Abstract
Hydrophobic stop-transfer sequences generally serve to halt the translocation of polypeptide chains across the endoplasmic reticulum membrane and become integrated as transmembrane α-helices. Using engineered glycosylation sites as topology reporters, we show that the length of the nascent chain between a hydrophobic segment and the carboxy terminus of the protein can affect stop-transfer efficiency. We also show that glycosylation sites located close to a protein's C terminus are modified in two distinct kinetic phases, one fast and one slow. Our findings suggest that membrane integration of a hydrophobic segment is not simply a question of thermodynamic equilibrium, but can be influenced by details of the translocation mechanism.
Introduction
The dominating topogenic signals in most integral membrane proteins consist of stretches of hydrophobic residues, often flanked by charged residues (von Heijne, 2000). In singlespanning type I membrane proteins, a hydrophobic stop-transfer segment interrupts the translocation of the polypeptide chain across the membrane and becomes embedded in the lipid bilayer as a transmembrane α-helix (Blobel, 1980). Stop-transfer efficiency is determined by the overall hydrophobicity and length of the segment, as well as by charged residues at the carboxy-terminal end of the hydrophobic stretch (Chen & Kendall, 1995; Kuroiwa et al., 1991; Sääf et al., 1998). In Escherichia coli, it has further been shown that the stop-transfer function of a marginally hydrophobic segment depends on its translocation kinetics; that is, the longer the hydrophobic segment remains in the translocon, the more efficiently it promotes the formation of a transmembrane topology (Duong & Wickner, 1998).
While the importance of the hydrophobicity of the stop-transfer segment and the potentiating role of flanking charged residues is well established, it appeared to us that an additional variable might be the position of the stop-transfer segment relative to the C-terminal end of the protein. In particular, given that translocation kinetics seem to be important for the recognition of stop-transfer segments, and since there appears to be a close co-operation between the ribosome and the translocon during membrane insertion of a stop-transfer segment (Haigh & Johnson, 2002), we surmised that stop-transfer segments may behave differently if they are still attached to the ribosome via a growing nascent chain when they enter the translocon, or if they have at this point already been released from the ribosome by chain termination.
To address this possibility, we used a model protein containing engineered potential stop-transfer segments of varying hydrophobicity, composed either of 18 pseudo-random residues (Sääf et al., 1998), or of poly-Leu stretches of 5 to 10 residues with and without C-terminally flanking positively charged residues. The effect of the carboxy tail length, that is, the number of residues between the stop-transfer sequence and the C-terminal end of the protein, on the stop-transfer efficiency of the stop-transfer segments was studied in a eukaryotic in vitro transcription/translation system in the presence of dog pancreas rough microsomes (RMs). Our results show that the carboxy tail length indeed has an effect on the stop-transfer efficiency of marginally hydrophobic segments, suggesting that membrane integration of a hydrophobic stop-transfer segment is not simply a question of thermodynamic equilibrium, but can be influenced by details of the translocation mechanism. We also show that the kinetics of the glycosylation reaction are different depending on whether the glycosylation acceptor site is placed close to or far away from the C-terminal end of the protein.
Results
Construction and expression of model proteins
For these studies, we used the E. coli inner membrane protein leader peptidase (Lep) as a model protein (Fig. 1). Lep consists of two transmembrane segments (H1 and H2), a short cytoplasmic loop (P1), and a large periplasmic domain (P2). Lep inserts into RMs in vitro and adopts its native topology with both the N- and C-termini in the lumen (Nilsson & von Heijne, 1993). To study stop-transfer function, we introduced a collection of short segments (Ssegments) into the P2 domain, 92 residues away from the C terminus. As S-segments, we used either pseudo-random stretches of 18 predominantly hydrophobic amino acids chosen from a previous study (Sääf et al., 1998), see Fig. 1, or 5 to 10 amino acid-long poly-Leu stretches either with or without two extra Arg residues at their C-terminal end. The length of the carboxy tail was varied by introducing a stop codon at different positions downstream of the Ssegment.
Figure 1.
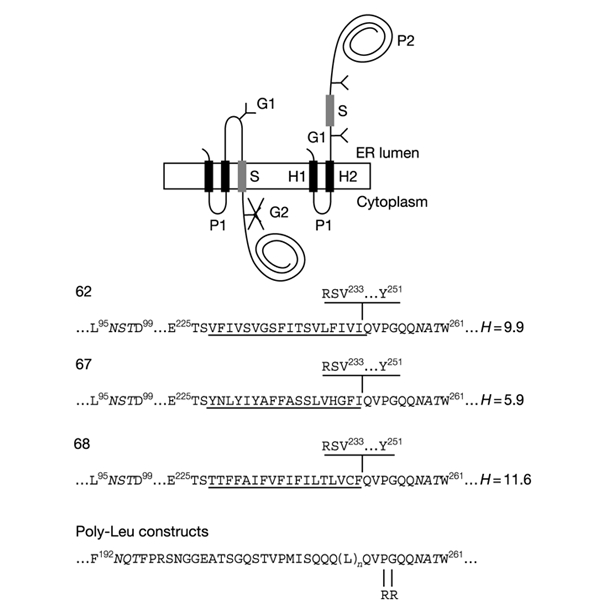
Model proteins. Lep has two transmembrane segments (H1, H2) and a large globular domain (P2). Different S-segments were inserted into the P2 domain in the location shown. Y indicates Asn-X-Thr glycosylation acceptor sites (G1, G2). Expressed proteins with a single glycosylated site (G1) have an S-segment with stop-transfer activity (top left), while those with two modified sites (G1, G2) have an S-segment that does not integrate into the membrane (top right). The amino-acid sequences of the relevant regions around the S-segments of the different constructs are shown at the bottom. Numbered residues indicate positions in the Lep wild-type protein. The G1 and G2 glycosylation sites are shown in italics. S-segments are underlined. The sequence RSV...Y shown above the sequences for clones 62, 67, and 68 is present only in the longest constructs tested (carboxy tail length = 92 residues). Carboxy tail lengths are counted from the first residue immediately downstream of the underlined S-segments (Gln in the case of all constructs except for those with a carboxy tail length of 92 residues, where counting starts from Arg). The mean hydrophobicities (H, calculated as described in Methods) of the S-segments in clones 62, 67, and 68 are also given. The Arg-Arg mutation in the poly-Leu constructs is shown.
To map the topology of the different constructs, we introduced two Asn-X-Thr acceptor sites for N-linked glycosylation: the G1 site between H2 and the S-segment, and the G2 site 7 residues downstream of the S-segment. The stop-transfer efficiency of a given S-segment was quantified as the fraction of singly glycosylated molecules to the sum of singly and doubly glycosylated molecules.
All constructs were cloned into the pGEM1 vector under control of the SP6 promoter, and were expressed using a reticulocyte lysate system in the absence and presence of RMs. Products were analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and quantified on a phosphorImager.
Stop-transfer function of pseudo-random segments
To measure the stop-transfer efficiency as a function of the carboxy tail length we used three constructs with different S-segments composed of 18 pseudo-random amino acids spanning a range of hydrophobicities and stop-transfer efficiencies (Fig. 1, bottom panel). The stop-transfer efficiency of these segments in the context of the full-length Lep protein was determined previously (Sääf et al., 1998). The Ssegments in clones 62 and 68 are efficient stop-transfer segments in the full-length context (that is, they are glycosylated only on the G1 site; see Fig. 6 in Sääf et al., 1998), whereas the Ssegment in clone 67 has a low mean hydrophobicity and lacks stop-transfer function in the context of the full-length protein (that is, it is glycosylated on both the G1 and G2 sites).
C-terminally truncated versions of clones 62, 67, and 68 were made by introducing stop codons at different positions downstream of the S-segments. As shown in Fig. 2, for the Ssegment in clone 62 the fraction of singly to singly and doubly glycosylated proteins, that is, the stop-transfer efficiency, increases strongly with the carboxy tail length and reaches its maximal value at a carboxy tail length of 40 to 50 residues. The more polar S-segment in clone 67, in contrast, is translocated across the membrane regardless of the carboxy tail length, and the more hydrophobic S-segment in clone 68 is an efficient stop-transfer segment at all carboxy tail lengths.
Figure 2.
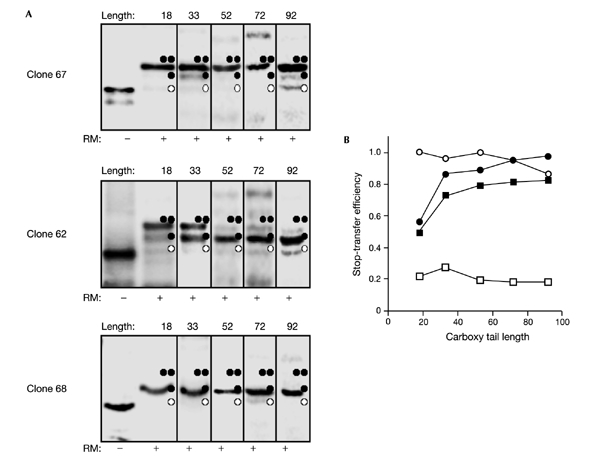
Stop-transfer efficiency increases with the carboxy tail length for a marginally hydrophobic S-segment. (A) Clones 62, 67, and 68 with different carboxy tail lengths were translated in the absence and presence of dog pancreas rough microsomes. Unglycosylated, singly glycosylated, and doubly glycosylated products are indicated by one white circle, one black circle, and two black circles, respectively. Carboxy tail lengths are shown above the lanes. (B) Stop-transfer efficiency (the quotient between the intensity of the singly glycosylated product to the summed intensities of the singly and doubly glycosylated products) as a function of carboxy tail length (the number of residues between the C-terminal end of the S-segment and the C terminus) for clones 62 (black squares), 67 (white squares), 68 (white circles), and 62-pPL (black circles). Each value is an average of at least two independent experiments.
To confirm the effect of carboxy tail length on stop-transfer efficiency, we tested the proteinase K sensitivity in RMs of short (18- residue carboxy tail) and long (72 residue carboxy tail) versions of clones 62, 67, and 68 where for simplicity the G1 and G2 sites had been removed (Fig. 3). As expected, when the carboxy tail length was 72 residues (top panel) construct 67(−G1, −G2) was cleaved only in the P1 loop giving rise to a protected H2–P2 fragment with a molecular mass of ∼30 kDa, whereas the P2 domain in constructs 62(−G1, −G2) and 68(−G1, −G2) was completely digested and hence not translocated into the lumen of the microsomes. When the carboxy tail length was only 18 residues (Fig. 3, bottom panel), protected H2–P2 fragments were seen for constructs 62(−G1, −G2) and 67(−G1, −G2), but not for 68(−G1, −G2), again consistent with the glycosylation results.
Figure 3.
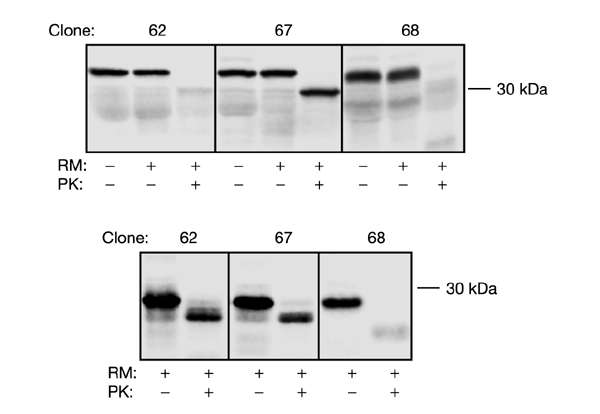
Proteinase K treatment of microsomes containing constructs 62(−G1, −G2), 67(−G1, −G2), and 68(−G1, −G2). mRNA was translated in the absence or presence of RMs, and samples were treated with proteinase K (PK) as indicated. The top panel is for a carboxy tail length of 72 residues, and the bottom panel for a carboxy tail length of 18 residues. Note that both the G1 and G2 glycosylation acceptor sites were removed in these constructs.
Although glycosylation of the G2 site takes place only after chain termination when the carboxy tail is short, we nevertheless wanted to test whether the rate of translation might affect the results. We therefore repeated the experiments in the presence of the translation inhibitor cycloheximide at a concentration of 0.18 μM. At this concentration, cycloheximide slows down, but does not completely block, protein synthesis; in our hands, the translation rate is reduced by approximately one-half (data not shown). We observed no significant change in the final levels of singly versus singly and doubly glycosylated molecules in the presence of cycloheximide for any of the clones with carboxy tail lengths of 18, 33, 52, and 72 residues (data not shown).
As a final control, we replaced the carboxy tail in clone 62 with an unrelated sequence from the secretory protein preprolactin (pPL). The stop-transfer efficiency was also found to increase with the carboxy tail length for the 62-pPL construct (Fig. 2B). The composition of the carboxy tail thus seems to be of little importance for the stop-transfer efficiency.
We conclude that the stop-transfer efficiency of the S-segment in clone 62 increases strongly when the carboxy tail length is increased from 18 to 52 residues, and then levels out. As shown previously (Sääf et al., 1998), the Ssegment in clone 62 is of intermediate hydrophobicity and close to the 'stop-transfer threshold' defined in that study. We further conclude that reducing the translation rate by cycloheximide treatment has no obvious effect on these results, and that the amino-acid composition of the carboxy tail is of little or no importance.
Stop-transfer function of poly-Leu segments
As an alternative to the pseudo-random S-segments, we also tested S-segments composed of five to ten Leu residues. Previous work has shown that stop-transfer function can be achieved with segments of seven to nine contiguous Leu residues (Kuroiwa et al., 1991). Again, truncated proteins were generated by introducing stop codons at various positions downstream of the Ssegment.
As shown in Fig. 4 (left panel), the effect of varying the carboxy tail length is similar for the Leu segments as for the pseudo-random Ssegment in clone 62: there is an increase in stop-transfer efficiency with carboxy tail length for the Leu8 and especially for the Leu9 S-segments, and little variation for the Leu5, Leu7, and Leu10 S-segments. The mean hydrophobicities (calculated as described in Methods) of the Leu5, Leu7, Leu8, Leu9, and Leu10 S-segments are 3.7, 7.4, 8.9, 10.3, and 11.1, respectively; thus the hydrophobicity of the S-segment in clone 62 is between those of the Leu8 and Leu9 segments (Fig. 1).
Figure 4.
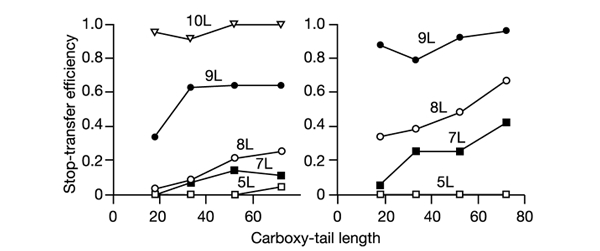
Stop-transfer efficiency increases with the carboxy tail length for marginally hydrophobic poly-Leu S-segments. Stop-transfer efficiency as a function of carboxy tail length for poly-Leu S-segments either without (left panel) or with (right panel) two C-terminal Arg residues. Each value is an average of two independent experiments.
Another parameter that is known to increase stop-transfer efficiency is the presence of positively-charged residues immediately downstream of a hydrophobic segment (Kuroiwa et al., 1991). We therefore added two extra Arg residues next to the poly-Leu segments, and repeated the stop-transfer efficiency measurements for different carboxy tail lengths, as shown in Fig. 4 (right panel). As expected, the stop-transfer efficiency is generally increased for all but the shortest Leu-based Ssegments, and the variation with the carboxy tail length is now seen for the Leu7 and Leu8 S-segments. Interestingly, for the S-segments close to the stop-transfer threshold (Leu7 to Leu9), the introduction of two C-terminal Arg residues increases stop-transfer efficiency by a similar amount as does the addition of one Leu residue (compare the two panels in Fig. 4).
Carboxy tail length affects the kinetics of glycosylation
The observation that the length of the carboxy tail affects the recognition of marginally hydrophobic S-segments prompted us to check whether the kinetics of glycosylation may also be different for glycosylation acceptor sites placed near to or far away from the C terminus of the protein.
The glycosylation kinetics of a given site can be measured by first adding the translation initiation inhibitor aurintricarboxylic acid (ATA) to the translation mix shortly after the addition of the messenger RNA, and then dissolving the microsomes by addition of the detergent Triton X-100 at later time points (Rothman & Lodish, 1977; Garoff et al., 1990). As treatment with Triton X-100 will efficiently prevent further glycosylation but will not block translation per se, full-length chains can be produced by continuing the incubation up to the standard 60-minute time-point, and the kinetics of glycosylation of any given acceptor site can be determined by varying the time of Triton X-100 addition.
In general, for all constructs, the glycosylation of the G1 and G2 sites commenced at the times expected from their positions in the nascent chain and the average translation rate in our system (∼0.5 residues s−1; data not shown). However, as seen in Fig. 5, while the G2 site in clone 67 is glycosylated rapidly when the carboxy tail is long, we observed two kinetic phases for the shortest carboxy tail. One is rapid and accounts for ∼30% of the final glycosylation level, and the second is very slow. A similar situation holds for clone 62 when the carboxy tail is short, where only ∼50% of the final glycosylation level is reached during the fast phase. The G1 site, which is 154 residues away from the C terminus in the shortest constructs, is fully glycosylated with fast-phase kinetics in all cases (data not shown).
Figure 5.
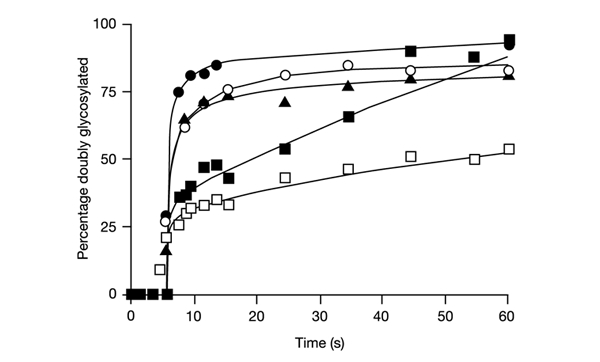
Two-phase kinetics of C-terminal glycosylation of acceptor sites. Clones 67 and 62 with different carboxy tail lengths were expressed in vitro in the presence of RMs. At 1.5 min after mRNA addition, ATA was added to block further initiation. Samples were removed at different time points and Triton X-100 was added to dissolve the microsomes and block further glycosylation. Translation was then allowed to continue up to a total time of 60 min. The time-course for the appearance of the second glycosylation event (site G2) is shown. n denotes the number of residues between the Asn in the G2 glycosylation acceptor site and the C-terminal end of the protein (see Fig. 1). 67 black squares (n=12); 67, black triangles (n=28); 67, white circles (n=46); 67, black circles (n=66); 62, white squares (n=12).
Discussion
Using a eukaryotic in vitro system, we have tested the effect on stop-transfer efficiency of the length of the carboxy tail downstream of a stop-transfer segment for three sets of polypeptide segments (S-segments): a set of pseudo-random 18-residue segments selected from a previous screen for stop-transfer function, a set of poly-Leu segments composed of five to ten contiguous Leu residues, and the same set of poly-Leu segments but with two extra Arg residues at their C-terminal end.
For all three sets, the trend is the same: the stop-transfer efficiency increases with the carboxy tail length for marginally hydrophobic S-segments. The effect is particularly striking for the marginally hydrophobic pseudo-random S-segment in clone 62: this sequence has only weak stop-transfer function when the carboxy tail is 18 residues long, but has nearly full stop-transfer function for carboxy tail lengths longer than 40–50 residues. This is the range for which the length of the carboxy tail would be expected to affect the time spent by the hydrophobic segment in the translocation channel, and our results are thus compatible with a kinetic partitioning model where the stop-transfer segment has to be recognized as such by the translocon during a time interval set either by the rate of translation (for long carboxy tails), or the rate with which a nascent polypeptide no longer attached to the ribosome moves through the translocon (for short carboxy tails). The rate of translation in our in vitro system is of the order of 0.5 residues s−1 (Nilsson et al., 2000), and the translocation rate for an unattached nascent chain must therefore be considerably faster than this. An alternative possibility, suggested by the observation that a lowering of the translation rate has little effect on the glycosylation efficiency, is that the translocon reacts to chain termination on the ribosome by a conformational change (Haigh & Johnson, 2002) that reduces the efficiency with which marginally hydrophobic segments are recognized as stop-transfer sequences.
By following the kinetics of glycosylation, we have further found that a glycosylation acceptor site placed near the C-terminus of the protein is modified in two distinct kinetic phases, one fast and one very slow. The fast phase is similar to what is seen for acceptor sites placed far away from the C-terminus and most likely represents molecules that get glycosylated during their initial passage through the endoplasmic reticulum translocon, whereas the slow phase probably represents molecules for which the C-terminal region is first released into the lumen before it gets modified. The slow phase is observed both for clone 67 (which has a non-hydrophobic S-segment) and for clone 62 (which has a marginally hydrophobic S-segment) when the C-terminal tail is short. Possible explanations for this phenomenon are (i) that the nascent chain is released from the ribosome well before the G2 site reaches the oligosaccharyltransferase (OST) when the carboxy tail is short, and that it will therefore pass by the OST too rapidly for fully efficient co-translocational glycosylation, or (ii) that the translocon undergoes a major conformational change upon chain termination (Haigh & Johnson, 2002), thereby preventing immediate access of the G2 site to the OST. In either case, the unglycosylated carboxy tail will be released to the luminal space and the G2 site can then only be modified in a much slower, post-translocational reaction.
A conceptually important implication of our findings is that the membrane integration of a transmembrane α-helix is not necessarily an equilibrium reaction; rather, the same segment may or may not become integrated depending on its location in the polypeptide chain. In a very real sense, membrane protein structure may thus—at least in certain cases—be kinetically rather than thermodynamically controlled.
Methods
Enzymes and chemicals.
Unless otherwise stated, all enzymes, plasmid pGEM1, dithiothreitol (DTT), BSA, RNase inhibitor, and rabbit reticulocyte lysate were from Promega. T7 DNA polymerase, [35S]methionine, ribonucleotides, deoxyribonucleotides, dideoxyribonucleotides, and the cap analog 7mG(5′)ppp(5′)G were from Amersham-Pharmacia. Spermidine, ATA, cycloheximide, and Triton X-100 were from Sigma. Proteinase K was from Boehringer Mannheim. Oligonucleotides were from Cybergene.
DNA manipulations.
For cloning into and expression from the pGEM1 plasmid, the 5′ end of the lep gene was modified, first, by the introduction of an XbaI site and, second, by changing the context 5′ to the initiator ATG codon to a 'Kozak consensus' sequence (Kozak, 1999).
For clones 62, 67 and 68, S-segments coding for 18 pseudo-random amino acids were introduced between a SpeI site in codons 226–227 and a BglII site in codons 231–232 in the lep gene (Sääf et al., 1998). Site-directed mutagenesis was used to introduce Asn-X-Thr acceptor sites for N-linked glycosylation at positions 96–98 and 258–260. Constructs with a carboxy tail length of 72 amino acids were made by deleting codons 231–251, first using the polymerase chain reaction (PCR) to amplify a fragment from a XbaI site located on the 5′ side of the gene to a primer-introduced 3′ KpnI site at codon 231, and then ligating this fragment back into the vector between the 5′ XbaI site and a KpnI site already present in codons 252 and 253. Truncations were made from this construct by using 3′ PCR primers introducing a stop codon followed by a short poly-A tail (6 consecutive As) in positions 270, 285, and 304 corresponding to carboxy tail lengths of 18, 33 and 52 amino acids, respectively.
In construct 62(pPL), the carboxy tail sequence after the glycosylation site (codons 258–260) was substituted with an unrelated sequence from bovine preprolactin (codons 33–115) and truncations were made as above.
S-segments composed of leucine stretches were introduced between a BclI site in codon 214 and a KpnI site in codon 253 using PCR on constructs with a carboxy tail of 72 residues described earlier (Nilsson & von Heijne, 2000). Asn-X-Thr glycosylation acceptor sites were introduced with the Asn residue in positions 193 and 258. Truncations were made using the same 3′ PCR primers as for the constructs with pseudo-random Ssegments.
To make S-segments composed of leucine stretches followed by Arg-Arg, codons 255 and 256 were substituted by two Arg codons (AGG and AGA) using the QuickChange site-directed mutagenesis kit (Stratagene).
Expression in vitro.
Constructs in pGEM1 were amplified using a 5′ primer hybridizing upstream of the SP6 promoter and the 3′ primers described above. The PCR products were transcribed by SP6 RNA polymerase for 1 h at 37 °C. The transcription mixture was as follows: 1–5 μg DNA template, 5 μl 10 × SP6 H-buffer (400 mM HEPES-KOH (pH 7.4), 60 mM magnesium acetate, 20 mM spermidine-HCl), 5 μl BSA (1 mg ml−1), 5 μl 7mG(5′)ppp(5′)G (10 mM), 5 μl DTT (50 mM), 5 μl rNTP mix (10 mM ATP, 10 mM CTP, 10 mM UTP, 5 mM GTP), 18.5 μl H2O, 1.5 μl RNase inhibitor (50 units), 0.5 μl SP6 RNA polymerase (20 units).
Translation of 1 μl mRNA in 9 μl nuclease-treated reticulocyte lysate, 1 μl RNase inhibitor (40 units μl−1), 1 μl [35S] Met (10 μCi μl−1), 1 μl amino acids mix (1 mM of each amino acid except Met), 1 μl dog pancreas rough microsomes (2 units; one unit is defined as the amount of RM required for 50% translocation of in vitro synthesized preprolactin) was performed as described in Liljeström & Garoff (1991) at 30 °C for 1 h. For protease treatment of rough microsomes, proteinase K was used at a final concentration of 150 μg ml−1 and samples were incubated for 30 min on ice. To reduce the translation rate, cycloheximide was added to a final concentration of 0.18 μM to the translation mix.
To follow the kinetics of glycosylation, the translation mix was preincubated 4 min before [35S]Met and mRNA was added, and the translation initiation inhibitor ATA was added to a final concentration of 0.075 mM after an additional 1.5 min (Garoff et al., 1990). Samples were removed at different time points and were incubated further at 30 °C in the presence of 1% Triton X-100 until a total translation time of 60 min.
Translation products were analysed by SDS–PAGE and gels were quantified on a Fuji FLA-3000 phosphorimager using the Image Reader 8.1j software. The stop-transfer efficiency of a given mutant was calculated as the quotient between the intensity of the singly glycosylated band divided by the summed intensities of the doubly glycosylated and singly glycosylated bands (mean of at least two independent experiments). In general, the levels of glycosylation varied by no more than ±5% between different experiments.
Hydrophobicity analysis.
Hydrophobicity analysis of the pseudo-random and poly-Leu S-segments was carried out using the Membrane Protein Explorer software at http://blanco.biomol.uci.edu/mpex/ with default settings and the water–octanol hydrophobicity scale (Jayasinghe et al., 2001).
Acknowledgments
This work was supported by grants from the Swedish Research Council and the Swedish Cancer Foundation to G.vH.
References
- Blobel G. (1980) Intracellular protein topogenesis. Proc. Natl Acad. Sci. USA, 77, 1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.F. & Kendall D.A. (1995) Artificial transmembrane segments—Requirements for stop transfer and polypeptide orientation. J. Biol. Chem., 270, 14115–14122. [DOI] [PubMed] [Google Scholar]
- Duong F. & Wickner W. (1998) Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J., 17, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Huylebroeck D., Robinson A., Tillman U. & Liljeström P. (1990) The signal sequence of the p62-protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J. Cell Biol., 111, 867–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh N.G. & Johnson A.E. (2002) A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol., 156, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe S., Hristova K. & White S.H. (2001) Energetics, stability, and prediction of transmembrane helices. J. Mol. Biol., 312, 927–934. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- Kuroiwa T., Sakaguchi M., Mihara K. & Omura T. (1991) Systematic analysis of stop-transfer sequence for microsomal membrane. J. Biol. Chem., 266, 9251–9255. [PubMed] [Google Scholar]
- Liljeström P. & Garoff H. (1991) Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol., 65, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I. & von Heijne G. (1993) Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem., 268, 5798–5801. [PubMed] [Google Scholar]
- Nilsson I. & von Heijne G. (2000) Glycosylation efficiency of Asn-Xaa-Thr sequons depends both on the distance from the C terminus and on the presence of a downstream transmembrane segment. J. Biol. Chem., 275, 17338–17343. [DOI] [PubMed] [Google Scholar]
- Nilsson I., Witt S., Kiefer H., Mingarro I. & von Heijne G. (2000) Distant downstream sequence determinants can control N-tail translocation during protein insertion into the endoplasmic reticulum membrane. J. Biol. Chem., 275, 6207–6213. [DOI] [PubMed] [Google Scholar]
- Rothman J. & Lodish H. (1977) Synchronised transmembrane insertion and glycosylation of a nascent membrane protein. Nature, 269, 775–780. [DOI] [PubMed] [Google Scholar]
- Sääf A., Wallin E. & von Heijne G. (1998) Stop-transfer function of pseudo-random amino acid segments during translocation across prokaryotic and eukaryotic membranes. Eur. J. Biochem., 251, 821–829. [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1999) Recent advances in the understanding of membrane protein assembly and structure. Q. Rev. Biophys., 32, 285–307. [DOI] [PubMed] [Google Scholar]


