Abstract
Deranged oxidative metabolism is a property of many tumour cells. Oxidation of the deoxynucleotide triphosphate (dNTP) pool, as well as DNA, is a major cause of genome instability. Here, we report that two Y-family DNA polymerases of the archaeon Sulfolobus solfataricus strains P1 and P2 incorporate oxidized dNTPs into nascent DNA in an erroneous manner: the polymerases exclusively incorporate 8-OH-dGTP opposite adenine in the template, and incorporate 2-OH-dATP opposite guanine more efficiently than opposite thymine. The rate of extension of the nascent DNA chain following on from these incorporated analogues is only slightly reduced. These DNA polymerases have been shown to bypass a variety of DNA lesions. Thus, our results suggest that the Y-family DNA polymerases promote mutagenesis through the erroneous incorporation of oxidized dNTPs during DNA synthesis, in addition to facilitating translesion DNA synthesis. We also report that human DNA polymerase η, a human Y-family DNA polymerase, incorporates the oxidized dNTPs in a similar erroneous manner.
Introduction
The recently identified Y-family DNA polymerases comprise proteins from different species, including members of the Bacteria, Eukarya and Archaea (Ohmori et al., 2001). The most distinct feature of this family of enzymes is their ability to bypass various lesions, such as ultraviolet light photoproducts, in template DNA (Friedberg et al., 2002). Some bypass reactions catalysed by these enzymes are error prone, whereas others are error free (Wang, 2001). Thus, this family of DNA polymerases seems to be involved in mutagenesis and DNA-damage tolerance. Structural analyses of two Y-family DNA polymerases from the archaeon Sulfolobus solfataricus strains P1 (Sso P1 DNA pol Y1, also known as Dbh) and P2 (Sso P2 DNA pol Y1, also known as Dpo4) reveal that these proteins have the same overall right-hand shape found in other DNA polymerases, but have several distinct features. These include the presence of a little-finger domain and a solvent-accessible base-pair-binding pocket, which might enable the polymerases to bypass lesions (Ling et al., 2001; Silvian et al., 2001; Zhou et al., 2001). We, and others, have shown that the archaeal Y-family DNA polymerases bypass a variety of DNA lesions (Gruz et al., 2001; Boudsocq et al., 2001).
Because the error rates of Y-family DNA polymerases are extremely high, and they seem to have active sites that are large enough to accommodate bulky lesions, we postulated that they might incorrectly incorporate structurally altered DNA precursors, such as oxidized deoxynucleotide triphosphates (dNTPs), into the nascent DNA strand. The dNTP pool and DNA itself are continuously exposed to various exogenous and endogenous damaging agents, including reactive oxygen species (ROS), and the incorporation of oxidized dNTPs into DNA is a major source of spontaneous mutagenesis and carcinogenesis (Ames & Gold, 1991). Indeed, the frequency of A:T to C:G transversion mutations increases more than a thousand-fold over the wild-type level in Escherichia coli mutT mutants, which are deficient in the ability to hydrolyse oxidized dGTP (8-OH-dGT, also known as 7,8-dihydro-8-oxo-dGTP) (Fig. 1A; Yanofsky et al., 1966; Maki & Sekiguchi, 1992). In agreement with this observation, 8-OH-dGTP leads to an A:T to C:G mutation when it is incorporated opposite adenine in the template (Cheng et al., 1992; Inoue et al., 1998; Kamiya & Kasai, 2000). In addition, spontaneous tumorigenesis in lung, liver and stomach is enhanced in mice that are deficient for MTH1, a mammalian homologue of E. coli MutT (Tsuzuki et al., 2001). MTH1 hydrolyses 2-OH-dATP (oxidized dATP; Fig. 1B) as well as 8-OH-dGTP (Fujikawa et al., 1999). Thus, it is of great interest to investigate the specificity of Y-family DNA polymerases incorporating oxidized dNTPs to the nascent DNA strand.
Figure 1.
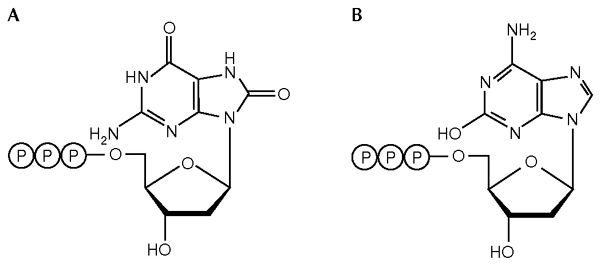
Structures of oxidized deoxynucleotide triphosphates (dNTPs) used in this study. The structures of 8-OH-dGTP (A) and 2-OH-dATP (B) are shown. P, phosphate group.
Here, we analyse the fidelity of Sso P1 DNA pol Y1 and Sso P2 DNA pol Y1 to incorporate 8-OH-dGTP and 2-OH-dATP into the nascent DNA strand. These enzymes were chosen because their crystal structures have been solved, as described above, and because hyperthermophiles such as S. solfataricus continuously face heat-induced damage to DNA and to the dNTP pool (Lopez-Garcia & Forterre, 2000). The spontaneous mutation rate in S. solfataricus is high, mainly because of the presence of an active transposable element (Martusewitsch et al., 2000). We refer to the polymerases as Sso P1 DNA pol Y1 and Sso P2 DNA pol Y1 based on the nomenclature system for DNA polymerases from Sulfolobus (Edgell et al., 1997). Strikingly, these two DNA polymerases incorporated oxidized dNTPs in an erroneous manner: they exclusively incorporated 8-OH-dGTP opposite adenine in the template, and incorporated 2-OH-dATP opposite guanine more efficiently than opposite thymine. In addition, human DNA polymerase η (hDNA pol η), a member of the human Y-family DNA polymerases (Masutani et al., 1999), also showed a similar specificity. These findings suggest the possible involvement of Y-family DNA polymerases in oxidative mutagenesis through misincorporation of altered nucleotides during DNA synthesis.
Results
Exclusive incorporation of 8-OH-dGTP opposite adenine
We analysed the incorporation of 8-OH-dGTP during in vitro DNA synthesis by S. solfataricus strains P1 and P2 Y1 DNA polymerases. We also analysed 8-OH-dGTP incorporation by hDNA pol η, the Klenow fragment of E. coli DNA polymerase I (KF exo−), which lacks 3′ to 5′ exonuclease activity, and Sso P2 DNA polymerase B1 (pol B1), which is responsible for the chromosomal replication of S. solfataricus strain P2. Incorporation of 8-OH-dGTP was analysed using primer extension assays, as described in the Methods section. Primers annealed to template 1A, which has adenine in the template N position, were elongated efficiently by P1 DNA pol Y1 and P2 DNA pol Y1 in the presence of 8-OH-dGTP (Fig. 2A and B). Similar results were obtained with hDNA pol η (Fig. 2C). No incorporation of 8-OH-dGTP was observed opposite cytosine, guanine or thymine residues. In contrast, the primers annealed to template 1A or template 1C were elongated by KF exo− and pol B1 (Fig. 2D and E). These results show that P1 and P2 DNA pol Y1 and hDNA pol η exclusively incorporated 8-OH-GTP opposite adenine, whereas KF exo− and pol B1 incorporated it opposite adenine and cytosine. The same results were obtained when another primer–template combination (template 2 and primer 2) was used (data not shown).
Figure 2.
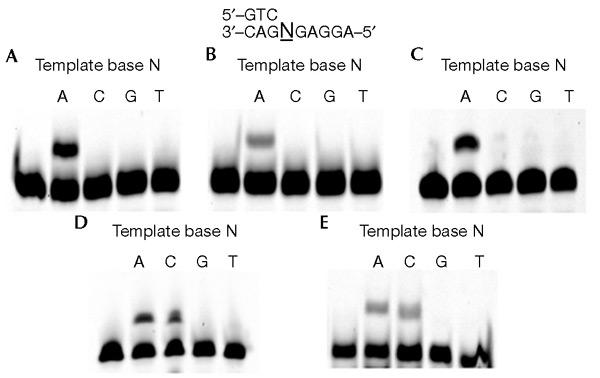
Incorporation of 8-OH-dGTP by DNA polymerases. The incorporation of 8-OH-dGTP into DNA by the following enzymes was assayed: P1 DNA polymerase Y1 (A), P2 DNA polymerase Y1 (B), hDNA polymerase η (C), KF exo− (D) and DNA polymerase B1 (E). Cy3-labelled-primer 1/template 1 (0.1 μM) was incubated with P1 DNA polymerase Y1 (200 nM), P2 DNA polymerase Y1 (25 nM), hDNA polymerase η (1 nM), DNA polymerase B1 (25 nM) or KF exo− (0.02 units) in the presence of 50 μM 8-OH-dGTP. The samples were analysed as described in the Methods section.
Next, we examined whether primers with 8-OH-dGMP at their termini could be extended by P1 and P2 DNA pol Y1 in the presence of other dNTPs. Primer 2 (an 18-base oligonucleotide) was elongated by these polymerases to yield 19-base oligonucleotides in the presence of 8-OH-dGTP (Fig. 3, lane 2 in both panels). The primers were elongated efficiently to produce 20-base oligonucleotides in the presence of 8-OH-dGTP and dGTP (Fig. 3, lane 3 in both panels), and further elongated to 22- and 23-base oligonucleotides in the presence of dCTP, dGTP and 8-OH-dGTP (Fig. 3, lane 4 in both panels). The primers could not be elongated to produce oligonucleotides longer than 23 bases when two adenines were present in the template strand (Fig. 3, lane 5 in both panels). These results show that a single incorporation of 8-OH-dGTP did not block further elongation by the polymerases, although incorporation of two 8-OH-dGTPs might have an inhibitory effect.
Figure 3.
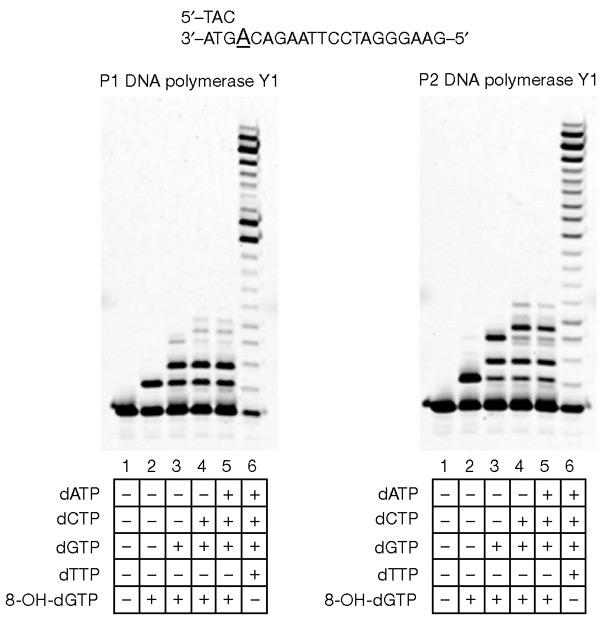
Primer extension by P1 DNA polymerase Y1 and P2 DNA polymerase Y1. Cy3-labelled-primer 2/template 2 (0.1 μM) was incubated with P1 DNA polymerase Y1 (200 nM) or P2 DNA polymerase Y1 (25 nM) in the presence of 50 μM each of dNTPs. The incubations were carried out in the presence of different combinations of dNTPs (lanes 3–6, both panels). The reaction mixtures were incubated for 30 min at 55 °C.
Preferential incorporation of 2-OH-dATP opposite guanine
2-OH-dATP is another mutagenic dNTP generated from dATP in the presence of oxygen radicals (Kamiya & Kasai, 1995). It pairs with guanine residues in the template, and induces G:C to T:A mutations (Inoue et al., 1998; Kamiya & Kasai, 2000). 2-OH-dATP was predominantly incorporated opposite guanine and thymine by P1 and P2 DNA pol Y1, and the frequnecy of incorporation opposite guanine was greater than or similar to that of incorporation opposite thymine (Fig. 4A and B). A similar incorporation pattern was observed with hDNA pol η (Fig. 4C). In contrast, KF exo− and pol B1 predominantly incorporated 2-OH-dATP opposite thymine (Fig. 4D and E). In particular, pol B1 almost exclusively incorporated 2-OH-dATP opposite thymine. The results for KF exo− are consistent with those of Kamiya et al. (2000).
Figure 4.
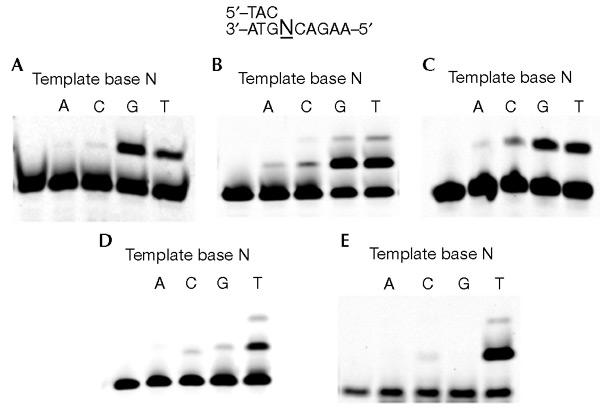
Incorporation of 2-OH-dATP by DNA polymerases. The incorporation of 2-OH-dATP into DNA by the following enzymes was assayed: P1 DNA polymerase Y1 (A), P2 DNA polymerase Y1 (B), hDNA polymerase η (C), KF exo− (D) and DNA polymerase B1 (E). Cy3-labelled-primer 2/template 2 (0.1 μM) was incubated with P1 DNA polymerase Y1 (200 nM), P2 DNA polymerase Y1 (25 nM), hDNA polymerase η (1 nM), DNA polymerase B1 (25 nM) or KF exo− (0.02 units) in the presence of 50 μM 2-OH-dATP.
We then examined the extension by the archaeal polymerases of primers with 2-OH-dAMP at the termini. As P1 and P2 DNA pol Y1 incorporate 2-OH-dATP opposite guanine and thymine, we used a version of template 2 with guanine at the N position followed by the sequence CAT (reading in the 3′ to 5′ direction. Primer 2 (18 bases) was first elongated to a 19-base oligonucleotide by the incorporation of 2-OH-dATP opposite guanine (Fig. 5, lane 2 in both panels). These primers were elongated to 20 bases in the presence of 2-OH-dATP and dGTP (Fig. 5, lane 3 in both panels), and further elongated to 24 or 25 bases in the presence of dTTP, dGTP and 2-OH-dATP (Fig. 5, lane 4 in both panels). When 2-OH-dATP and all dNTPs except for dCTP were present, the primers were elongated up to 31 or 32 bases when runs of three guanine residues were present in the template strands (Fig. 5, lane 5 in both panels). These results indicate that primers with 2-OH-dAMP at the termini could be elongated by these polymerases, although incorporation of two or three 2-OH-dATPs might have inhibitory effects.
Figure 5.
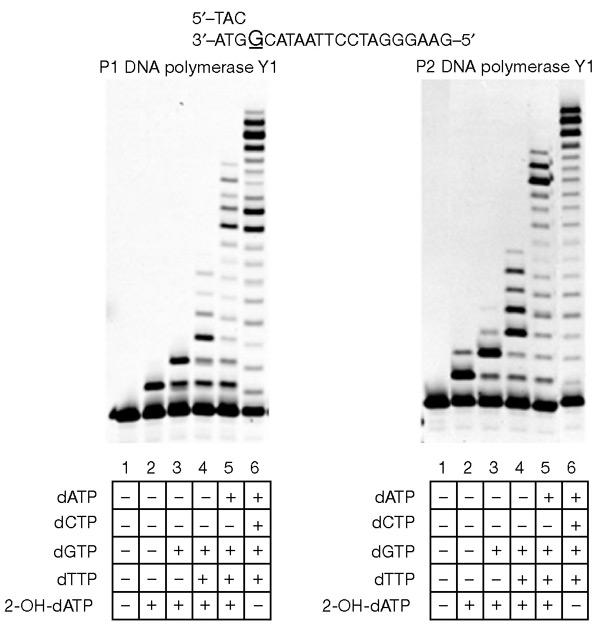
Primer extension by P1 DNA polymerase Y1 and P2 DNA polymerase Y1. The reaction conditions are the same as those described in the legend of Fig. 3.
rNTP and oxidized dNTP incorporation by the F12A mutant
It is proposed that the phenylalanine residue at position 12 (Phe 12) in the P1 DNA pol Y1 protein functions to discriminate against ribonucleotide triphosphate (rNTP) incorporation by acting as a 'steric gate' that would obstruct a 2′-OH group on an incoming nucleotide (Zhou et al., 2001). We examined whether Phe 12 also affects the incorporation of 8-OH-dGTP and 2-OH-dATP. Although no detectable incorporation of rNTP was observed with the wild-type enzyme, a mutant form of P1 DNA pol Y1 with a substitution of Phe to Ala at position 12 (F12A) efficiently incorporated and extended rNTP, even in the absence of Mn2+ ions (Fig. 6A and B). However, the mutation did not substantially affect the specificity of incorporation of oxidized dNTPs (Fig. 6C and D). These results suggested that Phe 12 senses the 2'-OH group on the nucleotide, but not OH groups on purine bases.
Figure 6.
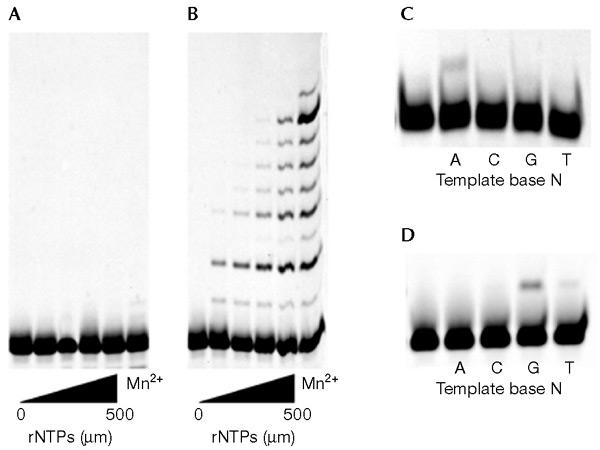
Incorporation of ribonucleotide triphosphates (rNTPs), 8-OH-dGTP and 2-OH-dATP by P1 DNA polymerase Y1 and the F12A mutant. Cy3-labelled-primer 2/template 2 (0.1 μM) was incubated with 200 nM of P1 DNA polymerase Y1 (A) or 400 nM of the F12A mutant enzyme (B) in the presence of rNTPs for 10 min at 55 °C. Manganese ions (1.5 mM) were also added to the reaction mixture containing 500 μM rNTP in the lanes labelled 'Mn2+'. The template/primer solution (0.1 μM) was incubated with the F12A enzyme (400 nM) in the presence of 50 μM 8-OH-dGTP (C) or 50 μM 2-OH-dATP (D) for 30 min at 55 °C.
Discussion
Here, we provide evidence that some Y-family DNA polymerases incorporate oxidized dNTPs into DNA in an erroneous manner (Figs 2 and 4). The incorporation of single-oxidized dNMPs did not inhibit further extension of the primer strands by archaeal DNA polymerases (Figs 3 and 5). Because the genome of S. solfataricus is continuously damaged by heat-induced ROS and depurination (Bruskov et al., 2002), and Y-family DNA polymerases are thought to act at stalled replication forks at DNA lesions (Friedberg et al., 2002), the archaeal DNA polymerases are likely to access the primer/templates very often. In addition, P2 DNA pol Y1 is about three times more abundant than pol B1 in strain P2 (F.M. Pisani, unpublished data), and P1 DNA pol Y1 is reported to become a processive enzyme on interaction with proliferating cell nuclear antigen and replication factor C (Gruz et al., 2001). Thus, we conclude that the erroneous incorporation of oxidized dNTPs by P1 and P2 DNA pol Y1 is relevant to genome instability in these organisms. To our knowledge, this is the first report that suggests the involvement of Y-family DNA polymerases in oxidative mutagenesis that proceeds through erroneous incorporation of 8-OH-dGTP and 2-OH-dATP.
P1 and P2 DNA pol Y1 can bypass a variety of lesions, such as abasic sites, 8-OH-guanine and a cis–syn thymine–thymine dimer, in template DNA (Boudsocq et al., 2001; Gruz et al., 2001). In addition, their replication fidelities are fairly low (10−3 to 10−4), even in the absence of DNA damage (Boudsocq et al., 2001; Silvian et al., 2001; Kokoska et al., 2002). Thus, the Y-family DNA polymerases seem to promote mutagenesis through three distinct pathways: misincorporation of the oxidized dNTPs during DNA synthesis; bypass synthesis across DNA lesions in template; and spontaneous replication errors.
As the DNA-replication apparatus of the Archaea is closer to that of the Eukarya than to that of the Bacteria, these findings could have implications for higher organisms, including humans. In fact, hDNA pol η was found to erroneously incorporate oxidized dNTPs into DNA (Figs 2 and 4). Interestingly, the mutator effect associated with human mismatch-repair deficiency is thought to be mainly a consequence of oxidative damage (Colussi et al., 2002). The majority of mutations in human cells that are deficient in mismatch repair do not arise from spontaneous replication errors, but from the incorporation of oxidized dNTPs.
In this study, we have revealed the erroneous incorporation of oxidized dNTPs into the nascent DNA strand by the Y-family DNA polymerases, and we propose that the main mutagenic activity of some Y-family DNA polymerases is incorporation of altered nucleotides, rather than lesion bypass.
Methods
Materials.
KF exo− was purchased from New England Biolabs. DNA pol B1, hDNA pol η and P1 DNA pol Y1 were purified as described (Pisani et al., 1998; Masutani et al., 2000; Gruz et al., 2001). The gene encoding the F12A mutant was constructed by site-directed mutagenesis, and the protein was purified as described for wild-type P1 DNA pol Y1 (Gruz et al., 2001). 8-OH-dGTP and 2-OH-dATP were prepared as described (Cheng et al., 1992; Kamiya & Kasai, 1995) and no discernible impurities were detected by high performance liquid chromatography (HPLC). FPLC-grade dNTPs and rNTPs were purchased from Amersham Pharmacia Biotech. Oligonucleotides, including those labelled with a Cy3 conjugate, were synthesized by BEX Corp., and were purified twice by HPLC.
Overexpression and purification of P2 DNA pol Y1.
The P2 dbh gene was amplified by PCR from the genome of S. solfataricus strain P2 and cloned into the vector pET-43a (Novagen), which was transformed into E. coli strain BL21-CodonPlus™ (DE3)-RIL (Novagen). The soluble protein fraction was precipitated at 70 °C for 20 min. After centrifugation, the protein was precipitated with ammonium sulphate (added to 58% saturation) and applied to a phenyl-sepharose HP column (Amersham Pharmacia Biotech.). Subsequent steps were identical to the method described previously (Wagner et al., 2000), except that the final sample was dialysed against a buffer containing 150 mM NaCl. The purity of the protein was confirmed by SDS–polyacrylamide gel electrophoresis, followed by Coomassie G250 staining.
Primer extension assay.
Standard reaction mixtures (10 μl) contained 30 mM potassium phosphate buffer (pH 7.4), 7.5 mM MgCl2, 1.25 mM β-mercaptoethanol, 5% glycerol, 50 μM dNTP(s), 200 nM P1 DNA pol Y1 or 25 nM P2 DNA pol Y1, and 100 nM Cy3-conjugated-primer annealed to the template in a 1:1 ratio. Reaction mixtures were incubated for 30 min (P1 pol Y1) or 15 min (P2 pol Y1) at 55 °C, unless otherwise indicated. When DNA pol B1 or KF exo− were used, the reaction was performed for 15 min at 55 °C, or for 15 min at 20 °C, respectively. The reaction mixture for hDNA pol η was the same as described (Masutani et al., 2000), except that 50 μM dNTP(s), 1 nM enzyme, 5 mM MgCl2 and 100 nM Cy3-labelled-primer/template were used. The reaction was performed for 15 min at 37 °C. Oligonucleotide sequences were 5′-Cy3-CGGAGCTCGGTCGGCGTCTGCGTC-3′ (primer 1, 24 bases); 5′-AGCCGCAGGAGNGACGCAGACGCCGACCGAGCTCCG-3′ (template 1, 36 bases); 5′-Cy3-CGCGCGAAGACCGGTTAC-3′ (primer 2, 18 bases); 5′-GAAGGGATCCTTAAYACNGTAACCGGTCTTCGCGCG-3' (template 2, 36 bases), where N represents either A, C, G, or T, and Y represents either G or T. A combination of primer 2/template 2 was used throughout the study, except for the experiments shown in Fig. 2. The reactions were terminated by adding 10 μl stop solution (98% formamide, 10 mM EDTA). Samples were heated at 100 °C for 10 min, separated on 15% denaturing polyacrylamide gels, and visualized using a Molecular Imager FX Pro System (BioRad).
RNA polymerase assay.
Reactions were carried out in 30 mM potassium phosphate buffer (pH 7.4), 7.5 mM MgCl2, 1.25 mM β-mercaptoethanol, 5% glycerol, 100 nM primer 2/template 2, all four rNTPs (50–500 μM each), and either wild-type P1 Pol Y1 (200 nM) or the F12A mutant enzyme (400 nM). The reactions were performed for 30 min at 55 °C and terminated by adding stop solution (composition as for that used in the primer extension assay).
Acknowledgments
This work was supported by grants-in-aid for Crossover Research from the Ministry of Education, Sports, Culture, Sciences and Technology, Japan, and an International Collaborative Research Grant from the Japan Health Science Foundation.
References
- Ames B.N. & Gold L.S. (1991) Endogenous mutagens and the causes of aging and cancer. Mutat. Res., 250, 3–16. [DOI] [PubMed] [Google Scholar]
- Boudsocq F., Iwai S., Hanaoka F. & Woodgate R. (2001) Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic pol η. Nucleic Acids Res., 29, 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruskov V.I., Malakhova L.V., Masalimov Z.K. & Chernikov A.V. (2002) Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res., 30, 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.C., Cahill D.S., Kasai H., Nishimura S. & Loeb L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G to T and A to C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- Colussi C., Parlanti E., Degan P., Aquilina G., Barnes D., Macpherson P., Karran P., Crescenzi M., Dogliotti E. & Bignami M. (2002) The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol., 12, 912–918. [DOI] [PubMed] [Google Scholar]
- Edgell D.R., Klenk H.P. & Doolittle W.F. (1997) Gene duplications in evolution of archaeal family B DNA polymerases. J. Bacteriol., 179, 2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Wagner R. & Radman M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science, 296, 1627–1630. [DOI] [PubMed] [Google Scholar]
- Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y. & Kasai H. (1999) The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem., 274, 18201–18205. [DOI] [PubMed] [Google Scholar]
- Gruz P., Pisani F.M., Shimizu M., Yamada M., Hayashi I., Morikawa K. & Nohmi T. (2001) Synthetic activity of Sso DNA polymerase Y1, an archaeal DinB-like DNA polymerase, is stimulated by processivity factors proliferating cell nuclear antigen and replication factor C. J. Biol. Chem., 276, 47394–47401. [DOI] [PubMed] [Google Scholar]
- Inoue M., Kamiya H., Fujikawa K., Ootsuyama Y., Murata-Kamiya N., Osaki T., Yasumoto K. & Kasai H. (1998) Induction of chromosomal gene mutations in Escherichia coli by direct incorporation of oxidatively damaged nucleotides. New evaluation method for mutagenesis by damaged DNA precursors in vivo. J. Biol. Chem., 273, 11069–11074. [DOI] [PubMed] [Google Scholar]
- Kamiya H. & Kasai H. (1995) Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steadystate kinetics of the incorporation. J. Biol. Chem., 270, 19446–19450. [DOI] [PubMed] [Google Scholar]
- Kamiya H. & Kasai H. (2000) 2-Hydroxy-dATP is incorporated opposite G by Escherichia coli DNA polymerase III resulting in high mutagenicity. Nucleic Acids Res., 28, 1640–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya H., Maki H. & Kasai H. (2000) Two DNA polymerases of Escherichia coli display distinct misinsertion specificities for 2-hydroxy-dATP during DNA synthesis. Biochemistry, 39, 9508–9513. [DOI] [PubMed] [Google Scholar]
- Kokoska R.J., Bebenek K., Boudsocq F., Woodgate R. & Kunkel T.A. (2002) Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. J. Biol. Chem., 277, 19633–19638. [DOI] [PubMed] [Google Scholar]
- Ling H., Boudsocq F., Woodgate R. & Yang W. (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell, 107, 91–102. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P. & Forterre P. (2000) Thermal stress in hyperthermophiles. In Bacterial Stress Response (eds Storz, G. & Hengge-Aronis, R.) 369–382. ASM, Washington, DC. [Google Scholar]
- Maki H. & Sekiguchi M. (1992) MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature, 355, 273–275. [DOI] [PubMed] [Google Scholar]
- Martusewitsch E., Sensen C.W. & Schleper C. (2000) High spontaneous mutation rate in the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by transposable elements. J. Bacteriol., 182, 2574–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K. & Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Iwai S. & Hanaoka F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, 8, 7–8. [DOI] [PubMed] [Google Scholar]
- Pisani F.M., De Felice M. & Rossi M. (1998) Amino acid residues involved in determining the processivity of the 3′–5′ exonuclease activity in a family B DNA polymerase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochemistry, 37, 15005–15012. [DOI] [PubMed] [Google Scholar]
- Silvian L.F., Toth E.A., Pham P., Goodman M.F. & Ellenberger T. (2001) Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol., 8, 984–989. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T., Egashira A. & Kura S. (2001) Analysis of MTH1 gene function in mice with targeted mutagenesis. Mutat. Res., 477, 71–78. [DOI] [PubMed] [Google Scholar]
- Wagner J., Fujii S., Gruz P., Nohmi T. & Fuchs R.P. (2000) The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep., 1, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. (2001) Translesion synthesis by the UmuC family of DNA polymerases. Mutat. Res., 486, 59–70. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Cox E.C. & Horn V. (1966) The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl Acad. Sci. USA, 55, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.L., Pata J.D. & Steitz T.A. (2001) Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell, 8, 427–437. [DOI] [PubMed] [Google Scholar]


