Summary
Conference on Bacterial Neural Networks
Introduction
We must, albeit reluctantly, dismiss the naive but comforting myth of the bacterial cell as a simple loner: a self-contained bag of uncompartmentalized enzymes, so exquisitely uncomplicated as to be understood outright and considered passé in this new millennium—a biological beaker for examining the more complicated eukaryotic versions of life. The recent EURESCO Conference on Bacterial Neural Networks heralded broadly the inescapable truth that bacteria cooperate in both uniform and mixed societies, sabotage the relatively mammoth creatures that they colonize, modify their own living spaces, and anticipate predictable changes in their surroundings. They achieve these feats with a clear sense of time and place, both within and outside their small but well-organized bodies, and send and receive messages across comparatively vast space.

This EURESCO conference (June 7–12, 2002) was set in the lovely Alsatian village of Obernai, France, and was attended by 109 participants spanning junior to senior research careers, and representing both academic and industrial institutions. Organizers Barry Holland (Paris, France) and Charles Dorman (Dublin, Ireland) led participants on an excursion to Riquewihr, a scenic village on the Wine Road, pictured here.
Major themes of the meeting were the interconnected phenomena of signalling, behaviour and development, with just enough emphasis on technological advancements to presage how much more bacterial complexity is yet to be discovered as the tools improve. Overall, the conference was characterized by rigorous science that was professionally delivered by an overwhelmingly European cast of speakers and other participants (plus a few privileged interlopers such as myself). A small sampling of the diverse topics that were covered during a stimulating four-day meeting is featured here, emphasizing those presentations that best highlight aspects of the neural network theme.
Sniffing out the way to go
At the heart of signal transduction mechanisms in bacteria is the two-component paradigm, in which a histidine protein kinase and a response regulator sense and transmit a signal from a cellular or extracellular cue to an effector, propagating the signal via phosphoryl transfer-induced conformational changes (Robinson et al., 2000). The canonical example of this phenomenon is bacterial chemotaxis. This is one area that bacteriologists thought we indeed understood outright, but even this well-worn path is yielding unexpected surprises. J. Armitage (Oxford, UK) reported that the purple photosynthetic bacterium Rhodobacter sphaeroides uses multiple partial and overlapping sets of chemotaxis components (che genes) in a non-redundant way (Wadhams et al., 2002). Sequencing of the genome revealed the potential players, and herculean efforts to delete che genes cleanly in various combinations has allowed their roles to be scrutinized. The outcome was a picture of a 'smart' chemotaxis system that makes decisions about whether or not to move in response to an attractant stimulus after considering other aspects of the milieu, such as oxygen concentration or photosynthetic environment, which might be better in the current location than in the direction from which the seductive signal emanates. This picture has been developed further by determining the intracellular locations of many Che components with fluorescent reporters; for example, with respect to the related kinases CheA2 and CheA3, the former is localized to the cell pole, and the latter is found throughout the cytoplasm.
Even in Escherichia coli, for which the phosphorelay in the chemotaxis system has been best described, a second layer of modification seems to allow the cell to integrate chemotaxis signals with the metabolic state of the cell. M. Eisenbach (Rehovot, Israel) presented evidence for acetylation of the quintessential phosphoryl-accepting response regulator, CheY, by acetyl CoA synthase as a fine-tuning mechanism that might allow cells to compensate for variations in the levels of the kinase CheA and the phosphatase CheZ (Barak & Eisenbach, 2001).
Seeing the light
R. sphaeroides is not content to use only its sense of smell, but rather likes to see where its next meal is coming from. C. Bauer (Bloomington, Indiana, USA) described the multifunctional AppA protein, a blue-light-absorbing flavoprotein that is important for regulation of the photosynthetic apparatus by light. AppA acts as an antirepressor of photosynthesis gene expression by breaking a disulphide bond in the redox-regulated repressor protein PpsR. However, absorption of blue light blocks the ability of AppA to form a stable complex with the repressor for antirepression (Masuda & Bauer, 2002). The coordinated regulation of photosynthesis gene expression by blue light and redox state allows the cell to avoid gearing up unnecessarily for conditions that are unfavourable for its photosynthetic function, which requires anerobic conditions and red-enriched light.
Bearing fruit
Some bacteria can be viewed as multicellular organisms with differentiated tissues at certain stages of their life cycles. L. Sogaard-Andersen (Odense, Denmark) described research to understand the C-signal, one of several informational prompts for the formation of Myxococcus xanthus multicellular fruiting bodies. C-signalling requires not only cell–cell contact but specifically a pole–pole handshake. Sogaard-Andersen showed evidence that the C-signal is a 17-kDa membrane-associated protein (P17) that is cleaved from a 25-kDa precursor (P25). Although P25 is predicted to bind NAD(P)+, the portion of the molecule responsible for this interaction is not present in P17. Proteolytic cleavage of P25 to generate the P17 C-signal might provide a regulatory checkpoint for the developmental process (Kruse et al., 2001).
Another striking example of multicellularity and cell-cycle complexity is Streptomyces coelicolor, which grows as a filamentous mat called a mycelium and, when starved, differentiates into specialized cell types to form aerially displayed spores, produce antibiotics or undergo apoptosis. J. Weiser (Prague, Czech Republic) presented a creative solution to a common technical barrier, namely that the use of agar plates to provide a solid surface to support aerial hyphae and spore formation is not favourable for sampling over time in complex protocols for proteomics. Weiser and his co-workers obtained synchronously differentiating three-dimensional colonies on liquid-medium-surrounded glass bead matrices designed to mimic the soil habitat of the organism (Kofronova et al., 2002) (Fig. 1). Small-column systems using these beads allowed liquid medium to flow in and out for sampling and nutrient manipulation, and thus made the analysis of the Streptomyces proteome and metabolome easy.
Figure 1.
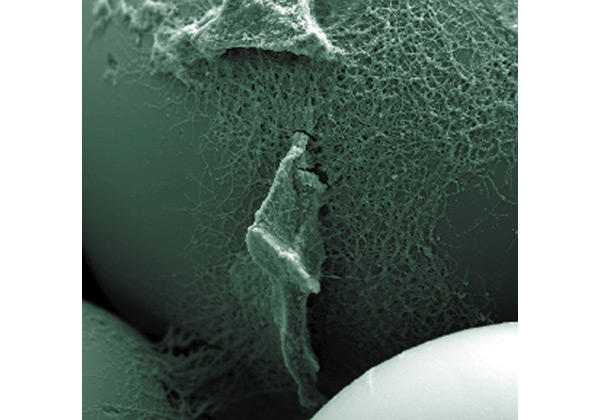
A network of vegetative hyphae of Streptomyces coelicolor growing on the surface of glass beads (smooth surface) takes up nutrients from the liquid medium that fills the inter-bead space. (Image courtesy of J. Weiser.)
Mob control
Among many visually satisfying images of bacteria engaged in multicellular collective behaviour were videos, presented by J. Shapiro (Chicago, Illinois, USA), of swarming colonies of Proteus mirabilis (Shapiro, 1998). The time-lapse films showed a predictable colony expansion pattern that can be modelled mathematically by accounting for population dynamics in terms of the fraction of cells that have the ability to 'swarm', the lifespan of such swarmer cells, and density thresholds for collective migration. The rules governing these processes create periodicity in Proteus expansion on an agar substrate, so that the colony morphology achieves a characteristic terraced pattern.
The writing on the wall
R. VanBogelen (Ann Arbor, Michigan, USA) focused on the biological aspects of what is typically thought of as a technical and cataloguing pursuit: profiles of the proteome. In her approach, proteomic signatures are identified by mining these cataloguing efforts and are used to diagnose cellular metabolic states and the mechanism of drug action (VanBogelen et al., 1999). For example, when two new antibacterial drugs of unknown function are tested on a pathogen and the proteomic profiles are compared with those obtained using known inhibitors, signatures of trauma to the ribosome or DNA synthesis machinery might be evident, pointing to the inhibitory mechanism that should be investigated. VanBogelen's talk emphasized the vastness of the datasets that can be generated by this technique, and the need for algorithms that allow seamless access by the user, making it possible to handle highly dimensional, multivariant information to exploit the potential of the technology.
Prokaryotic ninjas
B. Kenny (Bristol, UK) provided a vivid depiction of the extent to which bacteria undermine the processes in eukaryotic cells to provide a cushy landing for their own arrival (Kenny, 2002). Enteropathic E. coli injects several proteins into the host before forming a tight non-invasive association with the membrane of the gut epithelia. One of these, Tir, is essential for virulence and functions as a host plasma membrane receptor that mediates the intimate attachment of the bacteria. To execute its role, it must be modified by a host kinase, after which it induces the cytoskeletal rearrangements associated with disease.
A second protein, Map (mitochondrial-associated protein), has both mitochondrial-dependent and mitochondrial-independent functions, with the latter activating the small GTPase protein Cdc42 to induce filopodial formation (Fig. 2). This activity is inhibitory to Tir-mediated cytoskeletal rearrangements and is downregulated in a Tir-dependent manner. These findings demonstrate that pathogens have evolved both multifunctional effector molecules and complex regulatory mechanisms to exploit host cellular processes.
Figure 2.
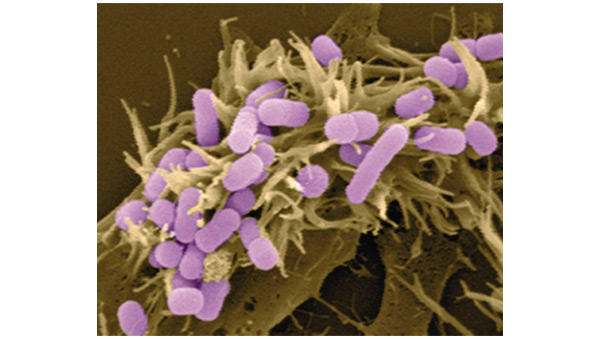
Enhanced filopodial formation at the site of host–pathogen interaction. (Image courtesy of B. Kenny and M. Jepson.) This phen-omenon results from the expression of plasmid-encoded Map (mitochondrial-associated protein) injected by a strain of E. coli.
'If I only had a brain'1
The title of the meeting may seem tongue-in-cheek because a bacterial cell is a fraction of the size of a single neuron. However, the roots of the meeting lay in a proposal by K. Hellingwerf (Amsterdam, The Netherlands) that the combined phosphorelays in a single bacterial cell have similarities to neural transduction, and that specific 'rules' that govern the latter could be applied to test bacterial signal transduction as a 'phospho-neural' network (Hellingwerf et al., 1995). These properties of neural networks include: multiple pathways acting in parallel (for which there are bacterial signalling precedents); logical operations at nodes in the pathway (such as the integration when two kinases feed phosphoryl groups into the same response regulator); a mechanism for autoamplification (such as positive autoregulation of the expression of a two-component system once it has been activated); and cross-talk between pathways. Hellingwerf's presentation focused on the last criterion, for which clear data in wild-type situations in vivo are scarce. However, carefully controlled environmental conditions have made possible the analysis of four separate two-component signal transduction pathways in E. coli. The results indicated that specificity dominates over connectivity, and that cross-talk is minimal in vivo (Verhamme et al., 2002). By this criterion, then, the similarity with neural networks breaks down.
Despite the lack of neurons, some bacteria are as good as eukaryotes at anticipating predictable changes in their environments by setting an internal biological clock to wake them up in the morning. S. Golden (College Station, Texas, USA) presented the current understanding of the molecular mechanism of the cyanobacterial circadian clock with respect to the oscillator that keeps time, input pathways that set the clock to local time, and output pathways that convey temporal information to the machinery that controls gene expression (Golden et al., 1998). A 24-hour rhythm in gene expression can be tracked by luciferase reporters that give off light as a function of the activity of the promoters that drive their genes. Clock components identified through genetic screens comprise familiar groups of proteins, such as the CikA and SasA histidine protein kinases, as well as more unusual players, such as the three Kai proteins that seem to comprise the circadian oscillator (Williams et al., 2002).
Think like a bacterium
This conference highlighted the sophisticated regulatory systems that exquisitely adapt bacteria to their environments. To fully understand them, perhaps we should think of 'bacterial culture' not in the sense of something in a flask, but rather as a dynamic and cosmopolitan lifestyle shared by creatures of the microbial world, into which we have been allowed to peer.
Acknowledgments
I thank Barry Holland and the other meeting organizers for the opportunity to participate in the conference, and the speakers whose work is discussed here for their input on the draft and for providing figures. I extend apologies to the many participants whose excellent presentations are not covered here. The work on the cyanobacterial circadian clock is supported chiefly by grants to S.S.G. from the US National Science Foundation (MCB-9982852) and National Institutes of Health (P01-NS39546).
Footnotes
1Scarecrow, The Wizard of Oz (1939), MGM studios.
References
- Barak R. & Eisenbach M. (2001) Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40, 731–743. [DOI] [PubMed] [Google Scholar]
- Golden S.S., Johnson C.H. & Kondo T. (1998) The cyanobacterial circadian system: a clock apart. Curr. Opin. Microbiol. 1, 669–673. [DOI] [PubMed] [Google Scholar]
- Hellingwerf K.J., Postma P.W., Tommassen J. & Westerhoff H.V. (1995) Signal transduction in bacteria: phospho-neural network(s) in Escherichia coli? FEMS Microbiol. Rev., 16, 309–321. [DOI] [PubMed] [Google Scholar]
- Kenny B. (2002) Enteropathogenic Escherichia coli (EPEC)—a crafty subversive little bug. Microbiology 148, 1967–1978. [DOI] [PubMed] [Google Scholar]
- Kofronova O., Nguyen L.D., Weiser J. & Benada O. (2002) Streptomycetes cultured on glass beads: sample preparation for SEM. Microsc. Res. Tech. 58, 111–113. [DOI] [PubMed] [Google Scholar]
- Kruse T., Lobedanz S., Berthelsen N.M. & Sogaard-Andersen L. (2001) Csignal: a cell surface-associated morphogen that induces and co-ordinates multicellular fruiting body morphogenesis and sporulation in Myxococcus xanthus. Mol. Microbiol. 40, 156–168. [DOI] [PubMed] [Google Scholar]
- Masuda S. & Bauer C. (2002) AppA Is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110, 613–623. [DOI] [PubMed] [Google Scholar]
- Robinson V.L., Buckler D.R. & Stock A.M. (2000) A tale of two components: a novel kinase and a regulatory switch. Nature Struct. Biol. 7, 626–633. [DOI] [PubMed] [Google Scholar]
- Shapiro J.A. (1998) Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 52, 81–104. [DOI] [PubMed] [Google Scholar]
- VanBogelen R.A., Schiller E.E., Thomas J.D. & Neidhardt F.C. (1999) Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20, 2149–2159. [DOI] [PubMed] [Google Scholar]
- Verhamme D.T., Arents J.C., Postma P.W., Crielaard W. & Hellingwerf K.J. (2002) Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology 148, 69–78. [DOI] [PubMed] [Google Scholar]
- Wadhams G., Martin A., Porter S., Maddock J., Mantotta J., King H. & Armitage J. (2002) TlpC, a novel chemotaxis protein in Rhodobacter sphaeroides localises to a discrete region in the cytoplasm. Mol. Microbiol. (in the press). [DOI] [PubMed] [Google Scholar]
- Williams S.B., Vakonakis I., Golden S.S. & LiWang A.C. (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc. Natl Acad. Sci. USA 99, 15357–15362. [DOI] [PMC free article] [PubMed] [Google Scholar]


