Summary
Control of epidermal cell patterning in plants
A recent publication by Schellmann et al. (2002) in The EMBO Journal has provided important insights into the control of cell fate and pattern formation in plants. To study the underlying molecular mechanisms, the authors made use of the development of root hairs and trichomes in Arabidopsis thaliana. A. thaliana trichomes are large, unicellular structures that project out from the shoot epidermal surface and are thought to form a defence against herbivorous insects. Root hairs are specialized cells extending from the root epidermis and are important for water and mineral uptake. The positional cues that regulate the spacing of trichomes and root hairs are unique. In the root, positional information is derived from the location of the epidermal cells relative to underlying cortex cells (Fig. 1A). Epidermal cells that are located over a junction between two cortex cells become root hairs (H cells), whereas the other epidermal cells become non-root-hair cells (N cells). On the leaves, the trichome spacing pattern is determined largely by the position of the first trichome (Fig. 1B). After the leaf primordium reaches a length of ∼100 μm, a single epidermal cell at the leaf tip becomes a trichome. Thereafter, a regulated spacing pattern evolves in which newly initiated trichomes are approximately equal distance from one another. As older trichomes become separated by epidermal cell divisions, new intervening trichomes emerge (Fig. 1B).
Figure 1.
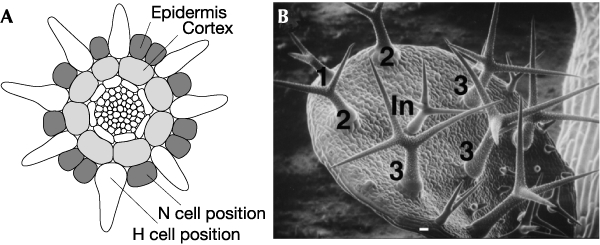
Epidermal patterning of roots and leaves. (A) Line drawing of a cross-section of a mature region of an Arabidopsis root. Epidermal and cortical cell layers are labelled. H, root-hair cell; N, non-root-hair cell. (B) Scanning electron micrograph of a young developing leaf. Trichomes are numbered according to their relative age. The mature trichome labelled 1 was the first to develop. The trichomes labeled In are intervening trichomes that have not fully developed.
Despite the obvious differences between these systems, extensive mutational analyses of root hair and trichome development have identified a partly shared set of genes encoding transcription factors that regulate the patterning of both cell types (see Table 1) (Szymanski et al., 2000; Dolan, 2001). These genes seem to act by promoting trichome development in the shoot, and inhibiting H-cell initiation in the root. They include the functionally equivalent Myb genes GLABROUS1 (GL1) and WEREWOLF (WER), which are expressed in the shoot and root, respectively; the WD-40 repeat-encoding gene TRANSPARENT TESTA GLABRA (TTG), which probably acts as a transcriptional scaffold in both the shoot and root; and the basic helix–loop–helix (bHLH) gene GLABRA3 (GL3). Although mutations in GL3 affect only trichome initiation, a closely related bHLH gene in A. thaliana is predicted to have overlapping functions with GL3 in both the root and shoot. Yeast interaction assays have shown that the GL3 protein can homodimerize and contains discrete domains that interact with the GL1 and TTG proteins (Payne et al., 2000). It has been proposed that a protein complex composed of GL1, GL3 and TTG functions as an activator of genes required for trichome formation and that a similar complex containing WER, TTG and a bHLH protein is important for N-cell identity. The homeodomain encoding GLABRA2 (GL2), which is required for both trichome differentiation and root hair repression, is likely to be a primary target of the shoot and root complexes.
Table 1.
Genes involved in trichome and root hair initiation
| Locus | Gene product | Wild-type expression profile | Function |
|---|---|---|---|
| GL1 | Myb transcription factor | Young leaf primordia and developing trichomes | Promotes trichome initiation |
| WER | Myb transcription factor | Non-root-hair (N) cells | Prevents root-hair (H)-cell initiation |
| GL3 | Myc transcription factor | Not well characterized | Promotes trichome initiation |
| TTG | WD-40 repeat | Not well characterized | Promotes trichome initiation and prevents H-cell initiation |
| CPC | Myb transcription factor | Young leaf primordia and developing trichomes and N cells | Limits trichome initiation and promotes H-cell initiation |
| TRY | Myb transcription factor | Young leaf primordia and developing trichomes and weakly expressed in roots | Prevents trichome clustering |
The expression patterns of WER, GL1 and GL2 have been well characterized. In the shoot, GL1 and GL2 are diffusely expressed throughout young leaf primordia before the emergence of trichomes, and increase sharply in incipient trichomes (Szymanski et al., 1998). In the root, the expression of WER and GL2 is largely restricted to the N cells (Lee & Schiefelbein, 1999).
So how is the spacing of trichomes controlled? The gene TRIPTYCHON (TRY) is a key player, because mutations in TRY result in an abnormally high frequency of clustered trichomes (Hülskamp et al., 1994). Now, in this recent paper, Schellmann et al. report that TRY encodes another Myb-like transcription factor; however, unlike WER and GL1, which encode proteins with two Myb DNA-binding domains and putative acidic transcriptional activation domains, the predicted TRY protein contains only one Myb DNA-binding domain and seems to lack an activator domain. The expression pattern of TRY in the shoot mirrors the expression of GL1 and GL2. Given that the genes that positively influence trichome initiation (GL1 and GL2) along with the negative regulator, TRY, are all initially expressed at high levels in young leaf primordia, it is difficult to understand how the pattern of trichome initiation is established. Schellmann et al. propose a model in which the activator complex (for example, GL1–GL3–TTG in the shoot) has the ability to stimulate the production of a transmitted inhibitory signal. Subtle differences in the levels of the activator complex would create a cell that is capable of sending a stronger inhibitory signal to its neighbours, and this signal would prevent those neighbouring cells from entering the trichome pathway. The prediction is that the TRY gene is a target of the activator complex and is responsible for transmitting the inhibitory signal. The exact nature of this inhibitory signal is still unknown, although it is possible that it is encoded by the TRY gene itself. The predicted TRY protein is sufficiently small for it to be able to move from cell to cell through the plasmodesmata, and it might act as an inhibitor either by disrupting the formation of the activator complex or by binding to regulatory elements of genes regulated by the activator complex (see Fig. 2 and Schellmann et al. (2002) for possible models).
Figure 2.
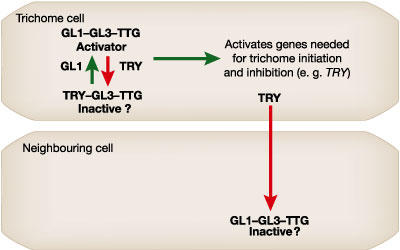
One possible model to explain the function of TRY. In the cell fated to become a trichome the proteins encoded by the GL1, GL3 and TTG genes form a transcriptional complex capable of activating genes required for trichome differentiation (such as GL2) and gene(s) needed to mediate the lateral inhibition signal (such as TRY) that prevents neighbouring cells from becoming trichomes. In the simplest case, the TRY protein is able to modulate the level of activator complex in the developing trichomes and also moves into the neighbouring cell to disrupt the activator complex. A complex containing TRY, GL3 and TTG is here predicted to be inactive; however, as an alternative this complex could activate the inhibition genes. A similar model could exist in the root in which GL1 and TRY are replaced by WER and CPC, and the activator complex level is highest in N (non-root-hair) cells.
Schellmann et al. also established that there is a relationship between TRY and CAPRICE (CPC), a closely related gene required for H-cell formation. CPC, like TRY, encodes a Myb-type factor that contains a single Myb domain and lacks an obvious activating domain (Wada et al., 1997). Previously, Lee & Schiefelbein (2002) presented evidence that CPC acts as a lateral inhibitor in the root epidermis to prevent WER from activating GL2 in H cells. Schellmann et al. extended these observations by providing evidence that TRY and CPC have overlapping functions in both the root and the shoot. They found that CPC expression overlaps TRY expression in the shoot and that the loss of both CPC and TRY activities greatly enhances trichome clustering. In addition, they showed that TRY is expressed at low levels in the root and that the cpc/try double mutant exhibits an even greater loss of H cells than the cpc mutant. Interestingly, cpc mutants do not have a trichome-clustering phenotype, nor do try mutants exhibit a root hair defect. Thus, we can speculate that the TRY and CPC genes, which probably arose from a fairly recent gene duplication event, are in the process of evolving distinct functions in Arabidopsis. It will be interesting to examine these genes in related species to compare the degree of this separation in function.


References
- Dolan L. (2001) How and where to build a root hair. Curr. Opin. Plant Biol., 4, 550–554. [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misra S. & Jürgens G. (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell, 76, 555–566. [DOI] [PubMed] [Google Scholar]
- Lee M.M. & Schiefelbein J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell, 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee M.M. & Schiefelbein J. (2002) Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell, 14, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C.T., Zhang F. & Lloyd A.M. (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics, 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S. et al. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J., 21, 5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D.B., Jilk R.A., Pollock S.M. & Marks M.D. (1998) Control of GL2 expression in Arabidopsis leaves and trichomes. Development, 125, 1161–1171. [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Lloyd A.M. & Marks M.D. (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci., 5, 214–219. [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y. & Okada K. (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science, 277, 1113–1116. [DOI] [PubMed] [Google Scholar]


