Summary
Although DNA clearly outclasses any silicon-based computer when it comes to information storage and processing speed, a DNA-based PC is still a long way off
On 28 February 2003 the scientific world will celebrate a very special anniversary. It was on this day fifty years ago that James Watson and Francis Crick discovered the structure of DNA — the very essence of life itself. Since then, research into DNA has given biologists a great understanding of life, and has also allowed them to create innumerable useful tools that have wide-ranging applications for both science and society. However, it was not until the early 1990s that researchers started exploring the possibility of utilising DNA's ability to store and process information outside the realms of biology. In 1994, an US proof-of-principle study showed that DNA could be used to solve mathematical problems, which attracted considerable interest from researchers hoping that DNA would one day replace silicon as the basis for a new wave of computers. But the initial excitement has since dampened down as scientists have realized that there are numerous problems inherent to DNA computing and that they would have to live with their silicon-based computers for quite a while yet. The field consequently changed its focus, and in essence, research into DNA computing is now chiefly concerned with “investigating processes in cells that can be viewed as logical computations and then looking to use these computations to our advantage,” as Martyn Amos from the University of Exeter, UK, described it.
A mix of 1,018 strands of DNA could operate at 10,000 times the speed of today's advanced supercomputers
It was Leonard Adleman, professor of computer science and molecular biology at the University of Southern California, USA, who pioneered the field when he built the first DNA based computer (L. M. Adleman, Science 266, 1021–102; 1994). Intrigued by the molecule's immense capacity to store information in a very small space, he set out to solve a classic puzzle in mathematics — the so-called Hamilton Path problem, better known as the Travelling Salesman problem. This seemingly simple puzzle — a salesman must visit a number of cities that are interconnected by a limited series of roads without passing through any city more than once–is actually quite a killer, and even the most advanced supercomputers would take years to calculate the optimal route for 50 cities. Adleman solved the problem for seven cities within a second, using DNA molecules in a standard reaction tube. He represented each of the seven cities as separate, singlestranded DNA molecules, 20 nucleotides long, and all possible paths between cities as DNA molecules composed of the last ten nucleotides of the departure city and the first ten nucleotides of the arrival city. Mixing the DNA strands with DNA ligase and adenosine triphosphate (ATP) resulted in the generation of all possible random paths through the cities. However, the majority of these paths were not applicable to the situation–they were either too long or too short, or they did not start or finish in the right city. Adleman then filtered out all the paths that neither started nor ended with the correct molecule and those that did not have the correct length and composition. Any remaining DNA molecules represented a solution to the problem.
The power contained in these tiny molecules caused a flurry of excitement in the computing world
The computation in Adleman's experiment chugged along at 1,014 operations per second, a rate of 100 Teraflops or 100 trillion floating point operations per second; the world's fastest supercomputer, Earth Simulator, owned by the NEC Corporation in Japan, runs at just 35.8 Teraflops. Clearly, computing with DNA has massive advantages over silicon-based machines. Whereas current technology rests on a highly linear principle of logic, and one computation must be completed before the next can begin, the use of DNA means that an enormous number of calculations can take place simultaneously. This parallel power is many times faster than that of traditional machines — a mix of 1,018 strands of DNA could operate at 10,000 times the speed of today's advanced supercomputers. The other major advantage is the potential for information storage. Whereas traditional storage media, such as videotapes, require 1012 cubic nanometres of space to store a single bit of information, DNA molecules require just one cubic nanometre per bit. Not surprisingly, the power contained in these tiny molecules caused a flurry of excitement in the computing world, and many hoped that “DNA computing could overtake silicon-based technology,” commented Ron Weiss, professor of electrical engineering at Princeton University, New Jersey, USA Fig. 1.
Figure 1.
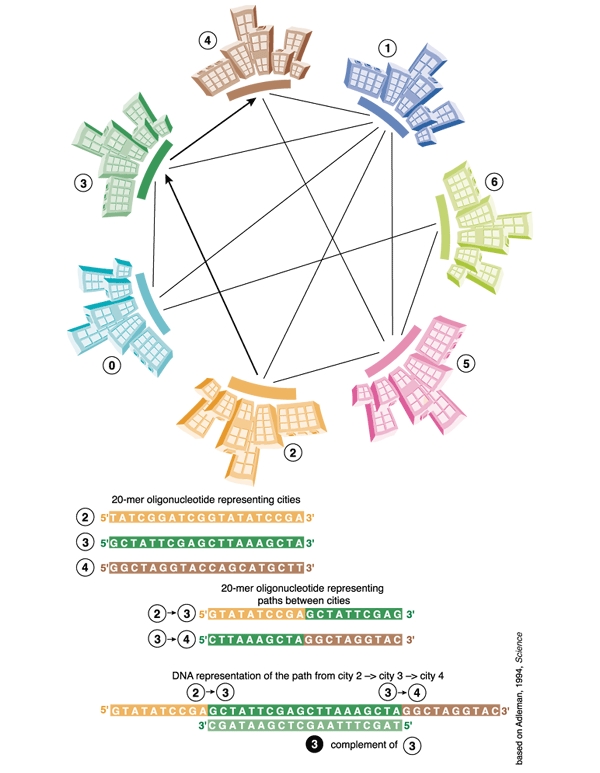
The principle of Leonard Adleman's DNA computer to solve the 'Travelling salesman' problem.
That was eight years ago, however, and although the potential of DNA computing seemed enormous, intervening research has shown that it is constrained by major limitations. Representing all the possible solutions to a problem as strands of DNA means the computation is completed quickly, however, said “You have to perform an exhaustive search to find a small needle in a large haystack”, said Amos, and it requires an exponential resource in terms of memory. Although DNA can store a trillion times more information than current storage media, the way in which the information is processed necessitates a massive amount of DNA if larger-scale problems are to be solved. “It has been estimated that if you scaled up the Hamilton Path Problem to 200 cities from Adleman's seven, then the weight of DNA required to represent all the possible solutions would exceed the weight of the earth,” said Amos. Furthermore, even although the computation process takes place at an awesome speed, the 'printout' of the result is excruciatingly sluggish, and involves many steps — it took Adleman a week of lab work to extract the potential solutions from his DNA cocktail.
The general consensus now is that DNA computing will never be able to compete directly with silicon-based technology
There are also problems concerning the accuracy of the process. DNA strand synthesis is liable to errors, such as mismatching pairs, and is highly dependent on the accuracy of the enzymes involved. Although this did not affect Adleman's work, he only dealt with less than 100 possibilities; a fully operational computer would need to perform thousands upon thousands of calculations, which means that the chance of errors increases exponentially. Furthermore, as more complicated molecules are required for more complicated procedures, the size of the molecules increases, as does their probability of shearing, again contributing to errors Fig. 2.
Figure 2.
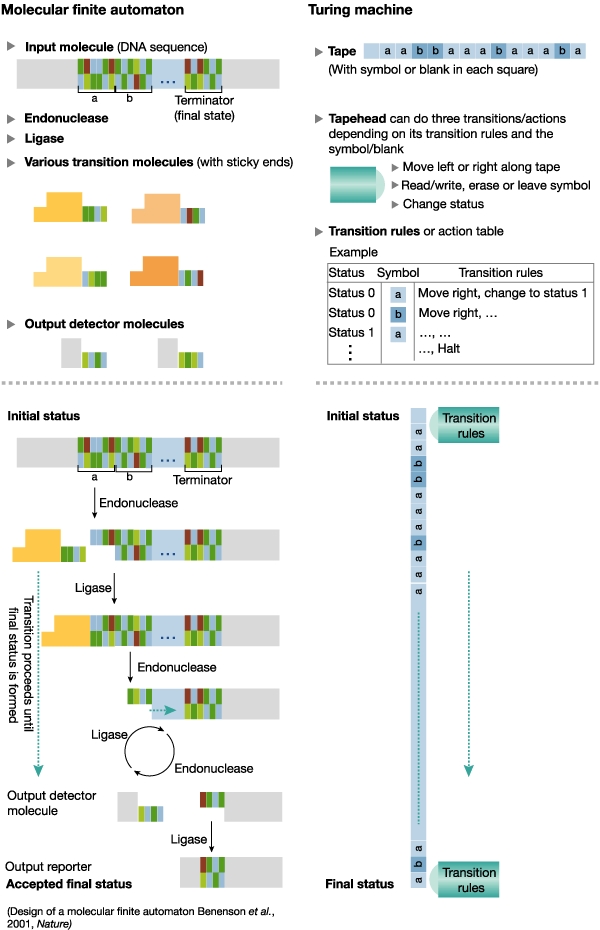
Ehud Shapiro's molecular Turing Machine.
Weiss is not confident about overcoming these technical issues, a sentiment echoed by others in the field. The general consensus now is that, as a result of these limitations, DNA computing will never be able to compete directly with silicon-based technology. This does not mean, however, that DNA computing is dead in the water – far from it. But the problems have forced a major rethink and “the emphasis has now shifted away from the original objective,” according to Amos. He thinks there is still great potential in DNA computing but for him “the rich potential of DNA computing lies in in vivo computing” — using the technology on a smaller scale, inside cells. For Weiss, the realistic aim is to “demonstrate control at a molecular level.”
One such demonstration of this aim was achieved two years ago by Ehud Shapiro's group at the Weizmann Institute in Israel (Y. Benenson et al. Nature 414, 430–434; 2001), who built a programmable and autonomous computing machine made of biomolecules. This 'automater' is similar to the hypothetical Turing Machine developed by the British mathematician Alan Turing (1912 to 54) in 1936, a device that converts information from one form into another and operates on a finite sequence of symbols — Shapiro's machine used two 'inputs'. Based on a series of transition rules, the machine changes its internal state according to the current state and input until it reaches a 'final state' when all inputs have been processed. Shapiro's automater uses restriction endonucleases and ligase as the 'hardware' to alter the state of the machine, and doublestranded DNA as the inputs and the transition rules. The DNA 'software' is continuously ligated and cut by the enzymes, until it reaches a final state — a defined sticky end — to which a 'reporter' DNA is ligated, thus terminating the computation. Shapiro hopes to be able to develop this very simple concept and build progressively more complicated models until he is able to construct a fully operational molecular Turing Machine. This would be quite an achievement as a Turing Machine is capable of performing all mathematical operations and is regarded as the basis of today's computers. He finds it hard to predict whether he will be able to complete his goal but “the direction is promising,” he added.
As Shapiro said, “A lot of information is available as biological molecules. If you can programme them and respond to the information then you can do a lot.” His long-term vision is “to create molecular computing machines that can analyse situations in cells, and then synthesize molecules to deal with them.” The potential applications of such technology are vast. The use of programmed cells as 'biological sentinels', as Weiss dubbed them, could have obvious applications in fighting diseases, by recognizing damaged cells or tissue and either reporting the problem or, even better, effecting the release of reparative molecules.
 Another promising direction is the molecular self-assembly of DNA to build complex molecular structures, which could have an impact on other fields, such as nanotechnology. Eric Winfree, from the California Institute of Technology, USA, has devoted considerable amounts of time to this topic, and has developed a method for building molecular 'tiles' — minute blocks of DNA. By programming the edges of these tiles, he has been able to force DNA to come together in tiny molecular patterns. He has so far only been able to build simple structures, however, and, he said, “we need to get to the point where we can construct complicated patterns.”
Another promising direction is the molecular self-assembly of DNA to build complex molecular structures, which could have an impact on other fields, such as nanotechnology. Eric Winfree, from the California Institute of Technology, USA, has devoted considerable amounts of time to this topic, and has developed a method for building molecular 'tiles' — minute blocks of DNA. By programming the edges of these tiles, he has been able to force DNA to come together in tiny molecular patterns. He has so far only been able to build simple structures, however, and, he said, “we need to get to the point where we can construct complicated patterns.”
Yet, as Amos pointed out, “this is all blue sky at the moment.” All of this research is still in the proof-of-principle stage, and any practical applications are at least five to ten years away. Clearly, DNA computing will not become a rival for today's silicon-based machines and “it will not affect the way you or I live,” said Weiss. However, the real excitement in the field lies in bringing together biologists, chemists, computer scientists and mathematicians to understand and simulate fundamental biological processes and algorithms taking place within cells. “We shouldn't be looking for competition with traditional machines, we should be looking outside the box for a niche for other applications.” said Amos. However, he added, “If I'm honest, biocomputing has yet to establish this niche.”


