Abstract
Mu bacteriophage inserts its DNA into the genome of host bacteria and is used as a model for DNA transposition events in other systems. The eukaryotic Ku protein has key roles in DNA repair and in certain transposition events. Here we show that the Gam protein of phage Mu is conserved in bacteria, has sequence homology with both subunits of Ku, and has the potential to adopt a similar architecture to the core DNA-binding region of Ku. Through biochemical studies, we demonstrate that Gam and the related protein of Haemophilus influenzae display DNA binding characteristics remarkably similar to those of human Ku. In addition, we show that Gam can interfere with Ty1 retrotransposition in Saccharomyces cerevisiae. These data reveal structural and functional parallels between bacteriophage Gam and eukaryotic Ku and suggest that their functions have been evolutionarily conserved.
Introduction
Ku is a heterodimer of two related subunits, Ku70 and Ku80 (Dynan & Yoo, 1998; Gell & Jackson, 1999). Orthologues of both subunits have been discovered in many eukaryotes. More recently, the genes for putative Ku orthologues have been identified in silico in certain bacterial genomes (Aravind & Koonin, 2001; Doherty et al., 2001). Eukaryotic Ku functions by binding to DNA ends and this property is essential for the efficient repair of DNA doublestrand breaks (DSBs) by the non-homologous end-joining (NHEJ) pathway. DSBs can be generated by exogenous DNA-damaging agents and during recombination events such as site-specific V(D)J recombination in immunoglobulin and T-cell receptor genes in mammals, and during retroviral DNA integration into the host. Consistent with there being mechanistic links between DNA DSB repair and these events, both V(D)J recombination and retroviral integration are potentiated by Ku (Smith & Jackson, 1999; Daniel et al., 1999; Li et al., 2001).
Mu phage is one of the most efficient transposable elements known. Its transposition mechanism shares many features with V(D)J recombination and retroviral integration (Craig, 1995). Mu can efficiently propagate itself in common laboratory Escherichia coli strains. We therefore speculated that a functional counterpart of Ku might operate in this system. Consistent with the fact that DNA DSB repair in E. coli is mediated by homologous recombination and that no evidence for NHEJ has been reported in this species, we failed to identify any apparent Ku orthologue in the E. coli genome sequence. However, we found that the Gam gene of Mu phage encodes a protein with homology to Ku. Previous work has indicated that Mu Gam facilitates Mu replication (Goosen et al., 1982) and that Gam can bind nonspecifically to DNA and protect linear Mu phage DNA from digestion with nuclease (Akroyd et al., 1984; Akroyd & Symonds, 1986; Abraham & Symonds, 1990). As described here, our further analysis of Gam indicates that it is indeed a functional counterpart of eukaryotic Ku.
Results and Discussion
Gam proteins have similarity to eukaryotic Ku
By using the sequence analysis software Clustal X, we discovered that the early gene Gam of bacteriophage Mu (hereafter termed MuGam) shares statistically significant sequence homology with both subunits of eukaryotic Ku (for example, 17% identity and 27% similarity with human Ku80, and 13% identity and 9% similarity with human Ku70; Fig. 1A). Subsequent BLAST searches of protein sequence databases identified MuGam orthologues in the bacterial genomes of Haemophilus influenzae, Salmonella typhi, Neisseria meningitidis and the enterohemorrhagic O157:H7 strain of E. coli (Fig. 1A, and data not shown). The genomic locations of these Gam-like genes indicated that they might be defective prophages (data not shown). In addition, we found that MuGam and the products of these Gam-like genes have homology with recently identified genes encoding Ku-like proteins present in the genomes of a different set of prokaryotes (Doherty et al., 2001; Weller et al., 2002).
Figure 1.
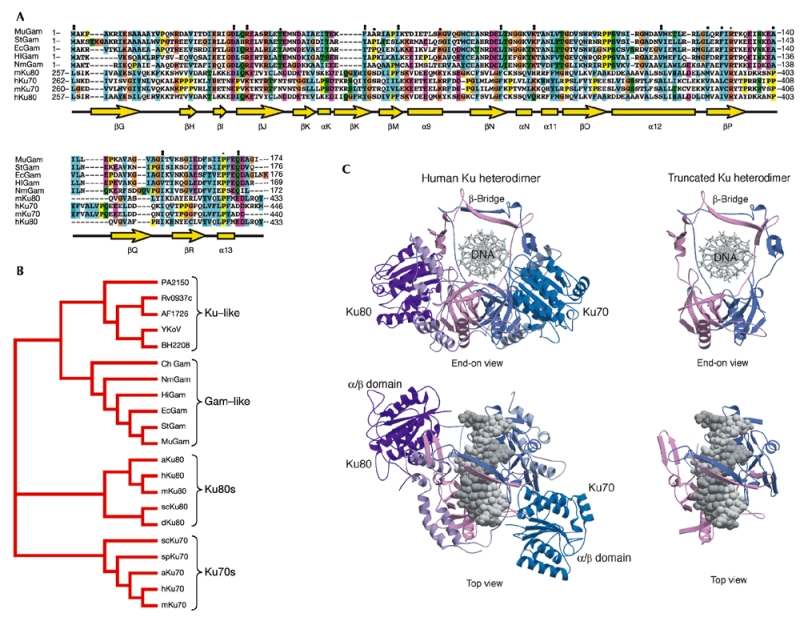
MuGam and Gam-like proteins have sequence and structural homology to eukaryotic Ku. (A) A structure-based sequence alignment of the Gam and Ku proteins. Secondary structural elements, from the crystal structure of the Ku heterodimer, are indicated below the sequences in yellow. Cylinders indicate α-helices, arrows β-sheets, asterisks identical residues in all sequences in the alignment, colons conserved substitutions and full stops semi-conserved substitutions. Ec, Escherichia coli O157:H7; Hi, Haemophilus influenzae; Mu, bacteriophage Mu; Nm, Neisseria meningitidis; St, Salmonella typhi; h, Homo sapiens; m, Mus musculus. (B) Phylogenetic analysis of Gam and Ku orthologues. Unrooted phylogenetic tree based on Ku and Gam orthologues from Saccharomyce cerevisiae (sc), Schizosaccharomyces pombe (sp), Arabidopsis thaliana (a), Archaeglobus fulgidus (AF), Bacillus subtilis (BS), Bacillus halodurans (Yk), Mycobacterium tuberculosis (RV), Pseudomonas aeruginosa (PA) and Campylobacter hyoilei (Ch). (C) Structure of the core region of human Ku dimer that aligns with Gam proteins.
Consistent with a proviral origin for the various bacterial Gam-like proteins, phylogenetic analysis revealed that they are more related to MuGam than they are to bacterial Ku-like proteins or to eukaryotic Ku subunits (Fig. 1B).
The recent determination of the crystal structure of the human Ku shows that the two subunits form a pseudo-symmetrical complex (Walker et al., 2001; Doherty & Jackson, 2001). In this structure, Ku70 and Ku80 have a common three-domain architecture composed of an amino-terminal α/β domain, a central β-barrel domain and a helical carboxy-terminal arm structure (Fig. 1C). The complex has an open ring-like shape with the DNA threaded through the aperture. These structural features allow Ku to recognize DNA ends irrespective of their sequence or structure, and permit Ku to then slide along the DNA. Strikingly, the region of homology of MuGam and the other Gam-like proteins with Ku corresponds to the central DNA-binding and dimerization core of Ku and lacks the N- and C-terminal domains of eukaryotic Ku, which have been postulated to mediate further interactions (Dynan & Yoo, 1998; Gell & Jackson, 1999). Indeed, our structure-based sequence analysis indicates that the Gam proteins might equate to the bare minimum DNA-binding and dimerization region of Ku (Fig. 1C). It has been reported that equivalent truncated versions of eukaryotic Ku retain both hetero-dimerization and DNA end-binding activities (Wu & Lieber, 1996; Wang et al., 1998).
MuGam and HiGam interact with DNA similarly to Ku
In view of the above, we decided to test whether MuGam and its H. influenzae orthologue (hereafter called HiGam) can dimerize and bind to DNA. To do this, we overexpressed MuGam and HiGam in E. coli and purified them to homogeneity. Eukaryotic Ku70 and Ku80 form a stable heterodimeric complex in solution. Analysis of purified recombinant MuGam and HiGam by analytical gel-filtration chromatography indicated that these proteins exist as a homodimer in solution (Fig. 2, and data not shown), because the protein eluted from the column as a single peak with an apparent molecular mass of ∼45 kDa (MuGam monomer ∼21 kDa; insert in Fig. 2). The dimeric state of MuGam was confirmed by sedimentation equilibrium ultracentrifugation analysis, which indicated a molecular mass of 39 kDa for MuGam in solution (see Supplementary Information).
Figure 2.
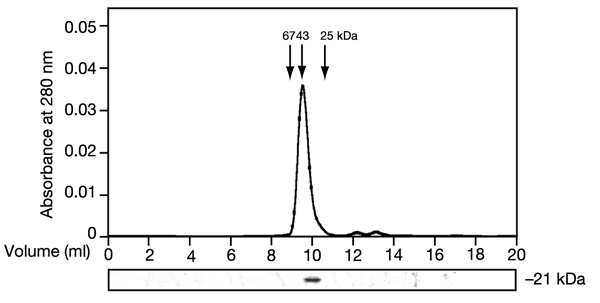
MuGam exists as a homodimer in solution. MuGam was eluted from a Superdex S-75 gel filtration column as a single peak with an apparent molecular mass of ∼45 kDa (upper panel). The relative elution points for the molecular mass standards are also indicated. SDS–PAGE lower panel) confirmed the presence of MuGam in the peak fraction and also its apparent molecular mass.
The most distinctive biochemical feature of Ku is its strong preference for linear as opposed to circular DNA. We therefore tested the DNA-binding ability of MuGam and HiGam, together with human Ku as a control, in electrophoretic mobility-shift assays (EMSAs). As shown in Fig. 3A, both MuGam and HiGam, along with Ku, were able to form complexes with the end-labelled 33-base-pair (bp) doublestranded (ds) oligonucleotide TL4 used in these studies. As has been reported for Ku, and in agreement with its ability to bind DNA like a bead on a thread, we observed that a sufficiently long fragment of DNA (longer than ∼20 bp) could accommodate more than a single molecule of MuGam or HiGam, giving rise to more than one retarded band (Fig. 3A, B, and data not shown). To examine the ability of these proteins to distinguish between circular DNA and linear DNA fragments, we performed competition assays in which increasing amounts of circular or linearized plasmid DNA were added to the reaction containing the labelled oligonucleotide, before the addition of protein. Significantly, although circular plasmid had little effect on the ability of MuGam, HiGam or Ku to recognize the labelled oligonucleotide and generate a retarded complex (Fig. 3A, lanes 3–5, 10–12 and 17–19), linearized plasmid DNA competed effectively for binding and led to the disappearance of the retarded complex (lanes 6–8, 13–15 and 20–22). Taken together, these data indicate that, like eukaryotic Ku, MuGam and HiGam bind preferentially to linear ds DNA.
Figure 3.
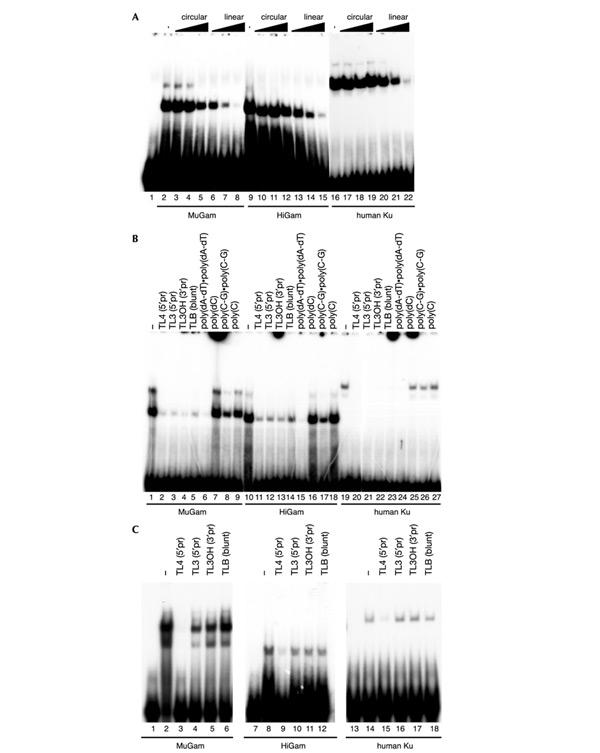
MuGam, HiGam and human Ku bind specifically to DNA ends. EMSAs were employed to characterize the DNA-binding abilities of MuGam, HiGam and human Ku. (A) An end-labelled ds oligonucleotide (TL4) was mixed with increasing amounts (5–250-fold molar excess) of circular (lanes 3–5, 10–12 and 17–19) or linearized (lanes 6–8, 13–15 and 20–22) pUC plasmid. Purified MuGam, HiGam or Ku was added last. (B) End-labelled ds oligonucleotide (TL4) was mixed with a 20-fold molar excess of unlabelled competitor ds oligonucleotides with the same core sequence but carrying different ends—5′ protruding (5′pr), 3′ protruding (3′pr) and blunt ends (lanes 2–5, 11–14 and 20–23)—or a set of synthetic DNA or RNA polynucleotides (lanes 6–9, 15–18 and 24–27); purified MuGam, HiGam or Ku was added last. (C) MuGam, HiGam and Ku were first allowed to bind to TL4 end-labelled ds oligonucleotide. Subsequently, these complexes were challenged with the same set of unlabelled ds oligonucleotides as used in B. MuGam, HiGam and Ku could translocate only onto that oligonucleotide (TL4; lanes 3, 9 and 15) that bore homologous DNA ends with matched bases.
We then extended our EMSA competition studies to examine the ability of MuGam and HiGam to bind to various forms of linear nucleic acid. Thus, we performed EMSAs with a labelled 33-bp ds DNA (TL4) that had been mixed with a series of unlabelled competitor ds DNA oligonucleotides that had the same core sequence but differed in their end sequences so that they possessed 5′ or 3′ protruding overhangs or blunt termini (see Bliss & Lane (1977) for their sequences). As shown in Fig. 3B, all of these DNA molecules competed effectively, indicating that MuGam, HiGam and human Ku bind to linear ds DNA irrespective of the precise architecture of the DNA terminus. In addition, in competition assays with a series of synthetic DNA or RNA polynucleotides in their singlestranded (ss) or ds form, ds DNA molecules such as poly(dA-dT)·poly(dA-dT) bound MuGam, HiGam or Ku effectively, whereas poly(dC), which can exist only in ss form, did not. Furthermore, as for eukaryotic Ku, no competition for MuGam or HiGam binding was observed with a range of ss or ds RNA molecules. Taken together, these data show that MuGam and HiGam, like eukaryotic Ku, bind effectively to ds DNA ends but not to RNA (ds or ss) or to ss DNA.
Previous work has established that eukaryotic Ku has the special ability to translocate from one DNA fragment to another if, and only if, the two DNA molecules bear complementary, cohesive termini (Bliss & Lane, 1997). To investigate whether this property is shared by MuGam and HiGam, we allowed these proteins or human Ku to bind to a labelled ds oligonucleotide with a 5′ overhanging terminus, and we then challenged this complex with a series of competitor oligonucleotides that did or did not possess complementary cohesive termini to the probe (Fig. 3C). Transfer of MuGam, HiGam or Ku onto the unlabelled competitor took place only when the competitor had a perfectly complementary cohesive end to the probe (Fig. 3C, lanes 3, 9 and 15). By contrast, transfer did not occur when the competitor possessed a single nucleotide gap in the complementarity of its 5′ overhang (Fig. 3C lanes 4, 10 and 16) or when the competitor had 3′ overhanging (lanes 5, 11 and 17) or blunt (lanes 6, 12 and 18) termini. The DNA binding properties of MuGam and HiGam are therefore remarkably similar to those of eukaryotic Ku.
Once bound to DNA, Ku has been shown to form intimate contacts with DNA ends and to be capable of protecting them from attack by exonuclease (de Vries et al., 1989). We therefore assessed the ability of MuGam and HiGam to prevent exonuclease III-mediated degradation of DNA. As shown in Fig. 4, the inclusion of human Ku, MuGam or HiGam led to a marked slowing of the rate of exonucleolytic degradation of DNA compared with that observed in the presence of BSA or the sequencespecific DNA-binding factor SP1. The extent of this protection mirrors the stability of the DNA–protein complexes as observed in EMSAs and in other assays (Fig. 3, and data not shown). Thus, like Ku, MuGam and HiGam can protect the extremities of linear DNA fragments from attack by exonuclease.
Figure 4.
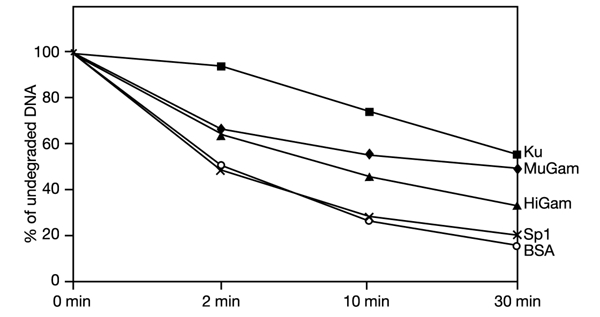
MuGam, HiGam and human Ku protect DNA ends from exonuclease attack. Uniformly labelled linear DNA was incubated with the indicated proteins, and exonuclease III was added subsequently. Samples were taken at the indicated time points. The amount of undegraded DNA was measured at the indicated time points.
MuGam affects Ku-dependent retrotransposition in yeast
Ku has previously been shown to be important in retroviral integration and transposition in eukaryotic cells (Daniel et al., 1999; Downs & Jackson, 1999). Saccharomyces cerevisiae Ty elements are retrotransposons that transpose by means of an RNA intermediate that is converted to linear ds DNA and then inserted into the host genome. By the use of an inducible Ty1 system, we have previously shown that Ty1 retrotransposition rates are decreased in the absence of Ku (Downs & Jackson, 1999). We therefore decided to test whether the expression of MuGam in yeast might influence the Ty1 retrotransposition process. As shown in Fig. 5, in wild-type yeast Ty1 transposition rates were not affected by the expression of MuGam in S. cerevisiae from a yeast vector. By contrast, expression of MuGam in Ku-deficient yeast cells led to a further marked decrease in Ty1 retrotransposition rates. These data suggest that, in the presence of functional Ku, MuGam is unable to gain access to Ty1 DNA and consequently is unable to influence the retrotransposition process. However, when Ku is absent the linear Ty1 DNA can now be bound by MuGam in a manner that interferes with retrotransposition, most probably by impairing access by cellular factors that are necessary for completion of the retrotransposition process.
Figure 5.
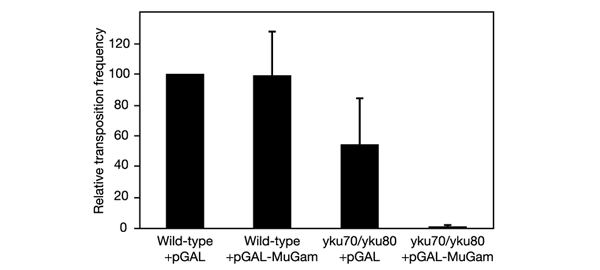
Expression of MuGam impairs Ty1 retrotransposition in S. cerevisae lacking Ku. Yeast cells carrying an empty vector or a vector expressing MuGam were tested for Ty1 transposition (see Methods). Frequencies are shown relative to that of wild-type yeast not expressing MuGam, which had a frequency of 1.1 × 10−4. Error bars indicate s.d.
In summary, we have discovered that the Gam protein of Mu phage and a set of related bacterial Gam-like polypeptides have significant sequence and structural homology with eukaryotic Ku. Moreover, we have shown that MuGam and HiGam recognize DNA ends in a manner strikingly similar to eukaryotic Ku. In addition, we have shown that, in the absence of Ku, MuGam can interfere with Ty1 retrotransposition in yeast cells. We therefore conclude that MuGam and its related bacterial Gam-like proteins are functional orthologues of eukaryotic Ku.
Speculation
The biochemical properties of MuGam suggest that its prime function in bacteriophage replication is to bind to linear Mu DNA, prevent its degradation by bacterial exonucleases and thus favour its efficient transposition into the bacterial chromosome.
Interestingly, expression of MuGam in E. coli allows this bacterium to take up foreign linear DNA (Akroyd & Symonds, 1986). Thus, E. coli cells infected by Mu or bearing a portion of the prophage encompassing the Gam gene can be transformed with a linearized plasmid ∼100-fold more efficiently than uninfected cells. This activity is remarkably reminiscent of a property of Ku in S. cerevisae, namely that linear plasmids can be efficiently introduced and maintained in this yeast only when functional Ku is present (Boulton & Jackson, 1996). It is noteworthy that bacteria possessing Gam-like proteins, such as H. influenzae, N. meningitidis and the O157:H7 strain of E. coli, are naturally competent for transformation and have the capacity to take up DNA from their environment and integrate it into their chromosomes (Kroll et al., 1998; Ohnishi et al., 1999; Tettelin et al., 2000). Such events are thought to have a significant role in bacterial evolution, which, unlike eukaryotic evolution, relies predominantly on a lateral transfer of genes. It is tempting to speculate that Gam-like proteins have a role in facilitating these events. Finally, it is of interest that Mu phage, H. influenzae and S. typhi express Gam proteins and use recombination systems that employ sitespecific recombinases that have homology with mammalian V(D)J recombinase Rag-1 (Spanopoulou et al., 1996). It would therefore be interesting to determine whether the Gam-like proteins in these bacteria act in these processes in a manner similar to that of eukaryotic Ku in V(D)J recombination.
Methods
Phylogenetic analysis.
An unrooted phylogenetic tree based on Ku and Gam homologues was retrieved by iterative similarity searches obtained by maximum-likelihood analyses with the program PROTML.
Modelling of the Gam homodimer complex.
A structure-based sequence alignment was used to map the location of MuGam on the crystal structure of the human Ku. The extreme N- and C-terminal domains have been removed from the structure of the human Ku heterodimer complex (PDB ID: ) to leave a core Ku structure consisting of truncated Ku70 (amino-acid residues 266-446) and Ku80 (residues 257-433).
Protein purification.
MuGam and HiGam open reading frames were cloned in pET15b vector, expressed in E. coli strain BL21(DE30pLysS) and affinity-purified as His-tag fusion proteins. Human Ku was purified as described by Gell & Jackson (1999).
Gel filtration analysis.
Analytical gel filtration of purified MuGam and HiGam was performed on an S-75 HR 10/30 column (Amersham Biosciences).
Sedimentation equilibrium ultracentrifugation.
Sedimentation equilibrium experiments were conducted at 20 °C on a Beckman Optima XL-I analytical centrifuge. The sedimentation equilibrium properties of MuGam were analysed at a rotor speed of 20,000 r.p.m. with Raleigh Interference Optics. To extract molecular mass information, the data were analysed with the shareware programs Sednterp 1.06 and Winnl106.
EMSAs.
32P end-labelled TL4 oligonucleotide (0.2 ng) was incubated for 20 min at 20°C with purified MuGam, HiGam or human Ku in binding buffer containing 40 mM Tris/HCl pH 8, 20% glycerol, 0.1% Triton X-100 and 20 mM KCl. The complexes were resolved on a 5% polyacrylamide gel in 0.5 × Tris-borate-EDTA at 4 °C. Retarded bands of MuGam and HiGam could be supershifted by the addition of antibodies against their histidine tags (data not shown). All oligonucleotide sequences are as described by Bliss & Lane (1997).
Exonuclease assays.
32P random prime-labelled linear calf thymus DNA (4 ng) was incubated with equimolar amounts of purified proteins in binding buffer. Exonuclease III (4,000 U; New England Biolabs) was then added. Aliquots were taken at the indicated time points and the amount of undegraded DNA was quantified by liquid-scinitillation counting after washes with trichloroacetic acid.
Ty1 retrotransposition assays.
Experiments were performed essentially as described by Downs & Jackson (1999). Yeasts strains were as described by Downs & Jackson (1999). MuGam was expressed by the pRS414 plasmid under the GAL1/10 promoter as a fusion with a nuclear localization signal and a Flag (DYKDDDK) epitope tag. MuGam expression was monitored by detection with anti-Flag antibodies and its expression level was identical in both yeast strains. Experiments were performed in triplicate.
Supplementary Material
Molecular weight determination of the MuGam protein by analytical ultracentrifugation.
Acknowledgments
We thank J. Downs, G. Smith, D. Gell, T. Bliss, L. Pellegrini, M. Moncrieffe and D. Leach for sharing reagents and advice, J. Bradbury for editorial help, and L. Serpell for help with figures. F.d'A.d.F. is supported by Cancer Research UK grant C6/A1675. The S.P.J. laboratory is funded by Cancer Research UK. G.R.W. was supported by an MRC studentship. A.J.D. is a Royal Society University Research Fellow and the A.J.D. laboratory is supported by BBSRC, Cancer Research UK, AICR and The Royal Society.
References
- Abraham Z.H. & Symonds N. (1990) Purification of overexpressed gam gene protein from bacteriophage Mu by denaturation-renaturation techniques and a study of its DNA-binding properties. Biochem. J., 269, 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akroyd J. et al. (1984) Mapping and properties of the gam and sot genes of phage mu: their possible roles in recombination. Cold Spring Harb. Symp. Quant. Biol., 49, 261–266. [DOI] [PubMed] [Google Scholar]
- Akroyd J. & Symonds N. (1986) Localization of the gam gene of bacteriophage mu and characterisation of the gene product. Gene, 49, 273–282. [DOI] [PubMed] [Google Scholar]
- Aravind L. & Koonin E.V. (2001) Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic doublestrand break repair system. Genome Res., 11, 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss T.M. & Lane D.P. (1997) Ku selectively transfers between DNA molecules with homologous ends. J. Biol. Chem., 272, 5765–5773. [DOI] [PubMed] [Google Scholar]
- Boulton S.J. & Jackson S.P. (1996) Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double- strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J., 15, 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- Craig N.L. (1995) Unity in transposition reactions. Science, 270, 253–254. [DOI] [PubMed] [Google Scholar]
- Daniel R. et al. (1999) A role for DNA-PK in retroviral DNA integration. Science, 284, 644–647. [DOI] [PubMed] [Google Scholar]
- de Vries E. et al. (1989) HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol., 208, 65–78. [DOI] [PubMed] [Google Scholar]
- Doherty A.J. & Jackson S.R. (2001) DNA repair: how Ku makes ends meet. Curr. Biol., 11, R920–R924. [DOI] [PubMed] [Google Scholar]
- Doherty A.J. et al. (2001) Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett., 500, 186–188. [DOI] [PubMed] [Google Scholar]
- Downs J.A. & Jackson S.P. (1999) Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol. Cell. Biol., 19, 6260–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W.S. & Yoo S. (1998) Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res., 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell D. & Jackson S.P. (1999) Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res., 27, 3494–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen T. et al. (1982) Bacteriophage Mu DNA replication is stimulated by non-essential early functions. Mol. Gen. Genet., 186, 135–139. [DOI] [PubMed] [Google Scholar]
- Kroll J.S. et al. (1998) Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl Acad. Sci. USA, 95, 12381–12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. et al. (2001) Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J., 20, 3272–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi M. et al. (1999) Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res., 6, 361–368. [DOI] [PubMed] [Google Scholar]
- Smith G.C.M. & Jackson S.P. (1999) The DNA-dependent protein kinase. Genes Dev., 13, 916–934. [DOI] [PubMed] [Google Scholar]
- Spanopoulou E. et al. (1996) The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell, 87, 263–276. [DOI] [PubMed] [Google Scholar]
- Tettelin H. et al. (2000) Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science, 287, 1809–1815. [DOI] [PubMed] [Google Scholar]
- Walker J.R. et al. (2001) Structure of the Ku heterodimer bound to DNA and its implications for doublestrand break repair. Nature, 412, 607–614. [DOI] [PubMed] [Google Scholar]
- Wang J. et al. (1998) A model for Ku heterodimer assembly and interaction with DNA. Implications for the function of Ku antigen. J. Biol. Chem., 273, 31068–31074. [DOI] [PubMed] [Google Scholar]
- Weller G.R. et al. (2002) Identification of a DNA nonhomologous end-joining complex in bacteria. Science, 297, 1686–1689. [DOI] [PubMed] [Google Scholar]
- Wu X. & Lieber M.R. (1996) Protein–protein and protein–DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol. Cell. Biol., 16, 5186–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular weight determination of the MuGam protein by analytical ultracentrifugation.


