Abstract
The high-mobility-group B (HMGB) chromosomal proteins are characterized by the HMG box, a DNA-binding domain that both introduces a tight bend into DNA and binds preferentially to a variety of distorted DNA structures. The HMGB proteins seem to act primarily as architectural facilitators in the manipulation of nucleoprotein complexes; for example, in the assembly of complexes involved in recombination and transcription. Recent genetic and biochemical evidence suggests that these proteins can facilitate nucleosome remodelling. One mechanism by which HMGB proteins could prime the nucleosome for migration is to loosen the wrapped DNA and so enhance accessibility to chromatin-remodelling complexes and possibly also to transcription factors. By constraining a tight loop of untwisted DNA at the edge of a nucleosome, an HMGB protein could induce movements in the contacts between certain core histones that would result in an overall change in nucleosome structure.
Introduction
A major component of transcriptional regulation in the eukaryotic nucleus is the control of the accessibility of packaged DNA sequences in chromatin to transcription factors and chromatin-remodelling complexes. This control can be mediated at one level by altering the equilibrium between compact, folded chromatin, such as that condensed into 30 nm fibres, and a more open configuration. However, access to sequences wrapped around the histone octamer might also require disruption of the structure of the nucleosome itself. Nucleosomes are essentially dynamic structures, a property manifested in the transient unwrapping of the bound DNA and in their translational mobility from one position to another on the DNA sequence. Here I argue that the abundant chromosomal high-mobility-group B (HMGB) proteins, exemplified by HMGB1 and HMGB2 of vertebrates (reviewed by Bianchi & Beltrame, 2000; Thomas & Travers, 2001), have a key role in disrupting nucleosome structure.
Nucleosome unwrapping and mobility
The unwrapping of nucleosomal DNA is, in essence, a topological change in the configuration of DNA. One general model for nucleosome accessibility, proposed and documented by Widom and his colleagues (Widom, 2001) posits that the transient unwrapping of the bound DNA from an entry/exit point exposes previously occluded sequences (Fig. 1). This exposure allows transcription factors and DNA-processing enzymes to bind to specific sequences so that recognition of sequences close to the entry/exit point is strongly favoured relative to those closer to the dyad (Fig. 1).
Figure 1.
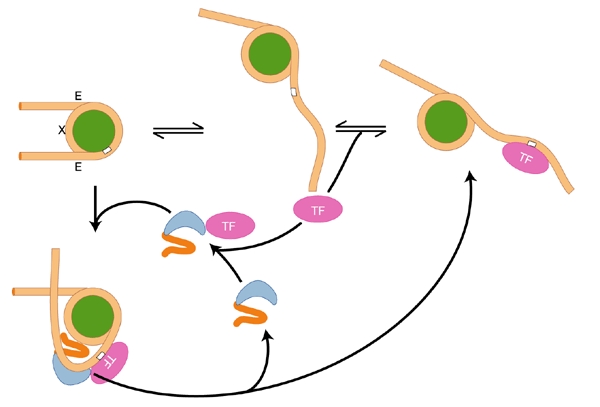
Proposed mechanism for transient unwrapping of histone-bound DNA and transcription factor binding (adapted from Widom, 2001). Stochastic unwrapping of DNA from one exit/entry point (E) can result in the exposure of a binding site (white box) for a sequencespecific DNA-binding protein. This model suggests one mechanism by which HMGB proteins (blue) might facilitate transcription factor (TF) binding. X indicates the nucleosome dyad.
Is this stochastic unwrapping sufficient to account for accessibility in vivo, or is the process facilitated by other mechanisms? Acetylation of the amino-terminal tails of the core histones certainly improves the accessibility of the bound DNA sequence to transcription factors in vitro, but only by a small amount (Anderson et al., 2001). Another process that affects accessibility is the migration of the histone octamer from one position to another along the DNA. As with transient unwrapping, migration involves the breakage and reformation of histone–DNA contacts. However, during transient unwrapping from an entry/exit point the histone–DNA contacts are ultimately reformed at the same DNA sequences, whereas migration requires that the histones contact different sites. In vitro, the intrinsic migration of octamers is strongly dependent on temperature, suggesting that the process might have a high activation energy (Pennings et al., 1991); in vivo, nucleosome migration is facilitated by ATP-dependent remodelling complexes that impart directionality, at least in a local sense (reviewed by Becker & Hörz, 2002). This high activation energy suggests that, in vivo, other proteins or complexes might be required to initiate remodelling by distorting the wrapped nucleosomal DNA.
Involvement of HMGB proteins in nucleosome mobility
One class of proteins whose members could perform this function is the HMGB family of chromosomal proteins. These proteins are highly abundant, with each mammalian nucleus containing on average 105 to 106 molecules (Duguet & de Recondo, 1978), equivalent to about 1 molecule for every 10 nucleosomes. They bind to linear DNA nonspecifically with a short residence time. Typically, molecules of this type contain at least one HMG domain, a basic region and a carboxy-terminal acidic tail. Whereas the vertebrate proteins contain two HMG domains, an A and a B domain, the principal fly and yeast counterparts contain only one, which is structurally homologous to the B domain. The HMG B domain untwists the DNA by binding in the minor groove and also introduces a sharp ∼90–100° bend within a single double-helical turn. This induced bend is stabilized by the basic region, which neutralizes the adjacent negative charges on the sugar-phosphate backbones in the opposing compressed major groove (Lnenicek-Allen et al., 1996; Payet & Travers, 1997). The role of the acidic tail, in contrast, remains an enigma. In vitro this tail usually decreases the affinity of the protein for DNA (Thomas & Travers, 2001). However, a key to its role might be the fact that such sequences are ideally suited to interact with the positive charges on histones—indeed, polyglutamate has long been used as a histone chaperone to facilitate nucleosome assembly in vitro (Stein et al., 1979). Therefore, in principle, the HMG domain and the basic region could potentially stabilize a DNA loop on the surface of the octamer while the acidic tail could bind to the core histones (Fig. 2).
Figure 2.
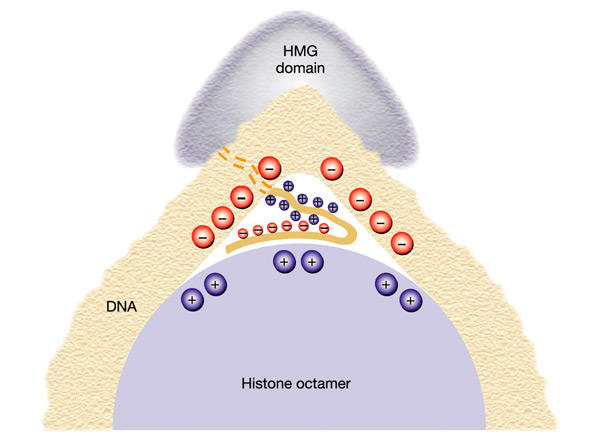
Proposed simultaneous interaction of a single high mobility group (HMG)-box HMGB protein with the histone octamer and associated DNA. The HMG domain binds on the outside of the DNA bend while the basic region neutralizes the negative charges on the DNA inside the bend. The positively charged acidic tail binds to negative charges on the histone octamer.
Is there any evidence that proteins of this class are involved in promoting nucleosome accessibility in vivo? The interaction of the abundant HMGB proteins with nucleosomal particles that lack the linker histone H1 is well documented: Jackson et al. (1979) observed that a soluble chromatin fraction released from mouse myeloma nuclei contained mononucleosomes associated with almost stoichiometric amounts of HMGB1 + HMGB2, but lacked histone H1. Furthermore, the yeast counterparts Nhp6a and Nhp6b bind directly to nucleosomal particles (Formosa et al., 2001), as does vertebrate HMGB1 (Nightingale et al., 1996). Early studies showed that HMGB1 and HMGB2 bound to nucleosomal particles containing 180-base-pair (bp) DNA but not 140-bp DNA (Schroter & Bode, 1982), suggesting that these proteins bind to DNA adjacent to, rather than that within, the nucleosome core particle. Subsequent footprinting studies revealed that the HMGB1 protein bound in close proximity to an exit/entry site on the core particle (Nightingale et al., 1996; An et al., 1999). Consistent with this binding location and a role in nucleosome mobility, the Drosophila HMGB protein HMG-D increases the nucleosome repeat length—the length of DNA bound to the histone octamer plus the linker DNA—from ∼165 bp to ∼185 bp in a cell-free assembly system (Ner et al., 2001). Consistent with the notion that HMGB proteins can bind to linker DNA in Saccharomyces cerevisiae is the observation that loss of the two-HMG-domain protein Hmo1 results in an average general increase in the sensitivity of chromatin to digestion by micrococcal nuclease (Lu et al., 1996). A similar phenotype is observed in strains that lack both Nhp6a and Nhp6b (M. Buttinelli and A.T., unpublished observations). Taken together, these results point to a general role of HMGB proteins in the maintenance of chromatin structure.
The phenotype of a strain lacking Nhp6 proteins raises a paradox: although the chromatin in this mutant is more accessible to endonucleolytic cleavage, general activation of transcription comparable to that observed when the level of histones is artificially reduced (Durrin et al., 1992) does not occur (Paull et al., 1996; Moreira & Holmberg, 2000). By contrast, although the chromatin of yeast strains lacking Sin4, a subunit of both the histone acetylase SAGA complex and the polymerase II mediator complex (Li et al., 1995), also has an enhanced sensitivity to endonucleases (Macatee et al., 1997), the mutant strain exhibits a pleiotropic phenotype similar to depletion of histones (Jiang et al., 1995). These contrasting observations suggest that enhanced accessibility is not necessarily indicative of the functional state of chromatin. Importantly, the reduction in expression of the yeast mating-type conversion gene, HO, on the loss of Nhp6 can be largely restored by mutations that increase nucleosome accessibility and mobility, but is further exacerbated by those with the opposite effect (Yu et al., 2000). Mutations both in SIN3 and RPD3, which encode components of a histone deacetylase complex and normally promote repression, and in SIN4 partly restore wild-type function in cells lacking both Nhp6a and Nhp6b. The loss of the histone acetylase Gcn5, also a component of the SAGA histone acetylase complex, in the same cells results in a more severe phenotype. Nevertheless, the strong growth defect of the triple mutant lacking both variants of Nhp6 as well as Gcn5 can also be partly rescued by a loss of SIN4 function. RPD3 and GCN5 have been shown to modulate the dynamic balance between histone acetylation and deacetylation (Verdone et al., 2002). Both histone acetylation and the Sin (switch independent) phenotype are correlated with chromatin unfolding (Tse et al., 1998; Horn et al., 2002) and/or enhanced nucleosome accessibility (Anderson et al., 2001), whereas histone deacetylation would be expected to favour folding. On the basis of this argument, one role of the Nhp6 proteins would be to antagonize folding, and possibly to promote nucleosome mobility. In other words, without Nhp6 the nucleosomes are less mobile and the wrapped DNA sequences are less accessible, even though the DNA in the linker between histone octamers is more accessible.
Evidence of a possible involvement of HMGB proteins in nucleosome mobility, and hence site accessibility, has recently become apparent from the observation that several chromatin remodelling complexes either contain or can associate with a polypeptide harbouring an HMG domain homologous to the B domain. Examples of such complexes include the SWI/SNF-like BAF complex (Wang et al., 1998) and the Drosophila BRM (brahma) complex (Papoulas et al., 2001) containing the HMG-domain proteins BAF57 and BAP111, respectively. Neither of these HMG-domain proteins seems to be essential for remodelling in vitro, but a loss of BAP111 results in a significant reduction of function (Papoulas et al., 2001). In vivo, mutations in the BAF57 subunit impair the function of the BAF complex in both the silencing of the CD4 locus and the activation of the CD8 locus (Chi et al., 2002). However, the BAF57 HMG domain is dispensable for tethering BAF complexes to the CD4 silencer or other chromatin loci, suggesting that BAF-dependent chromatin remodelling in vivo requires HMG-induced DNA bending (Chi et al., 2002). Additionally, the Drosophila CHD1 ATP-dependent remodelling protein co-localizes with the HMG-domain protein SSRP1 (Kelley et al., 1999). An HMG-domain protein is also a component of other, smaller, related remodelling complexes, the mammalian FACT complex (Orphanides et al., 1999) and the yeast SPN complex (Formosa et al., 2001), which facilitate nucleosome remodelling, primarily during transcription elongation. They contain the mammalian and yeast homologues, respectively, of Spt16 (a general effector of transcription) as well as an HMG-domain protein—SSRP1 in FACT (Orphanides et al., 1999) and Nhp6 in SPN (Brewster et al., 2001; Formosa et al., 2001). The SPN complex contains one additional subunit, Pob3, which has some homology to SSRP1 but lacks an HMG domain. In yet another example, repression of the CHA1 locus requires both Nhp6 and the RSC remodelling complex (Moreira & Holmberg, 1999, 2000). At this locus, one effect of loss of Nhp6a and Nhp6b is an increase in the basal level of transcription because the transition to the organized chromatin structure characteristic of the uninduced state is blocked. This genetic evidence is complemented by the recent important demonstration in vitro that HMGB1 promotes the binding of the ACF remodelling complex to a nucleosome and consequently facilitates remodelling itself (Bonaldi et al., 2002). However, the activity of the remodelling motor component of the complex, ISWI, is not stimulated by HMGB1, implying that another component of the complex might mediate this activation.
Priming nucleosome remodelling
The association of HMGB proteins with several classes of chromatin nucleosome remodelling complexes implies that their DNA-binding function might be used in this process. A characteristic of HMGB1 and HMGB2 and—with the exception of Nhp6a/b in S. cerevisiae—their abundant counterparts in other organisms is the juxtaposition of positively- and negatively-charged regions. Similarly, in the FACT complex SSRP1 also contains both positively- and negatively-charged regions. However, in the SPN complex, whereas Nhp6 contains only an HMG domain and a positively-charged region, both of the other subunits, Pob3 and Spt16, possess negatively-charged regions that could compensate for the lack of a comparable region in Nhp6. This same juxtaposition of differently-charged regions also occurs in certain chromatin-remodelling motor proteins, for example the Drosophila ATPase dMi-2. Although this parallel might be coincidental, it raises the possibility that these charged regions could interact with nucleosomes by simultaneously neutralizing the positive charges on the histones and the negative charges on any detached DNA, thus compensating for any broken histone–DNA contacts. The neutralization of the positive tail of the HMGB protein by the core histones could at the same time increase the affinity of the protein for the DNA.
An HMGB protein binding at the edge of the wrapped nucleosomal DNA would distort the DNA by imposing a tight DNA bend and also a short untwisted region. Both these distortions could facilitate unwrapping of the nucleosomal DNA: the bend by destabilizing the histone–DNA contacts immediately proximal to the bound HMGB protein (Längst & Becker, 2001), and the untwisting by inducing a compensating torque within the wrapped supercoil (Fig. 3B). Such a torque could loosen the left-handed wrapping and thus provide an entry site for a remodelling complex at a distant nucleosomal site. Torque transduction of this type requires the establishment of a short defined topological domain. This could be achieved by anchoring the HMGB protein to the nucleosome, for example by its acidic tail contacting histone H2A (Ner et al., 2001) (Fig. 3B). Strikingly, an HMGB1 derivative lacking the acidic tail no longer facilitates ACF-mediated remodelling in vitro (Bonaldi et al., 2002). This mechanism implies that a topological change in the wrapped DNA might be required for the initiation of nucleosome remodelling, but not necessarily for its subsequent propagation, and it would explain the observations that ATP-dependent nucleosome remodelling generates superhelical torsion (Havas et al., 2000), whereas relaxation of superhelicity facilitates the propagation of nucleosome sliding driven by the ISWI motor (Längst & Becker, 2001).
Figure 3.
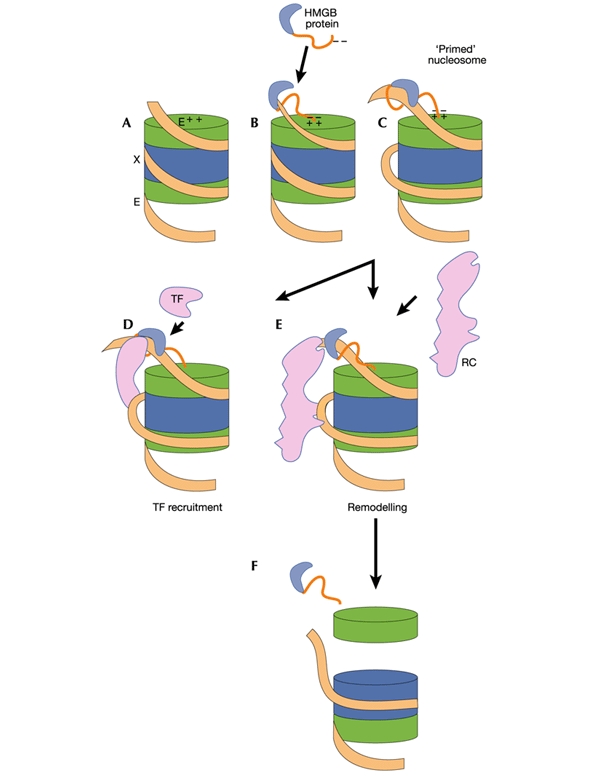
Proposed mechanism for priming nucleosomes by HMGB proteins. A, In an intact nucleosome the DNA is tightly wrapped, entering and leaving the structure at sites marked E. The dyad (X), or midpoint, is at the centre of the wrapped DNA. Histone H2A/H2B dimers and H3/H4 dimers are indicated in green and blue, respectively. B, Binding of an HMGB protein just outside one exit/entry point creates an untwisted bend in the DNA that alters the nucleosome structure. C, Previously inaccessible regions are exposed, which are potentially accessible to (D) transcription factors (TF) or to (E) chromatin remodelling complexes (RC). F, When associated with RNA polymerase II (as the FACT/SPN complex), the HMG-induced destabilization of nucleosomal structure could result in the dissociation of an H2A/H2B dimer (Kireeva et al., 2002).
In addition to their ability to bend DNA, the vertebrate HMGB1 and HMGB2 proteins can facilitate the binding of several eukaryotic transcription factors to their binding sites. The interactions that have been observed in vitro involve an HMGB protein and a single transcription factor, but it remains possible that in vivo, in a natural regulatory context, the bending of the DNA by HMGB1 and HMGB2 could allow the recruitment of a second transcription factor to the complex (Fig. 3D) in a manner analogous to the action of sequencespecific HMG-box transcription factors. In such cases it is inferred that the bend introduced by these proteins provides a DNA scaffold, allowing the assembly of a complex nucleoprotein structure (Giese et al., 1995). This might indeed happen in the assembly of the Epstein–Barr virus enhanceosome (Ellwood et al., 2000) and the human lymphocyte recombination complexes containing RAG1 and RAG2 (Fugmann et al., 2000).
Although the recruitment of transcription factors to their binding sites by HMGB proteins has been studied with naked DNA in vitro, these interactions occur in the context of DNA organized into nucleosomes in vivo. In this situation the binding of an HMGB protein to a nucleosome core particle close to an exit/entry point might facilitate the binding of a transcriptional activator or repressor to an internal site (Fig. 3D) by loosening the wrapping of histone-bound DNA. Both in this case and in the recruitment of a remodelling complex, the essential role of the HMGB protein would be to change nucleosome structure, and thereby to reduce the activation energy for the establishment of a complex with either the remodelling machinery or a transcription factor. This model does not exclude the possibility that during directed remodelling an HMGB protein itself could also migrate within a DNA loop around the histone octamer, incurring little or no energy penalty by the successive breakage and reformation of electrostatic contacts (a Manning walk).
Novel conformations of the nucleosome?
An induced alteration of nucleosomal structure by HMGB proteins implies that a major function of these proteins, in conjunction with the linker histone H1, is to modulate an equilibrium between alternative conformations of the nucleosome (Fig. 4). In this model, HMGB proteins would favour a generally more accessible form of the nucleosome, and histone H1 a more inaccessible form. The observed competition between these proteins (Ner et al., 2001) would thus influence not only gross chromatin compaction but also the mobility and overall accessibility of individual nucleosomes. The conformational modulations of nucleosome structure would involve changes in the contacts between individual histones in the octamer, and this internal mobility could be an intrinsic property of the particle (Negri et al., 2001). More specifically, substitutions of variant or modified histones, for example macroH2A (found in the Barr body) and H2A.X, for the normal core histones could alter the susceptibility of the particle to remodelling by modifying internal histone–histone contacts.
Figure 4.
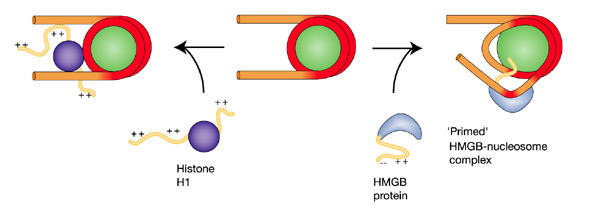
A general model for the function of HMGB proteins. An HMGB protein bound in the vicinity of an exit/entry point induces increased access to both DNA gyres in a spatially restricted patch. Elsewhere the HMGB protein lessens accessibility. The alternative, less accessible conformation of the nucleosome is stabilized by histone H1. Accessible and bound segments of DNA are indicated in orange and red, respectively.

References
- An W., van Holde K. & Zlatanova J. (1998) The non-histone chromatin protein HMG1 protects linker DNA on the side opposite to that protected by linker histones. J. Biol. Chem., 273, 26289–26291. [DOI] [PubMed] [Google Scholar]
- Anderson J.D., Lowary P.T. & Widom J. (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 307, 977–985. [DOI] [PubMed] [Google Scholar]
- Becker P.B. & Hörz W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. & Beltrame M. (2000) Upwardly mobile proteins. EMBO Rep., 1, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster N.K., Johnston G.C. & Singer R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldi T., Längst G., Strohner R., Becker P.B. & Bianchi M.E. (2002) The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J., 21, 6865–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T.H. et al. (2002) Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature, 418, 195–199. [DOI] [PubMed] [Google Scholar]
- Durrin L.K., Mann R.K. & Grunstein M. (1992) Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 12, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet M. & de Recondo A.M. (1978) A deoxyribonucleic acid unwinding protein isolated from regenerating rat liver. Physical and functional properties. J. Biol. Chem., 253, 1660–1666. [PubMed] [Google Scholar]
- Ellwood K.B., Yen Y.-M., Johnson R.C. & Carey M. (2000) Mechanism for specificity by HMG-1 in enhanceosome assembly. Mol. Cell. Biol., 20, 4359–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. et al. (2001) Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J., 20, 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J. & Schatz D.G. (2000) The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- Giese K., Kingsley C., Kirshner J.R. & Grosschedl R. (1995) Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev., 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- Havas K. et al. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- Horn P.J., Crowley K.A., Carruthers L.M., Hansen J.C. & Peterson C.L. (2002) The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nature Struct. Biol., 9, 167–171. [DOI] [PubMed] [Google Scholar]
- Jackson J.B., Pollock J.M. Jr & Rill R.L. (1979) Chromatin fractionation procedure that yields nucleosomes containing nearstoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry, 18, 3739–3748. [DOI] [PubMed] [Google Scholar]
- Jiang Y.W., Dohrmann P.R. & Stillman D.J. (1995) Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics, 140, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D.E., Stikes D.G. & Perry R.P. (1999) CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma, 108, 10–25. [DOI] [PubMed] [Google Scholar]
- Kireeva M.L. et al. (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell, 9, 541–552. [DOI] [PubMed] [Google Scholar]
- Längst G. & Becker P.B. (2001) ISWI induces nucleosome sliding on nicked DNA. Mol. Cell, 8, 1085–1092. [DOI] [PubMed] [Google Scholar]
- Li Y. et al. (1995) Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNApolymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 92, 10864–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lnenicek-Allen M., Read C.M. & Crane-Robinson C. (1996) The DNA bend angle and binding affinity of an HMG box increased by the presence of short terminal arms. Nucleic Acids Res., 24, 1047–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Kobayashi R. & Brill S.J. (1996) Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem., 271, 33678–33685. [DOI] [PubMed] [Google Scholar]
- Macatee T., Jiang Y.W., Stillman D.J. & Roth S.Y. (1997) Global alterations in chromatin accessibility associated with loss of SIN4 function. Nucleic Acids Res., 25, 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira J.M. & Holmberg S. (1999) Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J., 18, 2836–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira J.M. & Holmberg S. (2000) Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J., 19, 6804–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri R. et al. (2001) Sequence dependence of translational positioning of core nucleosomes. J. Mol. Biol., 307, 987–999. [DOI] [PubMed] [Google Scholar]
- Ner S.S. et al. (2001) HMG-D and histone H1 interplay during chromatin assembly and early embryogenesis. J. Biol. Chem., 276, 37569–37576. [DOI] [PubMed] [Google Scholar]
- Nightingale K., Dimitrov S., Reeves R. & Wolffe A.P. (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J., 15, 548–561. [PMC free article] [PubMed] [Google Scholar]
- Orphanides G., Wu W.-H., Lane W.S., Hampsey M. & Reinberg D. (1999) The chromatinspecific transcription factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- Papoulas O. et al. (2001) The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc. Natl Acad. Sci. USA, 98, 5728–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T.T., Carey M. & Johnson R.C. (1996) Yeast HMG proteins NHP6A/B potentiate promoterspecific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev., 10, 2769–2781. [DOI] [PubMed] [Google Scholar]
- Payet D. & Travers A.A. (1997) The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol., 266, 66–75. [DOI] [PubMed] [Google Scholar]
- Pennings S., Meersseman G. & Bradbury E.M. (1991) Mobility of positioned nucleosomes on 5 S rDNA. J. Mol. Biol., 220, 101–110. [DOI] [PubMed] [Google Scholar]
- Schroter H. & Bode J. (1982) The binding sites for large and small high-mobility-group (HMG) proteins. Studies on HMG–nucleosome interactions in vitro. Eur. J. Biochem., 127, 429–436. [DOI] [PubMed] [Google Scholar]
- Stein A., Whitlock J.P. Jr & Bina M. (1979) Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc. Natl Acad. Sci. USA, 76, 5000–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J.O. & Travers A.A. (2001) HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [DOI] [PubMed] [Google Scholar]
- Tse C., Sera T., Wolffe A.P. & Hansen J.C. (1998) Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol., 18, 4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdone L. et al. (2002) Hyperacetylation of chromatin at the ADH2 promoter allows Adr1 to bind in repressed conditions. EMBO J., 21, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. (1998) Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl Acad. Sci. USA, 95, 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J. (2001) Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys., 34, 269–324. [DOI] [PubMed] [Google Scholar]
- Yu Y., Eriksson P. & Stillman D.J. (2000) Architectural factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol., 20, 2350–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]


