Cre- splenocytes; and for males, (**) p > 0.05 versus S6wt/lox/CD4-Cre- splenocytes (Mann Whitney U-test). (B,D) FACS analysis of cells from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ spleens. (C,E) FACS analysis of cells from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ lymph nodes (n = 35, for each group). (*) p < 0.001 versus S6wt/lox/CD4-Cre- CD3+ cells (t-test). Genotypes are as indicated above the panels. Cell surface markers are shown as coordinates. The numbers in quadrants refer to percentage of respective cell population of live cells. (F) Southern blot analysis of genomic DNA from S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ T cells purified from lymph nodes of 8-wk-old mice using the S6 intron-specific probe. (G) Six-week-old S6wt/lox/CD4-Cre- and S6wt/lox/CD4-Cre+ mice were daily injected intraperitoneally with 100 μg/g of body weight 5-BrdU (Sigma) in 200 μL of PBS during the indicated time periods; 5-BrdU incorporation into DNA was determined by flow cytometry. The percentages of 5-BrdU-positive lymph node T cells are shown. The experiment is a representative of four independent experiments. (H) CFSE-labeled S6wt/lox/CD4-Cre+ or S6wt/lox/CD4-Cre+ T cells (10 × 106) were transferred into wild-type recipients by intravenous injection, and the percentage of CFSE-labeled T cells was determined in spleens. The total number of T cells in different recipients was similar. The experiment is a representative of three independent experiments.
The percentage of T cells in lymph nodes from S6wt/lox/CD4-Cre+ mice was reduced by ∼25% (Fig. 2C; data not shown). Interestingly, while a decreased number of T cells in the spleen resulted from loss of both CD4 and CD8 T-cell subsets (Fig. 2D), a decrease in the number of T cells in lymph nodes was mainly due to the reduction in CD8 cells (Fig. 2E). Similar reductions in the number of splenic and lymph node T cells were also observed in 1-, 2-, 3-, 5-, 6-, 7-, and 8-wk-old S6wt/lox/CD4-Cre+ mice (data not shown).
To exclude the possibility that the remaining T cells in the periphery are those in which Cre-mediated recombination was ineffective, Southern blot analysis of S6 alleles in purified T cells from lymph nodes was performed, and it revealed that all S6wt/lox/CD4-Cre+ lymph node T cells have an S6wt/del genotype (Fig. 2F). The similar results were obtained when genomic DNA from purified splenic T cells was analyzed for the presence of S6 alleles (data not shown). Forward light scatter analysis (FSC) of CD3, CD4, and CD8 positive cells from spleen and lymph nodes of S6wt/del T cells revealed their normal size (data not shown). These results indicate that conditional deletion of one allele of S6 has a substantial effect on the number of mature T cells in the periphery, while there was no effect on the cell size of resting T cells. To determine whether the number of mutant T cells in the secondary lymphoid organs is affected by homeostatic proliferation, we assessed the proliferative activity of T cells from S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- mice following in vivo labeling with BrdU for 7 d, starting at 6 wk of age. A similar percentage of labeled T cells was seen in both groups, indicating that the observed phenotype is not the result of slower homeostatic proliferation (Fig. 2G). Next, we set out to determine S6wt/del T-cell survival in vivo. Resting S6wt/del and S6wt/lox T cells were labeled with 5-6-carboxyfluorescein diacetate succinimidyl ester (CFSE) and then separately transferred into nonradiated syngenic wild-type hosts. The percentage of CFSE-labeled T cells was determined in the spleen and lymph nodes after 3 d by FACS analysis. The fraction of CFSE-labeled T cells in both spleen and lymph nodes was >50% lower in mice receiving S6wt/del than in mice receiving S6wt/lox T cells, indicating that mutant T cells have a shorter life span (Fig. 2H). The expression of the Eμ-bcl-2-36 transgene in S6wt/del T cells partially rescued their decreased number in the spleen, suggesting that increased Bcl-2-dependent apoptosis contributes to the phenotype (Strasser et al. 1994; Supplementary Fig. 1).
Inactivation of one S6 allele inhibits ribosome biogenesis, but not T-cell growth
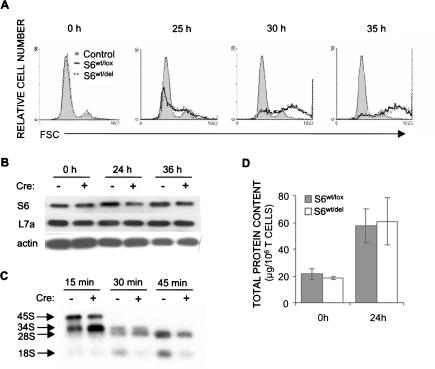
Following T-cell receptor stimulation, T cells increase in size before they enter the S phase of the cell cycle. To determine if S6wt/del T cells can mount this response, we stimulated these cells with anti-CD3 and anti-CD28 antibodies and measured forward light scatter parameter by flow cytometry at 25, 30, and 35 h following stimulation. There was no difference in kinetics or extent of cell growth between control and mutant T cells (Fig. 3A).
Figure 3.
T-cell receptor-stimulated ribosome biogenesis is decreased in S6wt/del T cells, but their growth is normal. (A) FSC analysis of S6wt/lox and S6wt/del lymph node T cells stimulated with soluble 1 μg/mL anti-CD3 and 0.1 μg/mL anti-CD28 in vitro was used to determine their size. (B) The protein expression of ribosomal proteins S6 and L7a in lysates from unstimulated (0 h) and stimulated (24 and 36 h) lymph node S6wt/del and S6wt/lox T cells was analyzed by Western blot. Reprobing with antibodies to actin served as a loading control. (C) To gain insight into rRNA processing, S6wt/del and S6wt/lox T cells were stimulated for 20 h with TCR and CD28 antibodies, pulse-labeled with L-[methyl-3H]methionine for 30 min, and chased in nonradioactive medium for the indicated time. Total RNA normalized to equal cell number was resolved on a formaldehyde-agarose gel, transferred to a nylon membrane, and visualized by autoradiography. The positions of major precursors and 28S and 18S rRNA species are indicated on the left. The experiment is a representative of five independent experiments. (D) Total cellular protein content per 1 × 106 S6wt/lox or S6wt/del lymph node T cells before stimulation and after 24 h of stimulation with antibodies against TCR and CD28. The experiment is a representative of six independent experiments.
The effect of the S6wt/del mutation on the protein expression of S6 in resting and stimulated T cells was determined by Western blot (Fig. 3B). S6wt/del and S6wt/lox T cells were analyzed unstimulated (0-h time point) or were analyzed following stimulation with anti-CD3 and anti-CD28 antibodies for 24 or 36 h. In unstimulated mutant and control T cells, the levels of S6 protein were indistinguishable, suggesting that one S6 allele is sufficient to support normal S6 protein production under such conditions. The S6 protein levels were substantially lower in stimulated S6wt/del T cells than in control, at 24 and 36 h following stimulation. In contrast, ribosomal protein L7a was produced at similar levels in stimulated mutant and control T cells. Since S6 ribosomal protein is an essential 40S ribosomal protein, and T-cell activation is accompanied by significant ribosome biogenesis, we wanted to determine whether deletion of one S6 allele in T cells affects this process (Fig. 3C). S6wt/del T cells were first stimulated with the combination of anti-CD3 and anti-CD28 antibodies for 20 h, then labeled with L-[methyl-3H]-methionine for 30 min and chased for indicated time periods. After 15 min, presumed 34S rRNA precursor accumulated in higher amounts in the mutant than in control T cells, suggesting an rRNA processing defect. At the 30-min time point, the S6wt/del mutation reduced 18S but not 28S rRNA labeling, while at the 45-min time point, a decrease in the amount of both 18S and 28S rRNA was observed in S6wt/del T cells, which closely reflects the amount of 40S and 60S ribosomal subunits, respectively. This is consistent with the decreased S6 protein levels in stimulated T cells shown in Figure 3B. Thus, conditional deletion of one S6 allele in stimulated T cells decreases the amount of both, 18S and 28S rRNA. Interestingly, the synthesis of ribosomal protein L7a continues normally.
The fact that TCR-stimulated cell growth was normal in S6wt/del T cells, even though ribosome biogenesis is affected, led us to investigate their general protein synthetic capacity (Fig. 3D). We compared the total cellular protein content per 1 × 106 of S6wt/lox and S6wt/del T cells prior to TCR/CD28 activation and 24 h after stimulation. There was no significant difference in the total cellular protein content in unstimulated or stimulated mutant and control T cells. These results suggest that a smaller number of ribosomes in S6wt/del T cells is sufficient for normal TCR/CD28-stimulated cell growth.
Analysis of proliferative defects in S6 heterozygous T cells
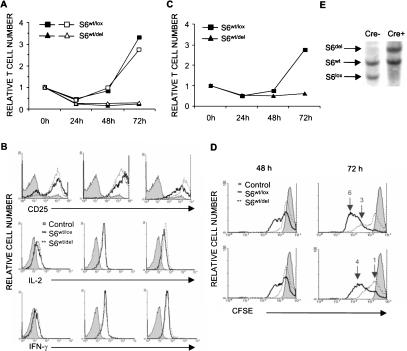
Although deletion of one allele of the S6 gene had no effect on TCR-stimulated cell growth, it was interesting to test the effects of an S6wt/del mutation on T-cell proliferation. Purified lymph node T cells from wild-type and mutant mice were stimulated with 0.1 μg/mL (open symbols) or 1 μg/mL (closed symbols) anti-CD3 and with a constant concentration of anti-CD28 antibodies (0.1 μg/mL) in vitro, and the relative number of live T cells was determined at 24, 48, and 72 h thereafter (Fig. 4A). The number of both control and mutant T cells decreased during the first 24 h of stimulation. There was a small increase in the number of S6wt/lox T cells at 48 h, while the dramatic increase was observed after 72 h of stimulation with either concentration of anti-CD3 antibodies. In contrast, no change in the number of S6wt/del T cells was seen after the initial drop that was observed at 24 h after stimulation.
Figure 4.
Stimulation of S6wt/del T cells does not increase their cell number. (A) Relative number of S6wt/lox and S6wt/del T cells stimulated with either 0.1 μg (open symbols) or 1 μg/mL (closed symbols) of anti-CD3 and fixed concentration of anti-CD28 antibodies (0.1 μg/mL) during 72 h in vitro. The experiment is a representative of 15 independent experiments. (B) FACS analysis of expression levels of CD25, intracellular IL-2, and IFN-γ in S6wt/lox and S6wt/del lymph node T cells stimulated for the indicated period of time in vitro. (C) Relative number of S6wt/lox and S6wt/del lymph node T cells stimulated with soluble anti-CD3 antibodies and saturating concentration of IL-2 containing MLA-144 supernatant (1:2 dilution). The experiment is a representative of three independent experiments. (D) FACS analysis of lymph node T cells that were stimulated with different concentrations of anti-CD3 (1 μg/mL, upper panel; 0.1 μg/mL, lower panel) and anti-CD28 antibodies (0.1 μg/mL) and labeled with CFSE. The filled area represents CFSE labeling of T cells before they divided for the first time. The number of cell divisions is indicated above the peaks. (E) Southern blot analysis of genomic DNA from S6wt/lox and S6wt/del lymph node T-cell cultures at 72 h following stimulation.
To understand how the S6wt/del mutation impairs TCR- and CD28-induced proliferation, we first analyzed activation events that precede cell cycle progression. Since proliferation of T cells is mediated by the interaction of IL-2 with the IL-2 receptor, we first analyzed the kinetics of their expression levels following stimulation with anti-TCR and anti-CD28 antibodies by flow cytometry (Fig. 4B). Despite dramatic differences in cell numbers after 48 or 72 h of stimulation, expression of IL-2 receptor and IL-2 following TCR and CD28 stimulation of T cells of the different genotypes was similar. Furthermore, TCR and CD28 stimulation induced equivalent increases in the expression of the activation marker CD69 and IFN-γ in mutant and control T cells (Fig. 4B). We also examined proliferation of mutant and control T cells in response to the exogenous IL-2 (Fig. 4C). Both cell types were stimulated with anti-CD3 antibodies in the absence of CD28 costimulation. Such treatment induced a similar level of the IL-2 receptor expression but did not induce IL-2 production and T-cell proliferation (data not shown). When a saturating concentration of IL-2 containing MLA-144 supernatant was added to these cultures, increases in the cell number were seen in S6wt/lox but not in S6wt/del T cells. These results suggest that proximal TCR signaling pathways are similar in S6wt/lox and S6wt/del T cells and that the S6wt/del mutation interferes with T-cell proliferation downstream of the IL-2 receptor.
The inability of stimulated S6wt/del T cells to proliferate could arise from impaired cell division, increased cell death, or both. We first analyzed division of individual S6wt/lox and S6wt/del T cells using CFSE labeling. This method is based on the approximately twofold decreases in CFSE fluorescence after each cell division. Lymph node T cells of the indicated genotype were labeled with CFSE and stimulated, as before, with different concentrations of anti-TCR and anti-CD28 antibodies for 48 or 72 h, and CFSE fluorescence was determined by flow cytometry (Fig. 4D). The majority of stimulated control cells progressed trough three to four divisions after 48 h. In contrast, the majority of mutant T cells did not proliferate at all during the same period, and those that did progressed through only one division. Analysis of CFSE fluorescence at 72 h following stimulation revealed that the proportion of mutant T cells remained undivided, while the rest went through a small number of divisions. In contrast, all control T cells progressed through three to six divisions. These data indicate that deletion of one S6 allele dramatically decreases both the percentage of T cells that proliferates in response to stimulation with anti-CD3 and anti-CD28 and the extent of proliferation. Southern blot analysis of DNA isolated from mutant T-cell culture at 72 h following stimulation confirmed the presence of S6wt/del T cells and the absence of T cells that did not delete one floxed S6 allele (Fig. 4E).
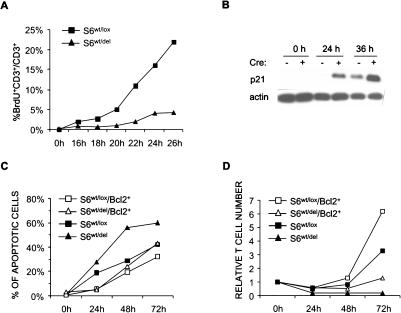
To determine why the doubling time of S6wt/del T cells is increased, we determined the percentage of mutant and control T cells that enter S phase at 2-h intervals, from 16 to 26 h after stimulation, by measuring BrdU incorporation using flow cytometry (Fig. 5A). The number of control T cells that enter S phase was much greater than the number of mutant cells at all time points. Thus, the deletion of one S6 allele in T cells results in G1/S cell cycle block. Passage through the cell cycle requires timely modulation of activities of the cell cycle regulators. Cell cycle progression can be inhibited by cyclin-dependent kinase inhibitors. To determine the molecular mechanism of the proliferative defect in S6wt/del T cells, we first analyzed the protein expression levels of p21 inhibitor at different time points following TCR stimulation (Fig. 5B). In unstimulated mutant and control T cells, p21 protein was undetectable. At 24 h after stimulation, the protein levels of p21 were up-regulated in mutant T cells but not in the control. At 36 h after stimulation, p21 protein levels were significantly higher in S6wt/del T cells than in the control. These results suggest that p21 induction could be responsible for a slower cell cycle of S6wt/del T cells and their inability to proliferate following stimulation.
Figure 5.
Defect in proliferation of S6wt/del T cells is due to p21-induced G1/S block. (A) BrdU incorporation into lymph node T cells stimulated with anti-CD3- and IL-2-containing MLA144 supernatant in vitro. The experiment is a representative of four independent experiments. (B) The protein expression of p21 cell cycle inhibitor in lysates from resting and stimulated lymph node S6wt/del and S6wt/lox T cells was analyzed by Western blot. Reprobing with antibodies to actin served as a loading control. (C) T cells of indicated genotypes were stimulated for the indicated period of time, labeled with propidium iodide, and analyzed by FACS. The percentage of apoptotic cells was determined by analyzing sub-G1 DNA content. The experiment is a representative of four independent experiments. (D) Relative number of S6wt/lox, S6wt/del, S6wt/lox/Bcl-2+, and S6wt/del/Bcl-2+ T cells stimulated with anti-CD3 and anti-CD28 antibodies for 24, 48, and 72 h. The experiment is a representative of three independent experiments.
To investigate whether apoptosis also contributes to the failure of S6wt/del T cells to increase in numbers following stimulation, mutant and control T cells were stimulated through TCR/CD28 for 24, 48, and 72 h and stained with propidium iodide, and DNA content was analyzed by flow cytometry (Fig. 5C). Cells having sub-G1 DNA content were considered to be apoptotic. A slightly higher percentage of S6wt/del than S6wt/lox T cells was apoptotic at all tested time points following TCR/CD28 stimulation. It is well known that anti-apoptotic protein Bcl-2 can inhibit some forms of apoptotic cell death (Strasser et al. 1994). The Bcl-2 transgene was introduced into S6wt/lox/CD4-Cre+ and S6wt/lox/CD4-Cre- mice by appropriate breeding (Strasser et al. 1994). In the presence of the anti-apoptotic Bcl-2 transgene, apoptosis was similar in TCR/CD28-stimulated S6wt/del and S6wt/lox T cells (Fig. 5C). Therefore, apoptosis contributes to the failure of S6wt/del T cells to proliferate after stimulation, and this defect can partially be reversed by anti-apoptotic protein Bcl-2.
To determine a relative contribution of apoptosis to the failure of mutant T cells to increase their numbers in response to TCR stimulation, S6wt/lox, S6wt/del, S6wt/lox/Bcl-2+, and S6wt/del/Bcl-2+ T cells were stimulated with the combination of anti-TCR and anti-CD28 antibodies in vitro for 24, 48, and 72 h. Even though apoptosis was similar in mutant and control T cells in the presence of the Bcl-2 transgene (Fig. 5C), increases in the cell numbers were only weakly rescued in mutant T cells (Fig. 5D).
These results show that the inability of S6wt/del T cells to proliferate following TCR-mediated stimulation is due to induction of G1/S cell cycle block and partially to increased apoptosis.
S6 inactivation induces a p53-dependent checkpoint response
The cell cycle inhibitor p21 is a transcriptional target of the p53 tumor suppressor. In addition to inhibition of cell cycle progression through induction of p21, p53 protein may induce apoptotic cell death (Vousden 2002). To examine the role of p53 in the observed in vivo and in vitro phenotypes, we decided to delete both flox alleles of p53 in combination with one or two floxed S6 alleles in the thymus using CD4-Cre transgenic mice (Sawada et al. 1994; Marino et al. 2000; Wolfer et al. 2001). Analysis of S6 and p53 alleles in thymi and peripheral T cells from compound S6wt/lox/p53lox/lox/CD4-Cre+ mice confirmed that S6lox and p53lox alleles are efficiently codeleted (Supplementary Fig. 2).
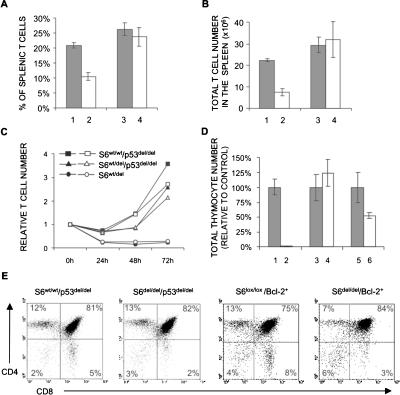
First, we set out to determine whether p53 deletion could correct the decreased number of S6wt/del T cells in the spleen. As shown in Figure 2, the number and proportion of T cells in the spleen from 4-wk-old S6wt/lox/CD4-Cre+ mice were significantly lower than in S6wt/lox/CD4-Cre- mice. However, the percentages (Fig. 6A) and total number (Fig. 6B) of T cells in spleens from S6wt/lox/p53lox/lox/CD4-Cre+ and S6wt/wt/p53lox/lox/CD4-Cre+ were similar, indicating that p53 is responsible for the decreased number of S6 heterozygous T cells in vivo.
Figure 6.
A p53-dependent checkpoint is activated following inactivation of one or two S6 alleles in T cells. The percentage (A) and total number (B) of T cells in spleens from S6wt/lox/CD4-Cre- (line 1), S6wt/lox/CD4-Cre+ (line 2), S6wt/wt/p53lox/lox/CD4-Cre+ (line 3), and S6wt/lox/p53lox/lox/CD4-Cre+ (line 4) 7-wk-old mice (n = 12 mice per each genotype). (C) Relative number of S6wt/lox/CD4-Cre+, S6wtwt/p53lox/lox/CD4-Cre+, and S6wt/lox/p53lox/lox/CD4-Cre+ T cells at 24, 48, and 72 h after stimulation with either 0.3 μg/mL (open symbols) or 1 μg/mL (closed symbols) of anti-CD3 and fixed concentration of anti-CD28 antibodies (0.1 μg/mL). The experiment is a representative of six independent experiments. (D) Total number of cells in thymi from S6lox/lox/CD4-Cre- (line 1), S6lox/lox/CD4-Cre+ (line 2), S6wt/wt/p53lox/lox/CD4-Cre+ (line 3), S6lox/lox/p53lox/lox/CD4-Cre+ (line 4), S6lox/lox/Bcl-2+/CD4-Cre- (line 5), and S6lox/lox/Bcl-2+/CD4-Cre+ (line 6) mice (n = 10 mice per each genotype). (E) S6wt/wt/p53lox/lox/CD4-Cre+, S6lox/lox/p53lox/lox/CD4-Cre+, S6lox/lox/Bcl-2+/CD4-Cre-, and S6lox/lox/Bcl-2+/CD4-Cre+ thymocytes were stained with anti-CD4 and anti-CD8 antibodies, and analyzed by FACS. Cell surface markers are shown as coordinates. The numbers in the quadrant refer to the percentage of a particular T-cell population of live cells.
To test if homozygous deletion of p53 can rescue the proliferative defect of S6 heterozygous T cells, S6wt/lox/CD4-Cre+, S6wt/wt/p53lox/lox/CD4-Cre+, and S6wt/lox/p53lox/lox/CD4-Cre+ were stimulated with anti-CD3 antibodies and anti-CD28 antibodies for 24, 48, and 72 h (Fig. 6C). Consistent with the results already shown, S6wt/lox/CD4-Cre+ T cells did not proliferate. In contrast, p53 inactivation almost completely rescued the proliferative defect of stimulated S6wt/lox/CD4-Cre+ T cells. Also, the percentage of apoptotic S6wt/wt/p53lox/lox/CD4-Cre+ and S6wt/lox/p53lox/lox/CD4-Cre+ T cells was similar at all tested time points following TCR/CD28 stimulation (data not shown). These results strongly suggest that a defect in ribosome biogenesis, induced by S6 inactivation, activates a p53-dependent checkpoint to prevent survival and expansion of defective T cells.
The thymi from the majority of S6lox/lox/CD4-Cre+ mice could not be recovered, and ones that were recovered manifested 100-fold decreases in the cell number relative to S6lox/lox/CD4-Cre- control mice (Fig. 1A). Surprisingly, inactivation of p53 gene completely rescued S6lox/lox/CD4-Cre+ thymocyte numbers (Fig. 6D). Also, FACS analysis of cells from S6lox/lox/p53lox/lox/CD4-Cre+ thymi revealed that T-cell populations defined by CD4, CD8 markers were essentially similar to control S6wt/wt/p53lox/lox/CD4-Cre+ thymi, except for a small decrease in the percentage of CD8 positive thymocytes (Fig. 6E). These results suggest that ribosomes synthesized before S6 deletion are sufficient to allow development of S6lox/lox/p53lox/lox/CD4-Cre+ T cells, and that inactivation of both S6 alleles abrogates T-cell development by activating a p53-dependent checkpoint.
To gain insight into the molecular mechanisms of p53 action in S6del/del thymocytes, we compared thymi from S6lox/lox/Bcl-2+/CD4-Cre+ and S6lox/lox/Bcl-2+/CD4-Cre- mice. The Bcl-2 transgene partially rescued a number of S6lox/lox/CD4-Cre+ thymocytes (Fig. 6D). The Bcl-2 transgene is expressed in CD4- CD8-, CD4+ CD8+, CD4+, and CD8+ thymocytes, and mature T cells. The CD4 promoter directs the expression of Cre recombinase in CD4+ CD8+, and to a lesser extent, in CD4- CD8- thymocytes. This argues against the possibility that thymocyte numbers in S6lox/lox/CD4-Cre+ mice are not completely rescued by the Bcl-2 transgene because of its delayed expression relative to CD4-Cre (Strasser et al. 1994). Additionally, FACS analysis revealed a small increase in the percentage of CD4+ CD8+ thymocytes and a decrease in CD4+ and CD8+ thymocytes, which is consistent with partial block in transition between double positive to single positive thymocytes (Fig. 6E). These results suggest that proapoptotic proteins that can be inhibited by Bcl-2 are p53 effectors in mediating a checkpoint response in S6lox/lox/CD4-Cre+ thymocytes, and they point to Bcl-2-independent p53 effectors.
Negligible numbers of S6lox/lox/p53lox/lox/CD4-Cre+ and S6lox/lox/Bcl-2+/CD4-Cre+ T cells were found in peripheral lymphoid organs, suggesting that ribosome biogenesis is necessary for their homeostatic expansion and survival (data not shown).
Discussion
In this study we conditionally deleted the S6 ribosomal protein gene in the thymus. Homozygous deletion of S6 induced a p53-dependent checkpoint response that abolished T-cell development. In contrast, heterozygous S6 deletion did not have any effect on T-cell development and TCR-stimulated cell growth response, but it decreased survival of T cells in the peripheral lymphoid organs and inhibited TCR-mediated increases in T-cell numbers in a p53-dependent manner.
The requirement for S6 in ribosome biogenesis
Conditional deletion of both S6 alleles in the liver using the interferon α-inducible Cre-loxP system abolished biogenesis of the 40S ribosomal subunit with no effect on the 60S ribosomal subunit (Volarevic et al. 2000). These results suggested that S6 is an essential component of the mouse ribosome. Here, we show that heterozygous deletion of S6 does not have any effect on the amount of 18S and 28S rRNA in the thymus, which closely reflects the amount of 40S and 60S ribosomal subunit, respectively. In parallel, the protein levels of S6 were normal, even though the amount of S6 mRNA was decreased by ∼50%. These findings suggest that one allele is sufficient to allow normal S6 protein production and ribosome biogenesis in developing thymus, explaining normal T-cell development.
The rate of ribosome biogenesis is very low in resting T cells. Activation of T cells induces ribosome biogenesis and increases protein output, causing T cells to grow rapidly within the first 24 h, and this growth phase is followed by a proliferative phase and increase in cell number (Rathmell et al. 2000; Frauwirth et al. 2002). Under such conditions, deletion of one S6 allele limits ribosome biogenesis in T cells.
Survival of mature S6 heterozygous T cells is impaired
Maturation of S6 heterozygous T cells in the thymus was normal, but the number of T cells in the spleen and lymph nodes was reduced. The number of T cells in the periphery in addition to the production in the thymus depends on the homeostatic T-cell proliferation and T-cell survival. Homeostatic proliferation of S6 heterozygous T cells was comparable to the control. This result suggests that one allele of S6 is sufficient to support relatively normal production of S6 protein and ribosome biogenesis during this slow proliferative response. The decreased number of mature T cells in S6 heterozygous mice is a result of p53 activation, as p53 inactivation restored normal T-cell numbers in the periphery. Bcl-2 transgene partially rescued the number of T cells in the spleen of S6wt/lox/CD4-Cre+ mice. These results show that p53/Bcl-2-dependent apoptosis contributes to a decreased number of S6wt/del T cells in the spleen. Bcl-2-independent p53 effectors could also regulate apoptosis of S6wt/del T cells in the periphery. However, we cannot formally rule out the possibility that decreased expansion of S6wt/del T cells contributes as well.
Normal T-cell growth and impaired proliferation of S6 heterozygous T cells
During the first 24 h after TCR stimulation in vitro, T cells grow but do not divide. As discussed earlier, deletion of one S6 allele decreased ribosome biogenesis in stimulated S6wt/del T cells, but total protein production and increase in cell size were comparable to control T cells. In sharp contrast to normal growth, proliferation of S6 heterozygous T cells in vitro was impaired. The lesion in proliferation does not seem to result from reduced translational capacity, but rather, activation of a p53-dependent cell cycle checkpoint response. However, our results cannot exclude an effect on translation of specific mRNAs under these conditions with absolute certainty.
A p53-dependent checkpoint response to a defect in ribosome biogenesis
Conditional deletion of both S6 alleles in CD4+ CD8+ thymocytes leads to a failure in T-cell development, most likely as a result of activation of a p53-dependent checkpoint, rather than defects in the protein translation. This conclusion is based on the fact that conditional inactivation of p53 completely rescues S6del/del thymocyte number. Surprisingly, even though ribosome biogenesis is blocked in S6lox/lox/p53lox/lox/CD4-Cre+ thymocytes, the remaining ribosomes are sufficient to allow T-cell development in the thymus. The rescued thymocytes have a normal expression of CD3, CD4, and CD8 markers, but it is not clear if they are normal by other criteria. Although a direct effect of S6 on p53 activation cannot be excluded, a p53-dependent checkpoint response is most likely induced by defective ribosome biogenesis that cannot support proper execution of the genetic program. S6lox/lox/p53lox/lox/CD4-Cre+ peripheral T cells are present in negligible numbers, probably as a consequence of translational deficiency that results from degradation of remaining ribosomes with time or inability to increase ribosome biogenesis after stimulation. Bcl-2 expression partially rescues T-cell development in S6lox/lox/CD4-Cre+ thymi. This suggests that a p53-dependent checkpoint response could be at least partially counteracted by Bcl-2. From our current results, we can rule out the existence of a p53-dependent, Bcl-2-independent apoptotic pathway(s) in S6del/del thymocytes. Furthermore, a possible role of p53-dependent nonapoptotic mechanisms in development of S6del/del thymocytes will require further investigation. One S6 allele is not sufficient to allow production of normal levels of S6 protein in TCR-stimulated peripheral T lymphocytes to support an increased rate of ribosome biogenesis, triggering a p53-dependent checkpoint response. Our results are consistent with p53-dependent checkpoint regulatory pathway activation by a defect in ribosome biogenesis. A similar p53-dependent defect in proliferation was observed in the LAP3 mouse cell line in which a defect in ribosome biogenesis was induced by the expression of a dominant-negative mutant of Bop1, a molecule that functions in ribosome biogenesis (Pestov et al. 2001). In TCR-stimulated S6 heterozygous T lymphocytes, a predominant outcome of p53 activation is induction of the p21 cell cycle inhibitor and, as a consequence, inhibition of the cell cycle progression. In that way, the cell cycle pace is adjusted to the capacity of the cell to synthesize ribosomal protein S6 and ribosomes, thereby preventing infidelity of the gene expression as a consequence of reduced translational capacity. We are currently testing the effect of p21 inactivation in vivo on the proliferation of S6 heterozygous T cells.
It has been previously proposed that the nucleolus is a stress sensor responsible for regulating the levels of p53, which is elevated when nucleolar function is impaired by different types of stresses (Rubbi and Milner 2003). In normal cells, p53 protein expression is maintained at low levels by MDM2-mediated ubiquitylation by MDM2, export to the cytoplasm, and degradation by proteasomes (Ryan et al. 2001). It is suggested that nucleolar proteins are important regulators of p53 levels under stress conditions in vitro (Olson 2004). The Arf protein, which binds to MDM2 and negatively regulates its activity, is under normal conditions sequestered in the nucleolus (Tao and Levine 1999; Zhang and Xiong 1999; Weber et al. 2000; Honda and Yasuda 1999). When the nucleolus is disrupted, Arf is released from the nucleolus to the nucleoplasm, where it binds to MDM2 and inhibits its activity and p53 degradation. Furthermore, Arf was found to suppress processing of pre-rRNA by interacting and inactivating the nucleolar protein B23, a nuclease that carries out early pre-rRNA cleavage (Itahana et al. 2003; Sugimoto et al. 2003). These findings suggest that the Arf-B23 interaction may function in coordinating cell growth with cell cycle progression. It has been shown recently that L5, L11, and L23 ribosomal proteins are released from the nucleolus following treatments that inhibit ribosome biogenesis—such as starvation for nutrients or incubation with a low concentration of actinomycin D in vitro—interact with MDM2, and inhibit its activity toward p53 (Marechal et al. 1994; Lohrum et al. 2003; Zhang et al. 2003; Bhat et al. 2004; Jin et al. 2004). It has been proposed that they couple cell growth to the cell cycle.
Here we demonstrate that deletion of the S6 gene, which is coding for a component of the 40S ribosomal subunit, induces a p53-dependent checkpoint response in vivo, most likely through a defect in ribosome biogenesis. Although, we cannot formally rule out the possibility that altered protein synthesis is responsible for the observed phenotypes, one argument against this is our observation that the protein translation inhibitor puromycin failed to up-regulate the protein levels of p53 in TCR/CD28-stimulated T cells (data not shown). In the same experiment, a low dose of actinomycin D and etoposide strongly induced p53. Impaired translation of mRNAs encoding a key DNA damage repair or replication proteins could lead to DNA damage and activation of a p53-dependent checkpoint. We did not observe any increase in phosphorylation of ATM/ATR substrates (phosphorylated SQ/TQ) and histone H2A.X (phospho-Ser139) in TCR/CD28-stimulated S6wt/del T cells relative to S6wt/lox T cells using immunofluorescence analysis (data not shown), suggesting that the DNA damage is not responsible for activation of a p53-dependent checkpoint in S6-deficient cells.
Ribosomal protein S6 is phosphorylated by S6 kinases 1 and 2 (S6K1/2) under growth-promoting conditions (Volarevic and Thomas 2001). Is a p53-dependent proliferation defect in TCR/CD28-stimulated S6wt/del T cells the result of decreased levels of phosphorylated S6? Good evidence against a positive role of S6 phosphorylation in cellular proliferation has been provided by the recent generation and analysis of knock-in mice, whose S6 contains alanine substitutions at all five phosphorylatable serines (Ruvinsky et al. 2005). These mice were indistinguishable from their wild-type littermates in development, appearance, and weight. Interestingly, mouse embryonic fibroblasts isolated from these mice proliferated even better than the wild-type control, implying that a decrease in S6 phosphorylation does not activate a p53-dependent checkpoint. Consistent with this notion, we have shown that rapamycin at concentrations that inhibit S6 phosphorylation does not induce the protein levels of p53 in TCR/CD28-stimulated T cells (data not shown).
Inducible deletion of both S6 alleles in the liver abrogated cell division in response to hepatectomy, which had been correlated with the inhibition of cyclin E expression (Volarevic et al. 2000). In contrast, TCR/CD28 stimulation of S6wt/del T cells normally up-regulated the protein levels of cyclin E (data not shown). What is the reason for such an apparent difference in these two models? It is possible that responses to deletion of one or two S6 alleles are different. Certainly, this issue needs to be addressed very carefully, before definitive conclusions can be drawn.
What are the physiological consequences of this checkpoint response? Since translation of specific mRNAs depends on their affinity for ribosomes, it is possible that mutations in the genes that decrease the number of ribosomes can change the accuracy of translation, altering the precise execution of the genetic program (Lodish 1974; Mauro and Edelman 2002; Ruggero and Pandolfi 2003; Holland et al. 2004). Although increases in ribosome biogenesis have been correlated with a malignant transformation in many instances, some ribosomal protein genes are suggested to be haploinsufficient tumor suppressors in Zebrafish, Drosophila, and humans (Watson et al. 1992; Stewart and Denell 1993; Draptchinskaia et al. 1999; Amsterdam et al. 2004). To uncover the possible pathogenic potential of the S6wt/del mutation in T cells, we will compare the incidence and severity of T-cell lymphomas in S6wt/lox/p53wt/lox/CD4-Cre+ and S6wt/wt/p53wt/lox/CD4-Cre+ mice and look for signs of autoimmune phenomena.
In addition to a potential role in malignant transformation and autoimmunity, activation of a p53-dependent checkpoint mechanism in response to defects in the translational machinery might play an important role in other fundamental biological processes such as senescence and aging.
Materials and methods
Mice
S6lox/lox mice were produced previously (Volarevic et al. 2000). CD4-Cre transgenic mice have been described earlier (Sawada et al. 1994; Wolfer et al. 2001). Bcl-2 transgenic mice were obtained from The Jackson Laboratory (Strasser et al. 1994). p53lox/lox mice were created by Marino (Marino et al. 2000). Mice were genotyped by PCR analysis of genomic DNA isolated from tail tips using specific primers for the Cre recombinase (5′-GTCCAATTTACTGACCGTACAC-3′ and 5′-CTGTCACTTGGTCGTGGCAGC-3′); for the wild-type S6 allele (5′-AGTGTAACTAGATAAATGAT-3′ and 5′-AGCCTTTTCTTTTAGCATA C-3′); for the floxed S6 allele (5′-TGCTCCTGCCGAGAAAGTATCCATCATGGC-3′ and 5′-GTGTTAACTCTTCTGCCAGA-3′); for the Bcl-2 transgene (5′-GTCCCGCCCCTAACTCCGCCCATCC-3′ and 5′-CCGCATCCCACTCGTAGCCCCTCTG-3′); for wild-type and floxed p53 allele (5′-AAGGGGTATGAGGGACAAGG-3′ and 5′-GAAGACAGAAAAGGGGAGGG-3′); and for the deleted p53 allele (5′-CACAAAAACAGGTTAAACCCA-3′ and 5′-GAAGACAGAAAAGGGGAGGG-3′) (Marino et al. 2000). All mice were housed in conventional animal facilities of the Medical School, University of Rijeka.
Southern and Northern blot analyses
Genomic DNA isolated from thymi, purified spleen, and lymph node T cells (R&D Systems) was genotyped by Southern blot analysis of HindIII-restricted fragments using intron-specific probes as described (Volarevic et al. 2000). Total RNA was isolated from thymi and T cells stimulated in vitro using Trizol (Invitrogen Life Technologies). RNA was separated on a 1% agarose/formaldehyde gel and transferred to a Hybond N+ membrane (Amersham Biosciences). Total RNA from thymi was hybridized with S6 and L11 cDNA probes labeled with the Alkphos Direct Kit (Amersham) according to the manufacturer's instructions.
Analysis of rRNA processing
For analysis of rRNA processing, T cells were stimulated for 20 h in vitro, starved for 15 min in methionine-free medium, labeled for 30 min with 25 μCi/mL [methyl-3H]methionine (Amersham Biosciences), and then chased in medium containing unlabeled methionine for indicated time periods. Total RNA, isolated from the same number of mutant and control T cells, was separated on a formaldehyde-agarose gel, blotted to the Hybond N+ membrane, which was dried and treated with EN3HANCE (New England Nuclear), and exposed to Kodak BioMax MS film at -80°C (Sigma).
T-cell lysis and protein determination
S6wt/lox and S6wt/del T cells were collected prior to TCR/CD28 activation and at different time points after stimulation. The same number of mutant and control T cells was dissolved in RIPA buffer (50 mM tris-Cl at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% Na-deoxycholate) containing protease and phosphatase inhibitors (1 μg/mL leupeptin, 1 μg/mL aprotinin, 50 μg/mL PMSF, 1 mM Na3VO4, 1 mM NaF), lysed on ice for 20 min, and centrifuged at 12,000 rpm for 30 min. Protein concentration was determined by the Bio-Rad protein assay according to the manufacturer's instructions (Bio-Rad).
Western blot analysis
RIPA total protein lysates (30 μg per lane) were separated by denaturing SDS-PAGE, transferred to Immobilon-P membrane (Millipore), and probed with mouse monoclonal anti-S6 and rabbit polyclonal anti-L7a antibodies and rabbit polyclonal anti-L11, raised against a synthetic peptide CIGAKHRISKEEAMRWFQQK. Primary antibodies were detected by using HRP-conjugated secondary antibodies to rabbit and mouse immunoglobulins (Santa Cruz Biotechnology) and the ECL Plus Western Blotting Detection System (Amersham Biosciences).
Flow cytometry
Single-cell suspensions from thymi, spleen, and lymph nodes were washed in PBS containing 2% FCS and 0.1% sodium azide at 4°C, and 1 × 106 cells were stained with monoclonal antibodies to CD3, CD45R (B220), CD4, CD8a, CD25, and CD69, all purchased from BD Biosciences. Flow cytometry was performed on a FACScan (Becton Dickinson) using Cell Quest software. Ten-thousand events were collected.
Stimulation of T lymphocytes in vitro
T lymphocytes from lymph nodes and spleen were purified using columns from R&D Systems. The purity of T cells was ∼90%. Purified T cells were plated at 1 × 106 cells per well in a flat-bottom six-well plate in RPMI 1640 medium containing 12% FCS, 2 mM L-glutamine, 5 μM 2-ME, 105 U/L penicillin, 0.1 g/L streptomycin, and 0.035 g/L gentamycin, and stimulated with various concentrations of soluble anti-CD3 (145-2C11; BD Biosciences) and anti-CD28 mAbs (37.51; BD Biosciences). In some experiments, T cells were stimulated with the combination of soluble anti-CD3 and IL2 containing MLA 144 supernatant.
Intracellular cytokine staining
Lymphocytes stimulated in vitro with anti-CD3 and anti-CD28 antibodies were incubated with 10 μg/mL brefeldin A (Sigma) during 3 h before harvesting. Cells were stained for T-cell surface markers, fixed in 2% paraformaldehyde in PBS at 4°C, permeabilized with 0.1% saponin (Sigma), and stained with anti-mouse IFN-γ-FITC or anti-mouse IL-2-FITC (BD Biosciences) in 0.05% saponin.
CFSE labeling
Cell division of stimulated T cells was analyzed using 2 mM 5-6-carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling (Molecular Probes). This method is based on the approximately twofold decreases in CFSE fluorescence after each cell division (Lyons and Parish 1994).
T cells from lymph nodes (1 × 107 cells/mL PBS) were stained with CFSE at 37°C for 10 min in a CO2 incubator, washed twice with PBS/2% FCS, and stimulated as described. For in vivo studies, 10 × 106 CFSE-labeled T cells were transferred intravenously into nonirradiated syngenic wild-type hosts. CFSE intensity was analyzed by flow cytometry.
BrdU staining and determination of apoptotic cells
For in vivo 5-bromo-2′-deoxyuridine (BrdU) labeling studies, mice were daily injected intraperitoneally with 100 μg/g of body weight BrdU (Sigma) in 200 μL of PBS during indicated time periods. BrdU uptake was measured in purified T cells from lymph nodes (R&D Systems). In vitro stimulated T lymphocytes were incubated with 10 μM BrdU for 2 h and stained with BrdU-FITC (BD Biosciences) according to the manufacturer's instructions. In brief, cells were washed with PBS, fixed in 70% ethanol at 4°C, resuspended in 2N HCl/0.5% Triton X-100, incubated for 30 min at room temperature, neutralized with 0.1 M tetraborate (pH 8.5), rinsed in PBS containing 1% BSA and 0.5% Tween 20, stained with the abovementioned anti-BrdU antibody, and then analyzed by FACS. For determination of apoptosis, T cells were stimulated, stained with propidium iodide at different time points after stimulation, and analyzed by flow cytometry. The percentage of sub-G1 cells was taken as the percentage of apoptotic cells.
Supplementary Material
Acknowledgments
We are grateful to Dr. George Thomas for his continuous support of our work; Jonathan Ashwell, Davor Solter, Nick Pullen, and Ivan Dikić for reading of the manuscript; Kristina Grabušić, Ines Mrakovčić Šutić, Astrid Krmpotić, and Bojan Polić for technical advice; Lidija Bilić-Zulle for help with statistical analysis; Anton Berns (The Netherlands Cancer Institute) for providing p53lox/lox mice; Dan Littman (New York University School of Medicine) for providing CD4-Cre mice; and Stefano Fumagalli and George Thomas (University of Cincinnati) for anti-S6 and anti-L7a antibodies. This work was supported by the Swiss National Science Foundation (SCOPES program) and by the Ministry of Science, Education, and Sports of the Republic of Croatia (to S.V.).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.359305.
References
- Amsterdam A., Sadler, K.C., Lai, K., Farrington, S., Bronson, R.T., Lees, J.A., and Hopkins, N. 2004. Many ribosomal protein genes are cancer genes in zebrafish. PloS. Biol. 2: 690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.P., Itahana, K., Jin, A., and Zhang, Y. 2004. Essential role for ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 23: 2402-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon I. and Raff, M. 1999. Size control in animal development. Cell 96: 235-244. [DOI] [PubMed] [Google Scholar]
- Conlon I., Dunn, G.A., Mudge, A.W., and Raff, M. 2001. Extracellular control of cell size. Nat. Cell Biol. 3: 918-921. [DOI] [PubMed] [Google Scholar]
- Draptchinskaia N., Gustavsson, P., Andersson, B., Pettersson, M., Willig, T.N., Dianzani, I., Ball, S., Tchernia, G., Klar, J., Matsson, H., et al. 1999. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 21: 169-175. [DOI] [PubMed] [Google Scholar]
- Du Y.C. and Stillman, B. 2002. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell 109: 835-848. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. 1996. Cell cycle checkpoints: Preventing an identity crisis. Science 274: 1664-1672. [DOI] [PubMed] [Google Scholar]
- Fatica A. and Tollervey, D. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14: 313-318. [DOI] [PubMed] [Google Scholar]
- Frauwirth K.A., Riley, J.L., Harris, M.H., Parry, R.V., Rathmell, J.C., Plas, D.R., Elstrom, R.L., June, C.H., and Thompson, C.B. 2002. The CD28 signaling pathway regulates glucose metabolism. Immunity 16: 769-777. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A.A. 1985. The nucleolus and ribosome biogenesis. Springer-Verlag, Wien, New York.
- Hafen E. 2004. Interplay between growth factor and nutrient signaling: Lessons from Drosophila TOR. Curr. Top. Microbiol. Immunol. 279: 153-167. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H. 1971. Genetic control of the cell division cycle in yeast: Genes controlling DNA replication and its initiation. J. Mol. Biol. 59: 183-194. [DOI] [PubMed] [Google Scholar]
- Holland E.J., Sonenberg, N., Pandolfi, P.P., and Thomas, G. 2004. Signaling control of mRNA translation in cancer pathogenesis. Oncogene 23: 3138-3144. [DOI] [PubMed] [Google Scholar]
- Honda R. and Yasuda, H. 1999. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18: 22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K., Bhat, K.P., Jin, A., Itahana, Y., Hawke, D., Kobayashi, R., and Zhang, Y. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 12: 1151-1164. [DOI] [PubMed] [Google Scholar]
- Jin A., Itahana, K., O'Keefe, K., and Zhang, Y. 2004. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24: 7669-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L.A. and Gallant, P. 2002. Control of growth and organ size in Drosophila. Bioessays 24: 54-64. [DOI] [PubMed] [Google Scholar]
- Johnston L.A., Prober, D.A., Edgar, B.A., Eisenman, R.N., and Gallant, P. 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.G., Plas, D.R., Kubek, S., Buzzai, M., Mu, J., Xu, Y., Birnbaum, M.J., and Thompson, C.B. 2005. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18: 283-293. [DOI] [PubMed] [Google Scholar]
- Jorgensen P. and Tyers, M. 2004. How cells coordinate growth and division. Curr. Biol. 14: R1014-R1027. [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Nishikawa, J.L., Breitkreutz, B.J., and Tyers, M. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395-400. [DOI] [PubMed] [Google Scholar]
- Lewis J.D. and Tollervey, D. 2000. Like attracts like: Getting RNA processing together in the nucleus. Science 288: 1385-1389. [DOI] [PubMed] [Google Scholar]
- Lodish H.F. 1974. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251: 385-388. [DOI] [PubMed] [Google Scholar]
- Lohrum M.A., Ludwig, R.L., Kubbutat, M.H., Hanlon, M., and Vousden, K.H. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577-587. [DOI] [PubMed] [Google Scholar]
- Lyons A.B. and Parish, C.R. 1994. Determination of lymphocyte division by flow cytometry. J. Immunol. Methods 171: 131-137. [DOI] [PubMed] [Google Scholar]
- Mamane Y., Petroulakis, E., Rong, L., Yoshida, K., Ler, L.W., and Sonenberg, N. 2004. eIF4E—From translation to transformation. Oncogene 23: 3172-3179. [DOI] [PubMed] [Google Scholar]
- Marechal V., Elenbaas, B., Piette, J., Nicolas, J.C., and Levine, A.J. 1994. The ribosomal protein L5 protein is associated with Mdm-2 and Mdm-2-p53 complexes. Mol. Cell. Biol. 14: 7414-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Vooijs, M., van Der Gulden, H., Jonkers, J., and Berns, A. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes & Dev. 14: 994-1004. [PMC free article] [PubMed] [Google Scholar]
- Mauro V.P. and Edelman, G.M. 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. 99: 12031-12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T. 2004. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 14: 210-217. [DOI] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz, A.F., Johnston, L.A., and Edgar, B.A. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183-1193. [DOI] [PubMed] [Google Scholar]
- Nurse P. 2000. A long twentieth century of the cell cycle and beyond. Cell 100: 71-78. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux, P., and Nasmyth, K. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146: 167-178. [DOI] [PubMed] [Google Scholar]
- O'Farrell P.H., Stumpff, J., and Su, T.T. 2004. Embryonic cleavage cycles: How is a mouse like a fly? Curr. Biol. 14: R35-R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.O.J. 2004. Sensing cellular stress: Another new function for the nucleolus? Sci. STKE 224: pe10. [DOI] [PubMed] [Google Scholar]
- Pardee A.B. 1989. G1 events and regulation of cell proliferation. Science 246: 603-608. [DOI] [PubMed] [Google Scholar]
- Pestov D.G., Strezoska, Z., and Lau, L.F. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21: 4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober D.A. and Edgar, B.A. 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100: 435-446. [DOI] [PubMed] [Google Scholar]
- Rajasekhar V.K., Viale, A., Socci, N.D., Wiedmann, M., Hu, X., and Holland, E.C. 2003. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12: 889-901. [DOI] [PubMed] [Google Scholar]
- Rathmell J.C., Vander Heiden, M.G., Harris, M.H., Frauwirth, K.A., and Thompson, C.B. 2000. In the absence of extrinsic signals, nutrient utilisation by lymphocytes is insufficient to maintain either cell size or viability. Mol. Cell 6: 683-692. [DOI] [PubMed] [Google Scholar]
- Rubbi C.P. and Milner, J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22: 6068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. and Pandolfi, P.P. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3: 179-192. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Sharon, N., Lerer, T., Cohen, H., Stolovich-Rain, M., Nir, T., Dor, Y., Zisman, P., and Meyuhas, O. 2005. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & Dev. 19: 2199-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.M., Phillips, A.C., and Vousden, K.H. 2001. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13: 332-337. [DOI] [PubMed] [Google Scholar]
- Sawada S., Scarborough, J.D., Killeen, N., and Littman, D.R. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77: 917-929. [DOI] [PubMed] [Google Scholar]
- Schmelzle T. and Hall, M.N. 2000. TOR, a central controller of cell growth. Cell 103: 253-262. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts, J.M. 1999. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes & Dev. 13: 1501-1512. [DOI] [PubMed] [Google Scholar]
- Stewart M.J. and Denell, R. 1993. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol. Cell. Biol. 13: 2524-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H. and Hafen, E. 2000. Genetic control of cell size. Curr. Opin. Genet. Dev. 10: 529-535. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris, A.W., Corcoran, L.M., and Cory, S. 1994. Bcl-2 expression promotes B- but not T-lymphoid development in scid mice. Nature 368: 457-460. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Kuo, M.L., Roussel, M.F., and Sherr C.J. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11: 415-424. [DOI] [PubMed] [Google Scholar]
- Sveiczer A., Novak, B., and Mitchison, J.M. 2004. Size control in growing yeast and mammalian cells. Theor. Biol. Med. Model. 1: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W. and Levine, A.J. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. 96: 6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2: E71-E72. [DOI] [PubMed] [Google Scholar]
- Volarevic S. and Thomas, G. 2001. Role of S6 phosphorylation and S6 kinase in cell growth. Prog. Nucleic Acid Res. Mol. Biol. 65: 101-127. [DOI] [PubMed] [Google Scholar]
- Volarevic S., Stewart, M.J., Ledermann, B., Zilberman, F., Terracciano, L., Montini, E., Grompe, M., Kozma, S.C., and Thomas, G. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288: 2045-2047. [DOI] [PubMed] [Google Scholar]
- Vousden K.H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602: 47-59. [DOI] [PubMed] [Google Scholar]
- Warner J.R., Vilardell, J., and Sohn, J.H. 2001. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 66: 567-574. [DOI] [PubMed] [Google Scholar]
- Watson K.L., Konrad, K.D., Woods, D.F., and Bryat, P.J. 1992. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. 89: 11302-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.D., Kuo, M.L., Bothner, B., DiGiammarino, E.L., Kriwacki, R.W., Roussel, M.F., and Sherr, C.J. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20: 2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinkove D. and Leevers, S.J. 2000. The genetic control of organ growth: Insight from Drosophila. Curr. Opin. Genet. Dev. 10: 75-80. [DOI] [PubMed] [Google Scholar]
- Weinkove D., Neufeld, T.P., Twardzik, T., Waterfield, M.D., and Leevers, S.J. 1999. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr. Biol. 9: 1019-1029. [DOI] [PubMed] [Google Scholar]
- Wolfer A., Bakker, T., Wilson, A., Nicolas, M., Ioannidis, V., Littman, D.R., Lee, P.P., Wilson, C.B., Held, W., MacDonald, H.R., et al. 2001. Inactivation of Notch1 in immature thymocytes does not perturb CD4 or CD8 T cell development. Nat. Immunol. 2: 235-241. [DOI] [PubMed] [Google Scholar]
- Zetterberg A. and Larsson, O. 1991. Coordination between cell growth and cell cycle transit in animal cells. Cold Spring Harb. Symp. Quant. Biol. 56: 137-147. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Xiong, Y. 1999. Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3: 579-591. [DOI] [PubMed] [Google Scholar]
- Zhang J., Schneider, C., Ottmers, L., Rodriguez, R., Day, A., Markwardt, J., and Schneider, B.L. 2002. Genomic scale mutant hunt identifies cell size homeostasis genes in Saccharomyces cerevisiae. Curr. Biol. 12: 1992-2001. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wolf, G.W., Bhat, K., Jin, A., Allio, T., Burkhat, W.A., and Xiong, Y. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23: 8902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.