Abstract
We propose an integrated model for eukaryotic DNA replication to explain the following problems: (1) How is DNA spooled through fixed sites of replication? (2) What and where are the helicases that unwind replicating DNA? (3) Why are the best candidates for replicative helicases, namely mini-chromosome maintenance (MCM) proteins, not concentrated at the replication fork? (4) How do MCM proteins spread away from loading sites at origins of replication? We draw on recent discoveries to argue that the MCM hexameric ring is a rotary motor that pumps DNA along its helical axis by simple rotation, such that the movement resembles that of a threaded bolt through a nut, and we propose that MCM proteins act at a distance from the replication fork to unwind DNA. This model would place DNA replication in a growing list of processes, such as recombination and virus packaging, that are mediated by ring-shaped ATPases pumping DNA by helical rotation.
Introduction
Precise replication of the eukaryotic genome presents a complex logistical challenge. There is a growing amount of information on the mechanisms that couple DNA replication to the cell cycle, ensuring that all DNA is replicated only once (Diffley & Labib, 2002), but major questions remain about how the extreme length of eukaryotic DNA is managed during replication.
Pulse labelling of newly replicated DNA has repeatedly suggested that hundreds of replication forks are clustered into replication foci or 'factories', rather than being uniformly distributed throughout the nucleus (Pardoll et al., 1980; Jackson & Cook, 1986; Nakamura et al., 1986; Mills et al., 1989; Nakayasu & Berezney, 1989; Hozak et al., 1993). Furthermore, pulse–chase experiments have shown that newly replicated DNA moves away from these foci. These observations have led to the repeated suggestion that DNA might spool through fixed sites of replication within the nucleus, although this interpretation is controversial and has been difficult to visualize at a molecular level.
The 'MCM paradox'
MCM (or mini-chromosome maintenance) proteins are required for the initiation of DNA replication throughout eukaryotes from yeasts to humans, and homologues also exist in Archaea (Yan et al., 1993; Madine et al., 1995a; Tye, 2000). They form heterohexameric complexes that have DNA helicase activity which, although weak, is sufficient to displace short oligonucleotides from complementary DNA circles (Ishimi, 1997; You et al., 1999; Chong et al., 2000; Lee & Hurwitz, 2000), and they are currently regarded as the prime candidates for the DNA helicases that unwind DNA at replication forks. However, their abundance and distribution suggest that they are more than just DNA helicases and that their site of action might not be the replication fork. Although, in yeast, it is possible to detect MCM proteins close to replication forks (Aparicio et al., 1997), it is not clear whether they are preferentially localized there. Furthermore, MCM proteins are not preferentially localized to sites of DNA replication in animal cells (Madine et al., 1995b; Krude et al., 1996; Dimitrova et al., 1999).
Before the onset of DNA replication in mammalian or Xenopus cells, the MCM proteins are bound to chromatin throughout the nucleus. However, after the initiation of DNA replication, the MCM proteins bind preferentially to unreplicated DNA, rather than to replicating or replicated DNA (Madine et al., 1995b; Krude et al., 1996; Dimitrova et al., 1999). Thus, they seem to mark unreplicated DNA and to contribute to the mechanism that prevents the reinitiation of replication within a single cell cycle. Chromatin-bound MCM complexes fail to co-localize with sites of DNA replication throughout S phase (Fig. 1A and B), in contrast to the essential replication protein RPA, which binds to singlestranded DNA at replication forks and co-localizes with sites of DNA replication throughout S phase (Fig. 1C). Another replication-fork-associated protein, proliferating cell nuclear antigen (PCNA), also differs from MCMs in that it co-localizes exactly with sites of DNA synthesis (Dimitrova et al., 1999). Strikingly similar patterns of MCM labelling are observed in a variety of somatic animal cells, both in vivo and in vitro (Madine et al., 1995b; Krude et al., 1996; Dimitrova et al., 1999) (Fig. 1). In all these cases, the MCMs fail to co-localize with sites of DNA replication, suggesting that this difference might be common in higher eukaryotes. This is further supported by experiments with human cells, which have shown that MCM and the origin recognition complex (ORC) proteins are separated by at least 0.5–1 kilobase of DNA (Ritzi et al., 1998). These observations present a dilemma: how can MCM complexes act as the replicative helicase yet fail to coincide spatially with sites of DNA replication?
Figure 1.
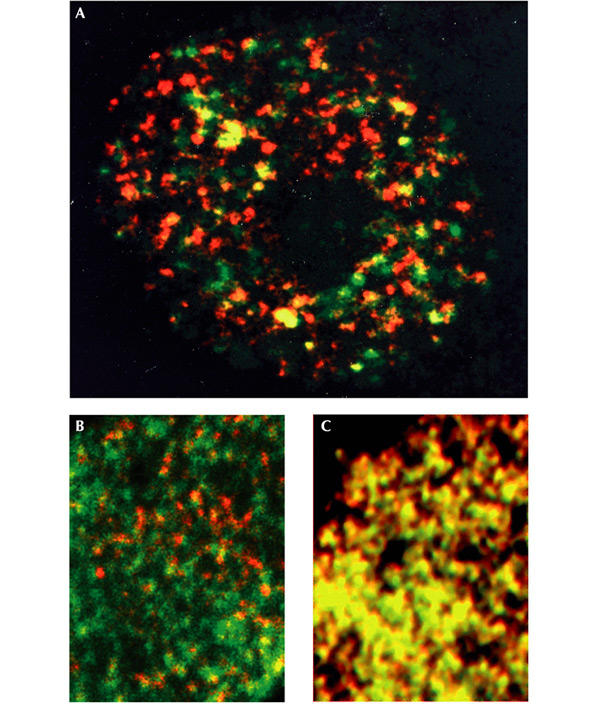
Mcm3 is not concentrated at sites of DNA replication, whereas RPA (replication protein A) is. (A) The nucleus of a single Xenopus cultured cell immunostained for newly replicated DNA (red) and Mcm3 (green). (B) Similarly, part of a Xenopus sperm nucleus replicating in Xenopus egg extract is stained for newly synthesized DNA (red), and Mcm3 (green). (C) Part of a Xenopus sperm nucleus replicating in Xenopus egg extract under identical conditions to those in (B), but stained for newly synthesized DNA (red) and RPA (green) for comparison, because RPA is known to be located at replication forks. Mcm3 staining (green) is not co-localized with newly replicated DNA (red) in A and B, whereas RPA staining is co-localized, producing yellow spots in C. B and C are reproduced from Madine et al. (1995b), with permission from Elsevier Science.
A further paradox lies in the abundance of the MCM proteins. In prokaryotes, origin-binding complexes bind to DNA and recruit additional proteins, including the replicative helicases. In Bacillus subtilis and Escherichia coli, two helicases per origin seem sufficient to allow bidirectional DNA replication, with each helicase acting at a single replication fork. Eukaryotes use a superficially similar loading mechanism, whereby ORC recruits Cdc6 and Cdt1, and these in turn recruit the MCM complexes (Maiorano et al., 2000; Nishitani et al., 2000; Tada et al., 2001; Diffley & Labib, 2002; Tanaka & Diffley, 2002). However, rather than two MCM complexes being loaded per ORC, the number is between 10 and 40 in animal cells (Mahbubani et al., 1997; Edwards et al., 2002). In yeast there seem to be more than 100 MCM complexes per replication origin, and at least one of the MCM subunits is limiting for the rate of DNA replication (Lei et al., 1996). These results illustrate that in eukaryotes about one MCM complex is bound per kilobase of DNA.
Interestingly, the MCM complexes seem to spread away from their loading sites on chromatin. Although they require ORC, Cdc6 and Cdt1 to load, they are not found only at sites of initiation, as with ORC (Romanowski et al., 1996; Edwards et al., 2002). These results emphasize the paradox between the spatial localization and function of MCM proteins. Moreover, they raise the question of how such a large excess of MCM complexes can be explained if they simply act as helicases.
Clues to a potential reconciliation of these observations come from a recent study of the Saccharomyces cerevisiae MCM complexes (Schwacha & Bell, 2001), which showed that their ATPase activities are remarkably similar to those of the mitochondrial F1-ATPase in terms of both kinetics and subunit behaviour (Boyer, 1993). Both of these complexes have ring-like hexamers composed of three ATPase catalytic subunits (MCMs 4, 6 and 7 in the MCM complex) interspersed with three regulatory subunits (MCMs 2, 3 and 5) (Boyer, 1993; Schwacha & Bell, 2001). The patterns of inactivating and suppressing mutations are also extraordinarily similar between the F1-ATPase and the MCM complex. In terms of mechanics, after ATP hydrolysis the F1-ATPase is able to rotate an additional component (the gamma subunit) that is bound in the central channel of its ring (Boyer, 1993). Schwacha & Bell (2001) have proposed that the MCM helicase acts in a similar way, rotating DNA through the central channel of its ring structure. Thus, ATP hydrolysis would drive rotational movement of the MCM complex, resulting in an unwinding of the DNA. Like previous proposals, Schwacha & Bell's involves MCM complex activity at the replication fork, where it encloses singlestranded DNA (Schwacha & Bell, 2001). However, image reconstruction from electron micrographs of MCM complexes reveals a central hole (Sato et al., 2000, Yu et al., 2002) which is apparently large enough to accommodate either singlestranded or double-stranded DNA.
A rotary pumping model for helicase action at a distance
We suggest that the existing data are far more consistent with an alternative model, namely that the MCM proteins are indeed rotary motors, but that they translocate DNA along its axis by helical rotation, causing it to unwind at distant, fixed replication forks (Fig. 2). This is analogous to a helical threaded 'bolt' rotating through 'nuts' (see below), and there are several precedents for such a rotational translocation of DNA. We envisage two steps, both using the same postulated rotary function. First, MCM complexes would load onto DNA at the origin of replication and move away from the origin by rotation along the helical thread of the DNA. In this way, MCMs would disperse along the unreplicated DNA. For this step, it is important that all of the hexamers on each side of ORC face in the same direction as their neighbours. We envisage that their orientation is specified by ORC in association with Cdc6 and Cdt1, all of which are required for MCM loading. Second, the dispersed MCMs would be anchored to an immobile nuclear structure and the identical rotary action would be repeated. Now only the DNA would be able to rotate, and this action would effectively pump DNA back towards the origin of replication (Fig. 2). This would provide a simple and efficient mechanism for spooling large quantities of DNA through 'anchored' sites of replication.
Figure 2.
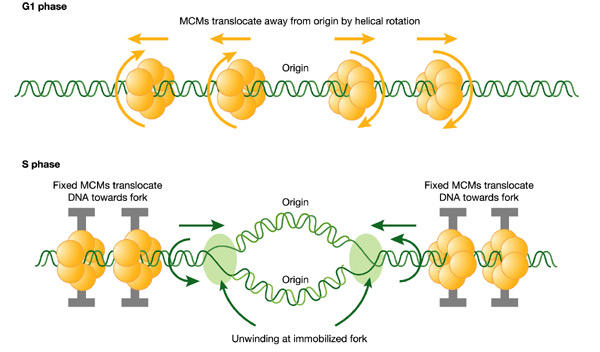
A hypothetical rotary pump model showing two stages in the distribution and function of MCM hexameric ATPase complexes. First, in G1 phase, MCM hexamers move spirally along the helical grooves of unreplicated DNA, away from ORC, which is required for their loading and orientation. Second, in S phase, MCMs become immobilized, so that exactly the same rotary mechanism moves the DNA instead of the MCM proteins. This would result in translocation of DNA towards the replication forks. As DNA is twisted by fixed MCMs in S phase, it would become unwound at the distant replication fork, which is itself immobilized in fixed clusters (see Fig. 3).
In addition to accounting for the observed distribution and behaviour of MCMs and replicating DNA, this two-step model has an important third consequence. Rotation of DNA in opposite directions from the two sides of the replication bubble would unwind the DNA at the replication fork (Fig. 2). This unwinding effect would be increased by the immobilization of replication forks. Thus, we propose that the MCM proteins are indeed replicative helicases, but that they unwind DNA by twisting it at sites that are remote from the replication fork. According to this model, the length of unreplicated DNA would shorten as replication proceeds, and the MCM complexes would be displaced from DNA. The model accounts for the immunofluorescent staining of the nucleus described above, and also for the observed excess of MCMs in higher eukaryotes. In addition, damage to the template in the form of a nick between the replication fork and the nearest MCM hexamer would automatically prevent further unwinding, causing a checkpoint delay.
Precedents for rotary motors that translocate DNA
Several other hexameric ATPases have been shown to act as DNA rotary translocation motors that exploit the helical structure of DNA to pump it along its fibre axis (Table 1; Egelman, 2001). For example, the bacteriophage φ29 gp 10 protein forms a hexameric ring that has been proposed to pump DNA into and out of the bacteriophage capsid in this way (Simpson et al., 2000). In addition, the bacterial proteins RuvB (Parsons et al., 1995), SpoIIIE (Bath et al., 2000) and TrwB (Gomis-Ruth et al., 2001) also seem to pump DNA through their central cavity by coupling ATP hydrolysis with rotational translocation of DNA. Interestingly, TrwB translocates singlestranded DNA whereas SpoIIIE, φ29 gp 10 and RuvB seem to translocate double-stranded DNA. The role of RuvB in recombination is a particularly relevant example, because two opposite-facing hexameric rings pump double-stranded DNA away from the space between them, unwinding the DNA as they do so (Parsons et al., 1995). Recently FtsK, a protein involved in linking chromosome segregation and cell division in E. coli, has been added to this growing list of ATP-dependent DNA translocases (Aussel et al., 2002). Furthermore, Kaplan & O'Donnell (2002) have now shown that the bacterial replicative helicase DnaB can also enclose and translocate along either singlestranded or double-stranded DNA.
Table 1.
DNA rotary motors
| DNA motor | Organism | Function | Reference |
|---|---|---|---|
| gp10 | Bacteriophage φ29 | Packaging double-stranded DNA into capsid | Simpson et al. (2000) |
| RuvB | Escherichia coli | Holliday junction branch migration | Parsons et al. (1995) |
| DnaB | Escherichia coli | Replication fork unwinding and Holliday junction branch migration | Kaplan & O'Donnell (2002) |
| SpoIIIE | Bacillus subtilis | Chromosome segregation | Bath et al. (2000) |
| TrwB | Escherichia coli | Conjugation | Gomis-Ruth et al. (2001) |
| FtsK | Escherichia coli | DNA supercoiling and chromosome segregation | Aussel et al. (2002) |
Features of the model
Whether the MCM ring complex wraps around double-stranded or single-stranded DNA or both remains an important question in determining its mechanism of action. However, it is difficult to explain the images shown in Fig. 1 if singlestranded DNA is the only binding site, whereas the concept of MCMs acting as a series of nuts on a bolt clearly illustrates how a cluster of 'rotary motors' can couple rotation with translocation to spool large quantities of DNA. Another analogy would be multiple rollers driving a conveyor belt, or many hands pulling on the same rope. This model would also explain the apparent difficulty in demonstrating helicase activity in the hexameric protein. This view predicts that immobilizing one end of a DNA molecule should substantially increase MCM helicase activity. Unfortunately, this is difficult to test directly at present, because MCM loading onto purified DNA remains inefficient in vitro, although a system described recently by Edwards et al. (2002) should allow this to be achieved in the near future.
An apparent problem with the model proposed here is one that has been likened to two people sucking the same strand of spaghetti: is DNA between two forks pumped in two opposing directions? Fortunately, an obvious answer to this problem is suggested by the radial loop structure of chromosomes and the observations that eukaryotic replication forks are grouped into foci (Pardoll et al., 1980; Jackson & Cook, 1986; Nakamura et al., 1986; Mills et al., 1989; Nakayasu & Berezney, 1989; Hozak et al., 1993). As shown in Fig. 3, adjacent forks within a cluster can be contiguous, so that loops of replicated DNA grow as unreplicated loops contract. The remaining problem, namely how the small sections of DNA between the focal clusters are replicated, is common to other models of replication and could possibly be solved by unidirectional fork movement between clusters. Similarly, the problem of nucleosome displacement during replication is common to other proposed mechanisms of DNA replication, and its solution awaits a better understanding of chromatin remodelling factors.
Figure 3.
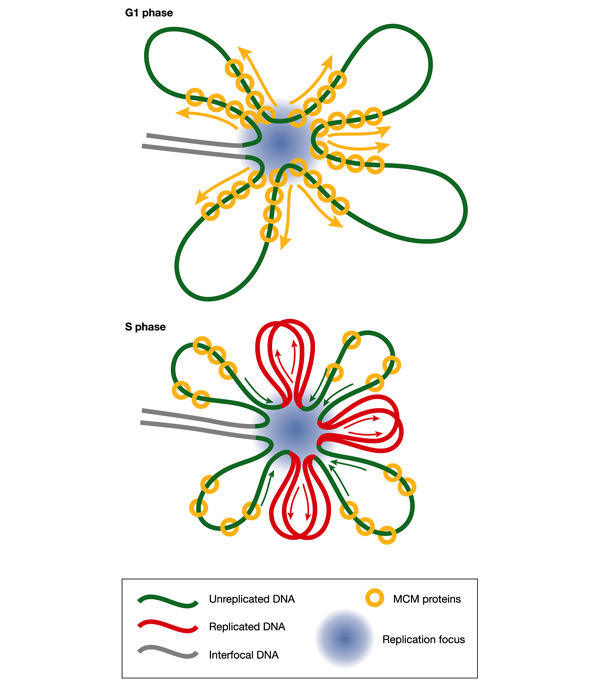
Implications of the rotary pump model for the clustering of several hundred replication forks into foci. Before DNA replication in G1 phase, MCM complexes (yellow) are distributed along radial loops of unreplicated DNA (green). During DNA replication in S phase, the forks remain clustered in foci (blue spot) and extrude replicated DNA loops (red). Arrows indicate growth and shrinkage of replicated and unreplicated loops, respectively, as replication proceeds.
In developing the model presented here, we have sought to explain the paradoxical distribution and abundance of the best candidates for eukaryotic replicative helicases, the MCM protein complexes. We propose that they are indeed replicative helicases, but that they act from a distance, by twisting DNA so that it unwinds at the constrained replication fork. This would explain their localization specifically to unreplicated DNA. In addition, it provides a plausible explanation for the abundant evidence that eukaryotic DNA is spooled through fixed sites of DNA replication. Thus we propose that spooling, like unwinding, would be achieved by MCM motors on the unreplicated DNA, rather than at the fork. This model adds DNA replication to the growing list of DNA manipulations mediated by ring-shaped rotary motors that translocate DNA along its fibre axis by a helical rotation mechanism.


Acknowledgments
We thank A. Klug, D. Sherratt, A. Venkitaraman, L. Ko-Ferrigno and all members of our research group for comments on the manuscript. We thank the Medical Research Council, Cancer Research UK and the Louis Jeantet Foundation for support.
References
- Aparicio O.M., Weinstein D.M. & Bell S.P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell, 91, 59–69. [DOI] [PubMed] [Google Scholar]
- Aussel L. et al. (2002) FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell, 108, 195–205. [DOI] [PubMed] [Google Scholar]
- Bath J., Wu L.J., Errington J. & Wang J.C. (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell–prespore division septum. Science, 290, 995–997. [DOI] [PubMed] [Google Scholar]
- Boyer P.D. (1993) The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim. Biophys. Acta, 1140, 215–250. [DOI] [PubMed] [Google Scholar]
- Chong J.P., Hayashi M.K., Simon M.N., Xu R.M. & Stillman B. (2000) A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl Acad. Sci. USA, 97, 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. & Labib K. (2002) The chromosome replication cycle. J. Cell Sci., 115, 869–872. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Todorov I.T., Melendy T. & Gilbert D.M. (1999) Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol., 146, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M.C. et al. (2002) MCM2–7 complexes bind chromatin in a distributed pattern surrounding ORC in Xenopus egg extracts. J. Biol. Chem., 277, 33049–33057. [DOI] [PubMed] [Google Scholar]
- Egelman E. (2001) Pumping DNA. Nature 409, 573–575. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth F.X. et al. (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature, 409, 637–641. [DOI] [PubMed] [Google Scholar]
- Hozak P., Hassan A.B., Jackson D.A. & Cook P.R. (1993) Visualization of replication factories attached to nucleoskeleton. Cell, 73, 361–373. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem., 272, 24508–24513. [DOI] [PubMed] [Google Scholar]
- Jackson D.A. & Cook P.R. (1986) Replication occurs at a nucleoskeleton. EMBO J., 5, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.L. & O'Donnell M. (2002) DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell, 10, 647–657. [DOI] [PubMed] [Google Scholar]
- Krude T., Musahl C., Laskey R.A. & Knippers R. (1996) Human replication proteins hCdc21, hCdc46 and P1Mcm3 bind chromatin uniformly before S-phase and are displaced locally during DNA replication. J. Cell Sci. 109, 309–318. [DOI] [PubMed] [Google Scholar]
- Lee J.K. & Hurwitz J. (2000) Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J. Biol. Chem., 275, 18871–18878. [DOI] [PubMed] [Google Scholar]
- Lei M., Kawasaki Y. & Tye B.K. (1996) Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5081–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine M.A., Khoo C.Y., Mills A.D. & Laskey R.A. (1995a) MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature, 375, 421–424. [DOI] [PubMed] [Google Scholar]
- Madine M.A., Khoo C.Y., Mills A.D., Musahl C. & Laskey R.A. (1995b) The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr. Biol., 5, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Mahbubani H.M., Chong J.P., Chevalier S., Thommes P. & Blow J.J. (1997) Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J. Cell Biol., 136, 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D., Moreau J. & Mechali M. (2000) XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature, 404, 622–625. [DOI] [PubMed] [Google Scholar]
- Mills A.D. et al. (1989) Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J. Cell Sci., 94, 471–477. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Morita T. & Sato C. (1986) Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res., 165, 291–297. [DOI] [PubMed] [Google Scholar]
- Nakayasu H. & Berezney R. (1989) Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol., 108 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T. & Nurse P. (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature, 404, 625–628. [DOI] [PubMed] [Google Scholar]
- Pardoll D.M., Vogelstein B. & Coffey D.S. (1980) A fixed site of DNA replication in eukaryotic cells. Cell, 19, 527–536. [DOI] [PubMed] [Google Scholar]
- Parsons C.A., Stasiak A., Bennett R.J. & West S.C. (1995) Structure of a multisubunit complex that promotes DNA branch migration. Nature, 374, 375–378. [DOI] [PubMed] [Google Scholar]
- Ritzi M. et al. (1998) Human minichromosome maintenance proteins and human origin recognition complex 2 protein on chromatin. J. Biol. Chem., 273, 24543–24549. [DOI] [PubMed] [Google Scholar]
- Romanowski P., Madine M.A., Rowles A., Blow J.J. & Laskey R.A. (1996) The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol., 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- Sato M. et al. (2000) Electron microscopic observation and singlestranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol., 300, 421–431. [DOI] [PubMed] [Google Scholar]
- Schwacha A. & Bell S.P. (2001) Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol. Cell, 8, 1093–1104. [DOI] [PubMed] [Google Scholar]
- Simpson A.A. et al. (2000) Structure of the bacteriophage φ29 DNA packaging motor. Nature, 408, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S., Li A., Maiorano D., Mechali M. & Blow J.J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nature Cell Biol., 3, 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. & Diffley J.F.X. (2002) Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2—7 during G1 phase. Nature Cell Biol., 4, 198–207. [DOI] [PubMed] [Google Scholar]
- Tye B.K. (2000) Insights into DNA replication from the third domain of life. Proc. Natl Acad. Sci. USA, 97, 2399–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Merchant A.M. & Tye B.K. (1993) Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev., 7, 2149–2160. [DOI] [PubMed] [Google Scholar]
- You Z., Komamura Y. & Ishimi Y. (1999) Biochemical analysis of the intrinsic Mcm4-Mcm6-Mcm7 DNA helicase activity. Mol. Cell. Biol., 19, 8003–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. et al. (2002) The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep., 3, 792–797. [DOI] [PMC free article] [PubMed] [Google Scholar]


