Abstract
Important progress in understanding messenger RNA export from the nucleus could be achieved by increasing the list of proteins that are involved in this process. Here, we present the identification of Gbp2 as a novel shuttling RNA-binding protein in Saccharomyces cerevisiae. Nuclear import of Gbp2 is dependent on the receptor Mtr10 and the serine/arginine-specific protein kinase Sky1. Deletion of the genes encoding both of these proteins or disruption of two of the arginine/serine repeats each shifts the steady-state localization of Gbp2 to the cytoplasm. Interestingly, deletion of MTR10 only also causes an increase in poly(A)+ RNA binding by Gbp2, suggesting a role of Mtr10 in the dissociation of Gbp2 from mRNA in the cytoplasm. The nuclear export of Gbp2 is always coupled to mRNA export and is dependent on continuous RNA polymerase II transcription and mRNA-export factors. Although GBP2 is not essential for normal cell growth, overexpression of this gene is toxic and causes a nuclear retention of bulk poly(A)+ RNA. Together, our findings clearly show an involvement of Gbp2 in mRNA transport. In addition, as a non-essential protein, Gbp2 also has the interesting potential to be spatially or temporally regulated.
Introduction
In all eukaryotes, the main sites of transcription and translation are separated by the nuclear envelope. Molecules can overcome this barrier by active nuclear transport. Messenger RNA export from the nucleus is dependent on several factors, reviewed in Dreyfuss et al. (2002). Among them are specific components of the nuclear pore complex (NPC), such as Nup159/NUP214 and proteins that dock the ribonucleoprotein particle (RNP) to the NPC. The latter may function as an mRNA-export receptor-complex comprised of Mex67 (TAP or NXF1 in metazoans) and Mtr2 (p15 or NXT in higher eukaryotes) (reviewed in Zenklusen & Stutz, 2001; Reed & Hurt, 2002). In addition, shuttling mRNA-binding proteins function in mRNA export. So far, several have been identified in yeast, including Pab1, Hrp1, Nab2, Yra1 and Npl3 (Anderson et al., 1993; Wilson et al., 1994; Zenklusen & Stutz, 2001). Only one of them, Npl3, contains a serine/arginine (SR)-rich domain, which is important for shuttling. The homology of Npl3 to mammalian SR-like shuttling proteins sets Npl3 apart from other RNA-binding proteins in yeast. Mutants of NPL3 show strong mRNA-export defects, which suggests a role in mRNA export from the nucleus (Lee et al., 1996). In fact, binding of Npl3 to mRNA seems to be crucial for the formation of an export-competent mRNP, as its dissociation results in an export-incompetent mRNP (Krebber et al., 1999). Npl3 is imported into the nucleus by the import receptor Mtr10 (Pemberton et al., 1997; Senger et al., 1998). The carboxy-terminal SR domain of Npl3 is phosphorylated on serine 411 by the only SRspecific protein kinase in yeast, Sky1 (Yun & Fu, 2000; Gilbert et al., 2001). It has been suggested that phosphorylation of Npl3 by Sky1 may increase the binding affinity of Npl3 to its import receptor, Mtr10 (Yun & Fu, 2000). However, in vitro binding studies resulted in no significant increase in binding of phosphorylated Npl3 to Mtr10 (Gilbert et al., 2001). Furthermore, it was proposed that phosphorylation of Npl3 is required for efficient release of mRNA on termination of export (Gilbert et al., 2001). So far, what happens on arrival of the RNP in the cytoplasm is still not clear, but two possible mechanisms by which SR proteins are dissociated from the exported mRNP complex are either binding of the import receptor or its phosphorylation.
Here, we present the identification of Gbp2 as a novel SR-like shuttling mRNA-binding protein in yeast. We characterize the requirements for its nuclear import and show the close relationship between mRNA export and the nuclear export of Gbp2. Interestingly, overexpression of Gbp2 is toxic and causes a nuclear retention of bulk poly(A)+ RNA. Conversely, because Gbp2 is non-essential, this novel SR protein has the potential to be spatially or temporally regulated.
Results
Gbp2 has features of an RNA-binding protein
Sequence analysis of Gbp2 revealed that the protein has features of an RNA-binding protein. It contains three RNA-recognition motifs (RRMs) and it is homologous to the well-characterized protein Npl3 (Fig. 1A) and to Hrb1 (not shown). Gbp2 and Npl3 show an overall identity of 27%. The first two RRMs in Gbp2 are conserved, whereas RRM3 is less conserved. The amino terminus of Gbp2 shows similarity to the C terminus of Npl3. It contains four Arg-Gly-Gly (RGG) repeat motifs and nine serine residues in the context of SR or arginine/serine (RS) motifs. In comparison, Npl3 contains 15 RGGs and 8 RS motifs. To determine the intracellular localization of Gbp2, we expressed the protein as a green fluorescent protein (GFP) fusion protein under the control of its own promoter. In wild-type cells, Gbp2 localizes to the nucleus (Fig. 1B).
Figure 1.
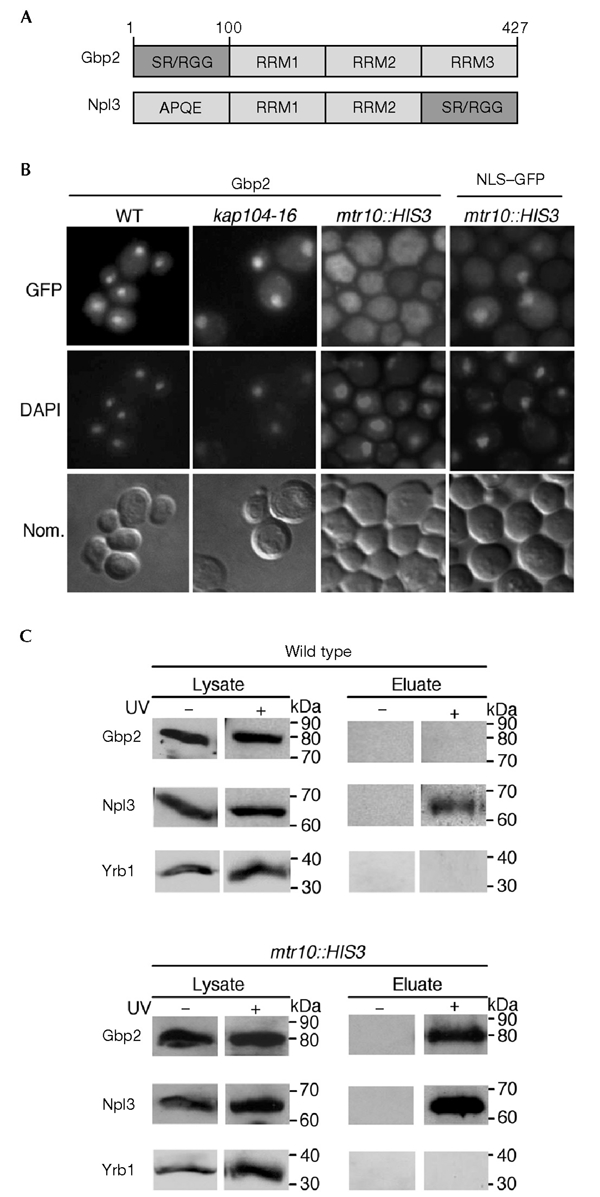
Gbp2 is a messenger RNA-binding protein that is transported into the nucleus by Mtr10. (A) Domain organization of Gbp2 and Npl3. Gbp2 consists of an amino-terminal domain with SR/RGG repeats followed by three RNA recognition motifs (RRMs). Npl3 contains two RRMs, a carboxy-terminal SR/RGG domain and an N-terminal domain with repeats of the amino-acid residues APQE. (B) Gbp2 is localized to the nucleus at steady state and accumulates in the cytoplasm of mtr10::HIS3 cells. Subcellular localization of Gbp2–GFP (green fluorescent protein) is shown in a wild-type strain (WT), in kap104-16 and in mtr10::HIS3 strains, shifted to 37 °C for 30 min. As a control, the localization of a nuclear localization signal–GFP reporter is shown under the same conditions in mtr10::HIS3 cells. The DNA was stained with DAPI (4,6-diamidino-2-phenylindole), and the cells were photographed using Nomarski optics (Nom.). (C) Ultraviolet (UV) crosslinking of Gbp2 to poly(A)+ RNA. Wild-type cells and mtr10::HIS3 cells that express Gbp2–GFP were grown to log phase and exposed to UV light for crosslinking (+ or − UV irradiation). The lysates were taken before the material was applied to an oligo(dT)-cellulose column. The eluates containing the crosslinked poly(A)+ RNA and bound proteins were treated with RNases. Proteins were analysed by SDS–polyacrylamide gel electrophoresis, followed by western blotting. The immunoblot was probed with anti-GFP antibodies and with anti-Npl3 antibodies. As a control for non-specific binding, anti-Yrb1 antibodies were used in a similar experiment.
Nuclear import of Gbp2 is mediated by Mtr10
The import of Gbp2 into the nucleus is mediated by Mtr10. Strains with mutations in genes encoding in different karyopherins were grown to log phase before they were shifted to 37 °C for 30 min. Gbp2 was mislocalized to the cytoplasm in both mtr10::HIS3 (Fig. 1B) and mtr10-1 (data not shown), whereas the localization in kap104-16 (Fig. 1B) and kap104::HIS3 cells (data not shown) was not affected. The localization was also unaltered in pse1-1 and kap123::HIS3 mutant strains (data not shown). The cytoplasmic mislocalization of Gbp2 in an mtr10 mutant is specific, as localization of a nuclear localization signal (NLS)–GFP reporter is unaffected (Fig. 1B). Furthermore, Gbp2 is a shuttling protein, as determined by a shuttling assay that took advantage of the mtr10-7 strain (see supplementary information online).
GBP2 is not essential and cells in which GBP2 is deleted show a growth rate similar to that of wild-type cells (data not shown). The overall structural features of Gbp2 indicate that it is an RNA-binding protein. To test its ability to bind to poly(A)+ RNA in vivo, we carried out ultraviolet (UV)-crosslinking experiments. Surprisingly, binding of Gbp2 to poly(A)+ RNA was barely detectable in wild-type yeast cells (Fig. 1C), whereas Npl3 binding in the same eluate was clearly visible. To investigate the possibility that Mtr10 is involved in the cytoplasmic release of Gbp2 from mRNA, UV-crosslinking experiments were carried out in the absence of Mtr10, which resulted in the appearance of a strong Gbp2 band (Fig. 1C). As expected, the binding of Npl3 to mRNA also increased (Fig. 1C), as was shown previously in Gilbert et al. (2001). The strong binding of both proteins to mRNA in the mtr10::HIS3 strain is specific, as binding of a cytoplasmic non-RNA-binding protein, Yrb1, to poly(A)+ RNA could not be detected (Fig. 1C). Together, the data indicate either that Mtr10 is indeed responsible for the release of Gbp2 from mRNA or that the cytoplasmic localization of Gbp2 leads to stronger binding of the protein to cytoplasmic mRNAs.
Phosphorylation is required forGbp2 nuclear localization
The N-terminal domain of Gbp2 might comprise the import signal, as a Gbp2–GFP fusion protein that lacks the first 100 amino-acid residues localizes to the cytoplasm (data not shown). In this region, Gbp2 contains several SR or RS motifs (Fig. 2A). Serine residues from these dipeptides are known to be potential phosphorylation sites for SRspecific protein kinases (SRPKs). Interestingly, a deletion mutant of the only SRPK in yeast, Sky1, leads to cytoplasmic mislocalization of Gbp2, whereas the nuclear localization of an NLS–GFP reporter protein is unaffected (Fig. 2B). To determine the potential phosphorylation sites in Gbp2, we applied rules that have been established for human SR protein kinase 2 in peptideselection experiments. This kinase has been shown to select strongly for arginine (R), histidine (H), glutamic acid (E) and proline (P), and against lysine (K), phenylalanine (F) and glycine (G) around the phosphorylation site (Yun & Fu, 2000). Using these rules, three out of all of the SR/RS motifs in Gbp2 remain candidate phosphorylation sites (Fig. 2A, top). In fact, the crystal structure of Sky1 revealed a binding 'pocket' in the molecule that has been suggested to recognize a consensus phosphorylation sequence in target proteins (Fig. 2A, bottom) (Nolen et al., 2001). Amino-acid residues 12–18 in Gbp2 match this consensus almost exactly. Using site-directed mutagenesis, we exchanged either all of these serine residues for alanine (gbp2s13/15/17A) or each residue individually (gbp2-S13A, gbp2-S15A and gbp2-S17A). All four gbp2 mutants were expressed as GFP fusion proteins. In contrast to wild-type Gbp2, which is found in the nucleus of log-phase yeast cells (Figs 1B and 2B), gbp2s13/15/17A was completely cytoplasmic (Fig. 2C). Strikingly, gbp2s15A and gbp2-S17A, but not gbp2-S13A, resulted in cytoplasmic mislocalization in wild-type cells (Fig. 2C). This mislocalization is not due to differences in expression levels, as determined by western blot analysis (data not shown). The nuclear localization of gbp2s13A is altered in sky1::TRP1 cells, in which the protein is completely cytoplasmic, similar to its wild-type counterpart. This indicates that it still has sensitivity to phosphorylation by Sky1 (see supplementary information online).
Figure 2.
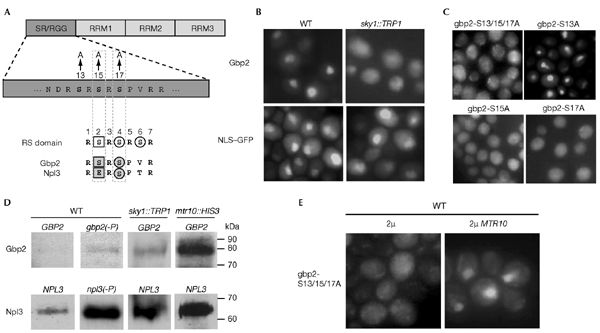
Sky1 is important for the nuclear localization of Gbp2 and for the efficiency of RNA release from Gbp2. (A) Domain organization of Gbp2 and indication of the mutations generated by site-directed mutagenesis. Bottom, an alignment of the phosphorylation sequence (RS domain) of SR-specific protein kinases (SRPKs) in mammals and the Sky1 phosphorylation recognition sequence in Npl3 and Gbp2 is shown. The white square indicates the position of an initial phosphorylation site that has been suggested to prime the activity of SRPKs, white circles indicate the phosphorylation sites for SRPKs. Shaded squares and circles indicate the corresponding residues in the yeast shuttling SR proteins. (B) Localization of Gbp2–GFP (green fluorescent protein) and NLS–GFP in logarithmic growing wild-type (WT) and sky1::TRP1 cells was detected by fluorescence microscopy. (C) Localization of the Gbp2 mutants in vivo. Wild-type strains that express mutant gbp2–GFP were grown to log phase and subcellular localization was determined by fluorescence microscopy. (D) Ultraviolet-crosslinking experiments were carried out in: a wild-type strain that expresses wild-type GBP2; a wild-type strain either expressing mutant GBP2 (gbp2-S13/15/17A, designated as gbp2(−P)) or mutant Npl3 (npl3-27, designated as npl3(−P)); sky1::TRP1 cells; and mtr10::HIS3 cells that express wild-type GBP2. All GBP2 constructs were expressed as GFP fusion proteins. Eluates were loaded in identical amounts in each lane. The western blot was probed with anti-GFP antibodies (top) and anti-Npl3 antibodies (bottom). (E) Wild-type strains that bear gbp2-S13/15/17A-GFP on a CEN plasmid and either MTR10 or no additional gene on a 2μ plasmid were grown to log phase and the subcellular localization of Gbp2 was determined by fluorescence microscopy. RRM, RNA recognition motif.
These results suggest that phosphorylation by Sky1 is important either for the nuclear import of Gbp2 or for its proper release from mRNA. To test if increased cytoplasmic localization of Gbp2 is always correlated with an increase in binding to mRNA (as is the case with Npl3), UV-crosslinking experiments were carried out with the cytoplasmically localized gbp2 mutant, gbp2-S13/15/17A. As shown in Fig. 2D, the binding of both Gbp2 and Npl3 to mRNA were, once again, significantly increased in the mtr10::HIS3 strain; however, gbp2s13/15/17A binding to mRNA in wild-type cells and Gbp2 mRNA binding in sky1::TRP1 cells was only slightly elevated. By contrast, a mutant of Npl3 that cannot be phosphorylated by Sky1, npl3-27 (Yun & Fu 2000), bound to poly(A)+ RNA to the same extent as its wild-type counterpart in sky1::TRP1 or mtr10::HIS3 cells. This indicates differences between the two SR proteins. Furthermore, it is clear that a cytoplasmic localization of Gbp2 is not equivalent to increased binding to mRNA.
Overexpression of MTR10 partially suppresses the mislocalization defect of gbp2-S13/15/17A. The reduced dissociation rate of Gbp2 from poly(A)+ RNA in mutant mtr10 cells and the cytoplasmic localization of Gbp2 in those strains suggests a physical interaction between these proteins that might be modulated by Sky1. Because we could not co-immunoprecipitate Gbp2 and Mtr10 to test this idea directly, we analysed the relationship between Mtr10 and Gbp2 phosphorylation in vivo by overexpressing MTR10 from a 2μ plasmid in gbp2-S13/15/17A cells. As shown in Fig. 2E, in the presence of 2μ MTR10, the nuclear localization of mutant gbp2 was partially restored; however, the nuclear signal was visible in only about 25% of the cells, as opposed to 100% of the cells for npl3-27 (Krebber et al., 1999).
Nuclear export of Gbp2 is dependent on mRNA export
Gbp2 accompanies mRNA to the cytoplasm, as determined by localization studies with the 'cytoplasmic version' of the protein in several mutants: gbp2-S15A was expressed in an rpb1-1 strain, expressing mutant RNA polymerase II. After a 30-min temperature shift to 37 °C, the export of gbp2-S15A was blocked and the protein was retained in the nucleus of all cells (Fig. 3A). These findings indicate that export of Gbp2 is dependent on continuous RNA polymerase II transcription. Mutants that are known to have strong mRNA export defects, such as rat7-1 and mex67-5, showed similar effects on gbp2s15A localization (Fig. 3A). Many cells (∼40%) show a strong nuclear signal after a 30-min shift to the restrictive temperature, pointing to a close relationship between Gbp2 and mRNA export. The same results were obtained using gbp2s17A in these experiments (data not shown). The localization of an NLS–NES–GFP reporter protein was not affected in these strains, indicating a specific export block for Gbp2 (Fig. 3A).
Figure 3.
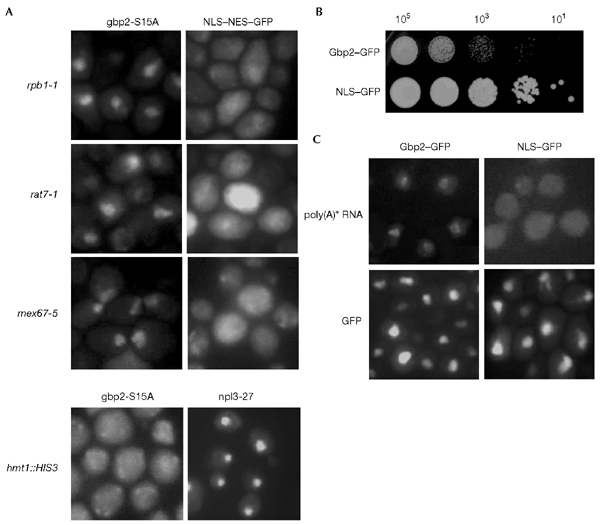
The export of Gbp2 is dependent on mRNA transport. (A) Gbp2 export is dependent on RNA polymerase II transcription and on the mRNA export factors Mex67 and Nup159/Rat7, but not on the arginine methyltransferase Hmt1. rpb1-1, mex67-5, rat7-1 or hmt1::HIS3 strains that express gbp2-S15A–GFP (green fluorescent protein) or nuclear localization signal–GFP were grown to log phase before they were shifted to 37 °C for 30 min. The intracellular localization of the GFP fusion proteins was analysed by fluorescence microscopy. The localization of npl3-27 was determined by immunofluorescence using anti-Npl3 antibodies. (B) Overexpression of gbp2 is toxic. Wild-type strains that express Gbp2–GFP under control of the strong GAL1 promoter or as a control NLS–GFP–GFP were spotted in 1:10 dilutions onto –uracil galactose plates. (C) Cells that overexpress Gbp2 accumulate poly(A)+ RNA in the nucleus. The same strains shown in (B) were grown to log phase in liquid –URA raffinose medium, shifted to galactose-containing medium for 4 h, followed by a 2-h repression in glucose-containing medium, before the intracellular mRNA localization was analysed by in situ hybridization using a digoxygenin-labelled oligo d(T)50 probe and rhodamine-labelled anti-digoxygenin antibodies. The subcellular localization of the GFP-fusion proteins was determined by fluorescence microscopy.
Export of Gbp2 is not dependent on arginine methylation by Hmt1. Many RNA-binding proteins have been shown to be substrates for arginine methylation. In S. cerevisiae, loss of HMT1 affects the dynamic localization of mRNA-binding proteins, such as Npl3 and Hrp1 (Shen et al., 1998). Whereas npl3-27p is trapped in the nucleus of hmt1::HIS3 cells, expression of gbp2s15A does not lead to a nuclear localization of the protein (Fig. 3A), indicating differences in the transport requirements of Npl3 and Gbp2.
Excess Gbp2 is toxic and causes mRNA-export defects
Although the absence of Gbp2 has no influence on cell viability, highly elevated levels of Gbp2 inhibit growth, as shown by growth tests (Fig. 3B). To find a potential cause for this toxicity, we analysed the distribution of bulk poly(A)+ RNA by in situ hybridization using an oligo d(T)50 probe in cells that overexpress GBP2 for 4 h. Approximately 25% of the cells showed mRNA-export defects in the presence of high levels of Gbp2 (Fig. 3C). By contrast, none of the cells that expressed NLS–GFP showed nuclear mRNA accumulation (Fig. 3C). The toxicity is not due to altered localization, as both of the GFP fusion proteins are, although highly expressed, nuclear at steady state (Fig. 3C).
Discussion
Only a few components of exported mRNPs have been identified so far. It is expected that several factors assemble on the mRNA to be exported, only some of which are involved in the export process itself. The others are thought to have different functions, such as marking exon–exon junctions to provide a key for the translation machinery or defining the time, efficiency or place of protein expression (Dreyfuss et al., 2002).
Here, we present the identification of Gbp2 as a novel shuttling mRNA-binding protein, involved in the delivery of mRNAs to the cytoplasm. The intracellular concentration of Gbp2 is crucial, because overexpression of the protein is toxic and results in mRNA-export defects. Interestingly, a similar phenotype has been described for Yra1 overexpression (Sträßer & Hurt, 2000). In fact, Gbp2 has been shown to be associated with the TREX ('transcription–export') complex, a multimeric complex that consists of several transcription factors and export factors, such as Yra1 and Sub2 (Sträßer et al., 2002).
By creating a mutant of Gbp2 that is localized in the cytoplasm at steady state, we were able to analyse the export requirements of the protein (Fig. 3A). gbp2s15A remains nuclear in a mutant of RNA polymerase II (in almost all cells), indicating the necessity of mRNA synthesis for its translocation to the cytoplasm. Surprisingly, the mislocalization defect in mutants of either the mRNA export receptor Mex67 or the nuclear porin Nup159/Rat7 is less pronounced. In both cases, gbp2-S15A is trapped in the nucleus in approximately 40% of the cells, even after a temperature shift for 3 h. Because both mutants are known for their instant and severe mRNA-export defects—which are visible within minutes of the temperature shift (Gorsch et al., 1995; Hurt et al., 2000)—one could speculate about the existence of other Gbp2/RNPspecific export factors, remaining to be identified.
Prevention of Gbp2 nuclear import or a significant reduction of the import rate is achieved by deleting either its import receptor, Mtr10, or the SRPK Sky1. Mammalian SRPKs were proposed to sequentially phosphorylate the serine residues of an RSRSRSR-consensus sequence, resulting in the phosphorylation of the second and third serine residues once an initial phosphorylation of the first serine residue has been primed by a different kinase (Nolen et al., 2001). This first phosphorylation event then serves as a nucleophile for phosphotransfer on serine 2 and the following serine residues that is accomplished by SRPKs. Amino-acid residues 12–18 in Gbp2 (RSRSRSP) match this consensus almost perfectly. Therefore, we were surprised to find that the first serine residue did not have any influence on the localization of Gbp2, in contrast to the second and third serine residues, which, when mutated, resulted in a cytoplasmic accumulation of the protein. Our results, and the alignment of the Gbp2 sequence (residues 14–20) with the Npl3 recognition site for Sky1 (Fig. 2A, bottom), suggest that Sky1 might phosphorylate only the serine residue at position 17 in Gbp2. Moreover, an alignment of Gbp2 and Npl3 suggests a Sky1specific recognition site with the consensus: R(E/S)RSP(T/V)R (Fig. 2A), which is slightly different to the mammalian SRPK recognition site. Npl3 has been shown to be phosphorylated on a single serine residue, indicated at position 4 in Fig. 2A. However, the glutamic-acid residue at position 2 has been shown to be essential for phosphorylation (Yun & Fu, 2000) and could mimic an initial phosphorylation, necessary for Sky1 to act. Taking the model of Nolen et al. (2001) into account, a mechanism could be suggested by which Sky1 could act on Npl3 at any time. By contrast, Gbp2 would require an initial phosphorylation of the serine residue at position 15 before Sky1 would be able to use the protein as a substrate. It is tempting to speculate that the kinase necessary for providing this first phosphorylation could determine the time and/or place of generation of a substrate for Sky1.
Our results show that cytoplasmic Gbp2 localization, due to deletion of its import receptor, Mtr10, results in a significantly increased binding of Gbp2 to poly(A)+ RNA, indicating that Mtr10 might be necessary for the dissociation of Gbp2 from mRNA. The same cytoplasmic phenotype is obtained by expressing either gbp2 with a mutant SR consensus sequence or wild-type GBP2 in sky1::TRP1 cells. Interestingly, however, UV-crosslinking experiments revealed that, although binding of mutant gbp2 to poly(A)+ RNA is increased in comparison to the wild type (similar to wild-type Gbp2 in sky1::TRP1 cells), it is significantly lower than in the mtr10::HIS3 strain (Fig. 2D). Thus, the amount of Gbp2-bound mRNA is not determined by the extent of the cytoplasmic localization. By contrast, expressing the non-phosphorylated SR mutant of NPL3 (or wild-type Npl3 in sky1::TRP1 cells) leads to a UV-crosslinking product comparable to that seen for wild-type Npl3 in mtr10::HIS3 cells (Fig. 2D), as was shown previously in Gilbert et al. (2001). For Npl3, a model was proposed in which Npl3 is exported bound to bulk mRNA and may function as a central component that is important for stabilization of the transport complex (Krebber et al., 1999). On arrival in the cytoplasm, Npl3 is phosphorylated by Sky1 and, as a consequence, is recognized by Mtr10 (Yun & Fu, 2000; Gilbert et al., 2001). For Npl3, no additional kinase would be necessary for Sky1 to act, because of the glutamic acid at position 2 (Fig. 2A, bottom). Here, Sky1 might simply be responsible for delaying the disassembling process to gain time for cytoplasmic factors, or the ribosome, to assemble on the delivered RNP.
Conversely, for Gbp2, dissociation from mRNA might be dependent on the type of mRNA. This means that if it guides bulk RNA to the cytoplasm, Gbp2 can be dissociated by Mtr10 on arrival in the cytoplasm. However, a tighter, possibly sequence-specific association to certain mRNAs might require phosphorylation of Gbp2 by Sky1 before Mtr10 can act. It is tempting to speculate that the potential initial phosphorylation site at serine residue 15 in Gbp2 (Fig. 2A, top), which might be required for Sky1 to act efficiently, could be part of a regulation cascade, signalling the mRNA–protein complex to disassemble at the right time or place. It will be challenging to identify specific Gbp2-bound mRNAs and thus learn more about potential cis-elements.
Many proteins accompany mRNAs on their way to the ribosomes and it is to be expected that association and dissociation of all proteins on the mRNP is tightly regulated. It is therefore of great interest to identify proteins that are part of this process and to unravel their functions. With the identification of Gbp2 as a novel component of transported mRNA–protein complexes, we were able to add one piece to the puzzle. It will be interesting to learn more about Gbp2-interacting proteins and the mRNAs to which it binds.
Methods
Plasmids and strains.
All plasmids were generated by PCR amplification of GBP2 from the genomic DNA of S. cerevisiae with the appropriate primers. All PCR products were sequenced. The promoter of GBP2 was cloned using restriction enzymes XbaI and ClaI into pHK12 (= pPS1372) (Taura et al., 1998) CEN URA3. In a second step, GBP2 (pHK367), truncated gbp2Δ1–100) (pHK389) and mutant gbp2 (pHK412 = gbp2s13/15/17A–GFP, pHK446 = gbp2-S13A–GFP, pHK431 = gbp2-S15A–GFP, pHK432 = gbp2-S17A–GFP) were generated by site-directed mutagenesis and cloned in the same vector with the appropriate primers and by using EcoRI and XhoI. The GAL1 promoter was amplified from genomic DNA and cloned in exchange for the GBP2 promoter into pHK367 using XbaI and EcoRI. The resulting plasmid was cut with NotI and KpnI to excise GAL1:Gbp2–GFP and the fragment was subcloned into pRS426, resulting in pHK422. The GFP reporter plasmids were described elsewhere (Taura et al., 1998).
The yeast strains hmt1::HIS3 npl3-27 (HKY52) and hmt1::HIS3 (HKY53) were generated by crossing hmt1::HIS3 (PSY865) (Henry & Silver, 1996) with npl3-27 (HKY16 = PSY1032) (Krebber et al., 1999), sporulating the diploid and consequent tetrad dissection and analysis. Wild type (HKY36), npl3-27 (HKY15), mtr10::HIS3 (HKY97), kap104-16 (HKY207), sky1::TRP1 (HKY267) and mex67-5 (HKY315) strains are described elsewhere (Winston et al., 1995; Aitchison et al., 1996; Segref et al., 1997; Krebber et al., 1999; Liu et al., 1999; Siebel et al., 1999).
Expression, GFP localization, in situ poly(A)+ RNA hybridization, immunofluorescence and UV-crosslinking experiments.
All procedures were carried out as described previously (Krebber et al., 1999). For fluorescent microscopy, strains were grown to logarithmic phase (1 × 107 to 5 × 107 cells ml−1) at 25 °C and then shifted to 37 °C for the indicated times. The GFP signal was analysed directly by fluorescence microscopy and pictures were taken from fixed cells. For this purpose, 5 ml of the cultures were mixed with 350 μl of 37% formaldehyde. Cells were collected immediately by centrifugation and washed once in 0.1 M K2HPO4–KH2PO4 at pH 6.5 and once in P solution (0.1 M K2HPO4–KH2PO4 at pH 6.5, 1.2 M Sorbitol).
Supplementary data are available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor763-s1.mov).
Supplementary Material
Supplementary information
Acknowledgments
We thank A. Rösser for technical assistance. We are grateful to C. Guthrie, E. Hurt, M. Rout, P. Silver and A. Tartakoff for providing strains, plasmids and antibodies. We thank H. Bastians, E. Hurt, M. Seedorf and K. Sträßer for helpful discussions. This work was funded by a grant from the Deutsche Forschungsgemeinschaft to H.K.
References
- Aitchison J.D., Blobel G. & Rout M.P. (1996) Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science, 274, 624–627. [DOI] [PubMed] [Google Scholar]
- Anderson J.T., Wilson S.M., Datar K.V. & Swanson M.S. (1993) NAB2: a yeast nuclear polyadenylated RNA-binding protein essential for cell viability. Mol. Cell. Biol., 13, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N. & Kataoka N. (2002) Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol., 3, 195–205. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Siebel C.W. & Guthrie C. (2001) Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA, 7, 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsch L.C., Dockendorff T.C. & Cole C.N. (1995) A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol., 129, 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M.F. & Silver P.A. (1996) A novel methyltransferase (Hmt1p) modifies poly(A)+-RNA-binding proteins. Mol. Cell. Biol., 16, 3668–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E., Sträßer K., Segref A., Bailer S., Schlaich N., Presutti C., Tollervey D. & Jansen R. (2000) Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem., 275, 8361–8368. [DOI] [PubMed] [Google Scholar]
- Krebber H., Taura T., Lee M.S. & Silver P.A. (1999) Uncoupling of the hnRNP Npl3p from mRNAs during the stress-induced block in mRNA export. Genes Dev., 13, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.S., Henry M. & Silver P.A. (1996) A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev., 10, 1233–1246. [DOI] [PubMed] [Google Scholar]
- Liu Y., Guo W., Tartakoff P.Y. & Tartakoff A.M. (1999) A Crm1p-independent nuclear export path for the mRNA-associated protein, Npl3p/Mtr13p. Proc. Natl Acad. Sci. USA, 96, 6739–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B., Yun C.Y., Wong C.F., McCammon J.A., Fu X.D. & Ghosh G. (2001) The structure of Sky1p reveals a novel mechanism for constitutive activity. Nature Struct. Biol., 8, 176–183. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum J.S. & Blobel G. (1997) A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol., 139, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R. & Hurt E. (2002) A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell, 108, 523–531. [DOI] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R. & Hurt E. (1997) Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J., 16, 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B., Simos G., Bischoff F.R., Podtelejnikov A., Mann M. & Hurt E. (1998) Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J., 17, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E.C., Henry M.F., Weiss V.H., Valentini S.R., Silver P.A. & Lee M.S. (1998) Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev., 12, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C.W., Feng L., Guthrie C. & Fu X.D. (1999) Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl Acad. Sci. USA, 96, 5440–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. & Hurt E. (2000) Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J., 19, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträßer K. et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature, 417, 304–308. [DOI] [PubMed] [Google Scholar]
- Taura T., Krebber H. & Silver P.A. (1998) A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc. Natl Acad. Sci. USA, 95, 7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.M., Datar K.V., Paddy M.R, Swedlow J.R. & Swanson M. (1994) Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol., 127, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C. & Ricupero-Hovasse S.L. (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast, 11, 53–55. [DOI] [PubMed] [Google Scholar]
- Yun C.Y. & Fu X.D. (2000) Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol., 150, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D. & Stutz F. (2001) Nuclear export of mRNA. FEBS Lett., 498, 150–156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


