Abstract
The reported draft human genome sequence includes many contigs that are separated by gaps of unknown sequence. These gaps may be due to chromosomal regions that are not present in the Escherichia coli libraries used for DNA sequencing because they cannot be cloned efficiently, if at all, in bacteria. Using a yeast artificial chromosome (YAC)/ bacterial artificial chromosome (BAC) library generated in yeast, we found that approximately 6% of human DNA sequences tested transformed E. coli cells less efficiently than yeast cells, and were less stable in E. coli than in yeast. When the ends of several YAC/BAC isolates cloned in yeast were sequenced and compared with the reported draft sequence, major inconsistencies were found with the sequences of those YAC/BAC isolates that transformed E. coli cells inefficiently. Two human genomic fragments were re-isolated from human DNA by transformation-associated recombination (TAR) cloning. Re-sequencing of these regions showed that the errors in the draft are the results of both missassembly and loss of specific DNA sequences during cloning in E. coli. These results show that TAR cloning might be a valuable method that could be widely used during the final stages of the Human Genome Project.
Introduction
Draft versions of the human genome sequence have recently been published, and completion and verification of the draft genome sequence (DGS) is the next important task. At present, the DGS consists of stretches of continuous sequence (contigs) separated by gaps, the exact sequences of which remain to be identified. In principle, the gaps in the draft could be filled by isolating and sequencing additional bacterial artificial chromosome (BAC) clones containing human DNA, if 100% of human DNA could be cloned in BACs. However, some gaps might correspond to genomic regions that are not present in BAC libraries. It is well documented that long inverted-repeats, AT-rich sequences, and sequences with structures such as Z-DNA are extremely unstable in Escherichia coli (Hagan & Warren, 1982; Schroth & Ho, 1995; Kang & Cox, 1996; Ravin & Ravin, 1999; Razin et al., 2001). These, and other sequences, may be under-represented or lost when cloned in E. coli. There are no quantitative data to show how much human genomic DNA may be lost or rearranged in clones propagated in E. coli. Here, we suggest an approach that may help both with the assessment of the magnitude of the problem, and with its resolution. It has been reported that AT-rich genomic fragments and long inverted-repeats are more stable in yeast than in E. coli (Hayashi et al., 1993; Gardner et al., 2002; Glockner et al., 2002). Transformation-associated recombination (TAR) cloning is a recently developed yeast-based method for the selective cloning of a specific chromosomal region from a complex genome. In the past few years, the TAR cloning method has been used successfully in the isolation of 12 single-copy genes from human and mouse genomes (Kouprina & Larionov, 1999; Leem et al., 2002, and references therein). In this study, we use TAR cloning to screen random clones to verify the amount of human genomic DNA that may not be represented in sequences derived from bacterial clones, and to recover genes and chromosome segments that have not been isolated in BAC libraries.
Results
Stability of KAI1 and Muc2 genomic regions
Experiments were carried out with the human prostate cancer metastasis-suppressor gene KAI1 (Dong et al., 1995) and the mouse mucin gene Muc2 (Kashiwagi et al., 2001). A full-length sequence of the coding region of Muc2 and a non-coding 3′-end region of KAI1 were absent from existing BAC libraries. However, we were able to selectively clone both regions as circular yeast artificial chromosomes (YACs) using vectors with a 5′ genespecific targeting-sequence (hook) and a common repeat (Alu for human DNA, or B1 for mouse DNA) as a second targeting sequence by TAR cloning (see Methods).
The integrity of these YACs and their stability during propagation in yeast was analysed. Exons of KAI1 and Muc2 were amplified by PCR (see supplementary information online). DNA was isolated from subclones carrying the KAI1 and Muc2 YACs, and analysed by contour-clamped homogeneous electric field (CHEF) electrophoresis. All subclones carried a circular YAC of similar size (∼200 kb; Fig. 1A), indicating that these clones were stable in yeast.
Figure 1.

Stability of the KAI1 genomic region in yeast and Escherichia coli. (A) Propagation in yeast of a 200-kb circular YAC/BAC (yeast artificial chromosome/bacterial artificial chromosome) containing a KAI1 insert. Chromosome-sized DNA was isolated from 12 yeast transformants carrying a KAI1 YAC/BAC. DNA was γ-irradiated to linearize circular YACs, separated by contour-clamped homogeneous electric field (CHEF) electrophoresis and blot-hybridized with a vector probe. (B) Deleted/rearranged isolates of a KAI1 YAC/BAC propagated in Escherichia coli. Chromosome-sized DNA was isolated from yeast and electroporated into E. coli. BAC DNA was isolated, digested with NotI and analysed by CHEF electrophoresis. The white arrowheads indicate the clones with the smallest inserts.
To compare the stability of these genomic sequences in yeast and E. coli, the KAI1 and Muc2 YACs were retrofitted into YAC/BACs by homologous recombination in yeast, and then transformed into a recA− DH10B E. coli strain that is used for the construction of genomic BAC libraries. Retrofitted YAC/BACs usually transform E. coli with high efficiency: 1 ng of YAC/BAC DNA gives 100–500 transformants (Kouprina & Larionov, 1999). In contrast, the YAC/BACs with KAI1 and Muc2 inserts transformed E. coli with very low efficiency, at least 100 times lower than the usual efficiency for YAC/BAC clones. This low transformation efficiency was observed with 10 different DNA preparations of these YAC/BACs. Twenty randomly selected E. coli clones that had been successfully transformed with the Muc2 YAC/BAC were characterized, and the insert length in the different clones was found to vary from 20 to 70 kb. Thus, none of these clones contained the entire 200-kb insert present in the parental YAC. PCR analysis showed that Muc2 exon sequences were lost in all of the BACs recovered from transformed E. coli, indicating that the coding region of Muc2 is unstable in E. coli. We also analysed 30 KAI1 YAC/BAC clones isolated after transformation of E. coli. Only three of these clones contained an insert of ∼200 kb that corresponds to the size of the original YAC insert (Fig. 1B); the other YAC/BACs contained inserts varying from 30 to 180 kb. Most of the BACs that had deletions contained KAI1 exons and transformed E. coli with high efficiency. This indicates that 3′-flanking KAI1 sequences rather than coding sequences are toxic to E. coli. These results suggest that some KAI1 and Muc2 genomic regions are difficult to clone in E. coli and are likely to contain deletions when cloning is successful.
Relative stability of human DNA in yeast and in E. coli
As some human DNA sequences, including coding regions, are unstable in E. coli, the question arises as to what extent human sequence information is under-represented or lost in E. coli libraries of human genomic DNA. To determine this, two human DNA libraries were generated by TAR cloning. The TAR vector contained a YAC/BAC cassette, so that each clone could be grown in yeast as a YAC clone, or in E. coli as a BAC clone. We have previously used the pNKBAC39 vector that contains Alu repeats as targeting sequences (Kouprina et al., 1998). The wide occurrence of Alu sequences in the human genome allows us to sample numerous genomic regions using this vector. One library was made in this vector using DNA from human chromosome 5; a second library contained random clones from the whole human genome.
We first investigated the stability of human genomic DNA clones in yeast, DNA was isolated from 400 randomly picked clones (250 from the genome-wide library and 150 from the chromosome-5-specific library), linearized by endonuclease digestion, separated by CHEF electrophoresis, and Southern blot-hybridized with the vector. DNA from 204 of the YAC clones from the genome-wide library, and from 96 of the chromosome-5-specific library clones, was detected as single linear fragments after digestion using restriction endonucleases. We conclude that the inserts from these clones were stable in yeast. Out of 204 of the genome-wide library clones, 192 transformed E. coli with high efficiency (100–500 colonies ng−1 of YAC/BAC DNA). The other 12 YAC/BACs (∼6%) transformed E. coli with very low efficiency, and produced few or no transformants (Table 1). These 12 YAC/BACs were re-isolated from yeast and retransformed into yeast cells at high efficiency by spheroplast transformation. The DNA inserts had the same size and Alu-profiles as those of the original clones.
Table 1.
Summary of results of Escherichia coli transformation with YAC/BACs containing random human inserts
| YAC/BAC clones1 | Yield of E. coli transformants2 | Size of inserts in YAC/BACs (kb) | 3Frequency of deletions in BACs |
|---|---|---|---|
| Unstable (group A) n = 19 | 0–20 | 50–200 | 161/190 |
| Stable (group B) n = 281 | 100–500 | 30–180 | 5/1,686 |
1Number of transformants ng−1 of YAC/BAC DNA.
2Two hundred and four YAC/BACs obtained from total human DNA and 96 YAC/BACs obtained from chromosome 5 DNA were transferred from yeast to E. coli. Twelve unstable clones from total DNA and 7 from chromosome 5 DNA were identified.
3For each YAC/BAC clone, 6–10 independent E. coli transformants were checked. For group A, most of the recovered BACs (85%) contained deletions. BAC, bacterial artificial chromosome; YAC, yeast artificial chromosome.
DNA from each individual YAC/BAC was isolated and analysed from 6–10 independent E. coli transformants (Table 1). More than 99% of the YAC/BAC clones that transformed E. coli with high efficiency had stable inserts. In contrast, inserts from YAC/BAC DNAs that transformed E. coli with low efficiency were variable in size, and carried deletions (data not shown). Similar results were observed with YAC/BAC clones carrying inserts from human chromosome 5 (Table 1).
These results suggest that about 6% of clones in a YAC/BAC library containing large inserts of human genomic DNA can be stably maintained in yeast, but cannot be efficiently transferred to or stably maintained in E. coli. It is likely, therefore, that BAC libraries of human DNA do not contain the entire human genome.
Analysis of clones that are difficult to maintain in E. coli
To examine further the possibility that a significant amount of human DNA is unstable when cloned in E. coli, we studied in greater detail five YAC/BAC clones that transformed E. coli with low efficiency (group A) and 11 YAC/BAC clones that transformed E. coli with high efficiency (group B). Fluorescence in situ hybridization analysis showed that each YAC/BAC clone yielded a single signal on one of the human chromosomes. The ends of the insert present in each YAC/BAC clone were sequenced, and the sequences compared with the human DGSs, published by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genome/guide), University of California at Santa Cruz (UCSC; http://genome.ucsc.edu) and Celera Genomics (http://www.celera.com). The results of this analysis are summarized in Table 2 and in supplementary information online. For four clones of group A, there was no complete match between the sequences obtained and the DGS. The sequences of both ends of the insert were present in the DGS, but the distance between the sequenced regions was different in the DGS and the YAC/BAC clones, and in two cases the end sequences had opposite orientation in the YAC/BAC and in the draft. This unexpectedly high degree of discrepancy between the DGS and group A clones may be due to poor clonability, rearrangement or loss of some regions of human DNA in E. coli clones used for the sequencing project.
Table 2.
Analysis of YAC/BAC end sequences
| YAC/BAC | Size of insert (kb)1 | Chromosome | Accession number | Distance between ends in drafts2 |
|
|---|---|---|---|---|---|
| Public DGS | Celera DGS | ||||
| Group A | |||||
| N23 | ∼170 | 5 | BH634639; BH634640 | 128 Mb, WO | 129 Mb |
| N21 | ∼140 | 12 | BH634655; BH634656 | 78 Mb | 100 Mb |
| N28 | ∼90 | 5 | BH634641; BH634642 | 10 kb, WO3 | 10 kb |
| N92 | ∼140 | 5 | BH634643; BH634644 | 139 kb | 135 kb |
| N11 | ∼200 | 11 | BH634657; BH634658 | 150 kb | 105 kb, WO |
| Group B | |||||
| N14 | ∼140 | X | BH634659; BH634660 | 149 kb | 149 kb |
| N96 | ∼70 | X | BH634661; BH634662 | 70 kb | 70 kb |
| N122 | ∼110 | 3 | BH634665; BH634666 | 106 kb | 106 kb |
| N45 | ∼130 | 2 | BH634667; BH634668 | 135 kb | 135 kb |
| N107 | ∼130 | 3 | BH634669; BH634670 | 130 kb | 131 kb |
| N151 | ∼140 | 4 | BH634671; BH634672 | 84 Mb, WO | 84 Mb |
| N101 | ∼30 | 5 | BH634645; BH634646 | 31 kb | 32 kb |
| N15 | ∼140 | 5 | BH634647; BH634648 | 0 kb | 0 kb |
| N19 | ∼20 | 5 | BH634649; BH634650 | 22 kb | 22 kb |
| N24 | ∼130 | 5 | BH634651; BH634652 | 137 kb, WO | 137 kb, WO |
| N54 | ∼100 | 5 | BH634653; BH634654 | 97 kb (39 kb4) | 97 kb |
1The average length of YAC/BAC end sequences was 467 bp.
2Where the distances between ends the YAC/BAC isolates analysed in this study differ from those in the published DGSs, this is indicated by highlighting the distances in bold in the table; WO indicates that ends are in the wrong orientation in the DGS.
3Orientation of the end sequences was different at the time of completion of this work.
4The YAC N54 end distance in the draft was 39 kb (UCSC, December 2001). The complete sequence of clone N54 has been submitted to GenBank and has been used to correct the sequence in the draft (UCSC, June 2002). BAC, bacterial artificial chromosome; DGS, draft genome sequence; YAC, yeast artificial chromosome.
Clones from group B transformed E. coli with high efficiency. The sequence of seven clones from this group (out of eleven tested) matched the DGS (Table 2). For three group B clones the distance between the end sequences was shorter in the public DGS than in our YAC/BAC clones, suggesting that the DGS might contain deletions. The ends of one group B clone were linked to different contigs in the draft sequence as compared with the YAC/BAC clone. In another case, the orientation of one of the end sequences in the draft and in YAC/BAC clone was different. Overall, sequences from group B clones were different from the DGS far less often than those from the group A clones.
Isolation of problematic regions in the draft sequence
Do the discrepancies between the YAC/BAC end sequences and the DGS result from cloning artefacts in our experiments? To rule out this possibility and to identify the underlying causes of differences from the draft, we decided to re-isolate sequences present in group A and group B clones by TAR cloning, and compared the new clones with the original isolates. The inserts present in clones N28 (group A) and N54 (group B) were used for this re-isolation. These inserts were much larger than predicted by a BLAST search of the DGS using sequence information from the end of inserts (Table 2; ∼97 kb in N54 compared with 39 kb in the DGS, and ∼90 kb in N28 compared with 10 kb in the DGS available at the time of completion of this work). Moreover, the orientation of the end sequences in the DGS differed from that in clone N28.
We constructed two different TAR cloning vectors, pVC-54 and pVC-28, with targeting sequences specific for the ends of clones N28 and N54, respectively. We then isolated the corresponding regions of human DNA by TAR cloning in yeast from human leukocytes and from a hybrid cell line carrying human chromosome 5 (see Methods). The sizes of the YACs isolated after TAR cloning were similar to those of the previously isolated circular YAC/BACs (∼100 kb for clone N28; ∼110 kb for clone N54). The Alu profiles of these YACs were indistinguishable from the profiles of the previously isolated clones N28 and N54 (Fig. 2). These results suggest that the re-isolated YACs, as well as the original clones N28 and N54, contain non-rearranged human genomic DNA. Because they were isolated using TAR vectors containing targeting sequences derived from the YAC ends, the orientation of the DGS sequence corresponding to clone N28 may need revision. From a broader perspective, our results suggest that TAR cloning may be useful for verification and completion of certain parts of the DGS, the sequence of which was compiled on the basis of clones propagated in E. coli.
Figure 2.
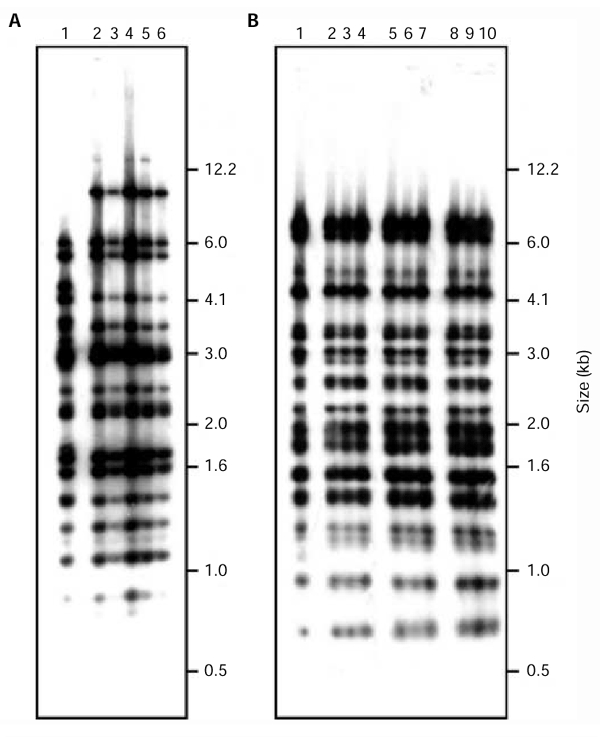
Alu profiles of the original YAC/BAC (yeast artificial chromosome/bacterial artificial chromosome) clones, N54 and N28, and clones re-isolated by transformation-associated recombination (TAR) cloning. TaqI digests of YAC DNAs were separated by gel electrophoresis, transferred to a nylon membrane and hybridized with an Alu probe. (A) Analysis of two YAC re-isolates with homology to the insert from YAC/BAC N54. Lane 1 is the original YAC/BAC N54. Lanes 2–6 are subclones of two independent re-isolates (three subclones for one of the isolates, and two subclones for the other isolate). The Alu profile of the re-isolates differs from the profile of the original clone, YAC/BAC N54, in three bands. This is due to differences in vector construction. (B) Three YAC re-isolates with homology to the insert from YAC/BAC N28. Lane 1 is the original YAC/BAC N28. Lanes 2–10 are subclones of three independent re-isolates (three subclones for each isolate).
Sequencing of genomic clones isolated by TAR cloning
To understand better the reasons for missing and misaligned sequences in the DGS, we sequenced YAC/BAC clone N54 and YAC/BAC clone N28 from the BAC form using the standard shotgun approach. Analysis of 8× read coverage for clone N54 (insert stable in E. coli) enabled assembly of a 97-kb sequence. The sequence was almost identical to that in the Celera DGS. Comparison of the N54 sequence with the public (NCBI and UCSC) DGS has shown that most of the reads contained in it were present in the draft, but were outside of the 39-kb contig deduced from the analysis of YAC/BAC ends. It is likely that the discrepancy with the public DGS is due to draft misassembly. After submission of the complete sequence of clone N54 to GenBank, the error was corrected (Table 2).
In contrast to the result with clone N54, shotgun sequencing did not allow us to assemble reads of YAC/BAC clone N28 (insert unstable in E. coli) into a contig with the predicted size. Assembly of 8× read coverage for clone N28 gave ∼10 kb of sequence different from that present in the drafts. Because of the failure of the shotgun approach to provide a contig with the predicted size, clone N28 was sequenced directly from BAC DNA (Polushin et al., 2001). This allowed us to assemble a 68-kb contig. Failure to assemble the complete N28 sequence of N28 by shotgun sequencing could be explained by the presence of numerous similar repeated sequences and AT-rich blocks in the insert that are difficult to sequence and assemble. Most of the sequences identified within the 68-kb contig are present in the draft of chromosome 5, but they are not contiguous, and are scattered over an ∼2.0-Mb region. This suggests that additional data will be required to refine the sequence of this genomic region.
Discussion
The Human Genome Project is now entering the final phase during which the sequence must be completed, corrected and finalized. During this phase of the project, gaps in the sequence must be closed, and the overall quality of the sequence improved. To do this, it will be necessary to collect extra sequence data. It is not clear if all of the sequences missing from the draft human genome sequence are represented in the bacterial libraries. The recent sequencing of two microbe genomes (Plasmodium falciparum and Dictyostelium discoideum) has shown that a high chromosomal AT content prevents the construction of large-insert BAC libraries (Gardner et al., 2002; Glockner et al., 2002). Only because large genomic fragments of these microbes are clonable in YACs did the authors succeed in constructing scaffolds of contiguous DNA sequence.
This study shows that a significant fraction of human DNA is poorly clonable in E. coli. However, these poorly clonable DNA sequences can be recovered in yeast using a selective TAR cloning method. Recently, sequences on chromosome 19 that lie within gaps in the DGS were cloned by TAR and found to be stable in yeast, but were toxic to bacterial cells (N.K., unpublished data). Sequencing of these regions requires non-standard approaches. Here, a clone containing an unstable region was retrofitted to a BAC, and analysed using a BAC direct-sequencing strategy (Polushin et al., 2001). Even if the insert in the retrofitted BAC has small rearrangements, the BAC may be used as starting material for DNA sequencing. As an alternative to transferring YAC clones to bacterial cells, where they undergo deletions and rearrangements, circular YAC DNA can be purified directly from yeast for sequencing (Strathern et al., 1979).
In this study, end-sequence analysis of YAC/BACs with random DNA inserts revealed differences from the published genome sequence in a significant fraction of clones, indicating potential errors in the corresponding contigs. Similar discrepancies have also been described in other reports (Aach et al., 2001; Katsanis et al., 2001; Christian et al., 2002; Semple et al., 2002). TAR cloning could be a powerful tool for the verification of contig assembly, as we showed for two contigs on chromosome 5.
In summary, this work, and other reports, indicate that new approaches may be needed to complete the final phase of the Human Genome Project. Alternative cloning systems and hosts may be essential for the success of this project. One of such approaches, TAR cloning in yeast, will allow for rapid and selective isolation of targeted regions of the human genome that cannot be verified or completed using clones generated and propagated in E. coli. This idea warrants further study and testing as the Human Genome Project enters its final stages. To assess the extent of the problem, we are now using TAR cloning to complete the sequencing of human chromosome 19.
Methods
Construction of TAR vectors and cloning by in vivo recombination in yeast.
The TAR vector pVC-KAI1, containing a 5′ region of the human KAI1 gene, was constructed from the vector pVC1 (Kouprina et al., 1998). The vector contains a 395-bp 5′ KAI1 sequence (nucleotide positions 381,329–381,719 in contig NT_009109.4 from NCBI), and an Alu repeat as a second hook. The pVC604-Muc2 TAR vector contains a 130-bp B1 repeat, and a 770-bp unique 5′ targeting sequence derived from the promoter region of the mouse Muc2 gene (nucleotide positions 1,483–2,265; GenBank accession number AF221746). To identify clones containing the KAI1 and Muc2 genes, yeast transformants were combined in pools and examined by PCR using a pair of primers specific for exon 1 of KAI1 or Muc2 (see supplementary information online). The TAR vectors pVC-54 and pVC-28 were constructed using the vector pVC604 (Kouprina & Larionov, 1999). pVC-54 contains 131-bp and 111-bp fragments corresponding to the 5′ and 3′ ends, respectively, of the YAC/BAC clone N54. pVC-28 contains 140-bp and 141-bp fragments corresponding to the 5′ and 3′ ends, repectively, of the YAC/BAC clone N28. The 5′ and 3′ targeting sequences in the pVC-54 vector correspond to nucleotide positions 1,391,074–1,401,352 in contig NT_006489.4 from NCBI. The 5′ and 3′ targeting sequences in the pVC-28 vector correspond to positions 294,790–333,360 in contig NT_006964.4. Primers used for vector construction, and diagnostic primers, are described in the supplementary information online. High molecular weight genomic DNA was prepared for TAR cloning from normal human leukocytes and the human/hamster monochromosomal somatic cell hybrid Q826, which contains human chromosome 5 (Kouprina et al., 1998). TAR cloning experiments, retrofitting of YACs into YAC/BACs and electroporation into DH10B-competent bacteria were performed as previously described (Kouprina & Larionov, 1999).
Physical analysis of YAC clones.
The size and Alu profiles of YACs was determined as previously described (Kouprina & Larionov, 1999). YAC ends were rescued and sequenced using standard protocols. Southern blot hybridization was performed as described by Kouprina et al. (1998) with 32P-labelled probes. Fluorescence in situ hybridization was performed as described elsewhere (Kouprina et al. 2003).
Sequencing.
Both ends of inserts in YAC/BACs were sequenced using vector-specific primers (described in the supplementary information online). For shotgun sequencing, BAC DNAs from clones N28 and N54 were purified, sonicated to produce 2-kb or 10-kb fragments, and cloned into the M13 vector. Clone N28, with an unstable insert, was also directly sequenced from BAC DNA (Polushin et al., 2001) by Fidelity Systems. The sequences of the inserts from clones N54 and N28 were deposited in the DDBJ/EMBL/GenBank database under accession numbers AF508802, AF510423 and AF510424, respectively.
Supplementary data are available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor766-s1.mov).
Supplementary Material
supplementary information
References
- Aach J., Bulyk M.L., Church G.M., Comander J., Derti A. & Shendure J. (2001) Computational comparison of two draft sequences of the human genome. Nature, 409, 856–859. [DOI] [PubMed] [Google Scholar]
- Christian S.L., McDonough J., Liu C., Shaikh Y.S., Vlamakis V., Badner J.A., Chakravarti A. & Gershon E.S. (2002) An evaluation of the assembly of an approximately 15-Mb region on human chromosome 13q32–q33 linked to bipolar disorder and schizophrenia. Genomics, 79, 635–656. [DOI] [PubMed] [Google Scholar]
- Dong J.T., Lamb P.W., Rinkerschaeffer C.W., Vukanovic J., Ichikawa T., Isaacs J.T. & Barrett J.C. (1995) KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science, 268, 884–886. [DOI] [PubMed] [Google Scholar]
- Gardner M.J. et al. (2002) Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature, 419, 531–534. [DOI] [PubMed] [Google Scholar]
- Glockner G. et al. (2002) Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature, 418, 79–85. [DOI] [PubMed] [Google Scholar]
- Hagan C.E. & Warren G.J. (1982) Lethality of palindromic DNA and its use in selection of recombinant plasmids. Gene, 19, 147–151. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Heard E. & Fried M. (1993) A large inverted duplicated DNA region associated with an amplified oncogene is stably maintained in a YAC. Hum. Mol. Genet., 2, 133–138. [DOI] [PubMed] [Google Scholar]
- Kang H.K. & Cox D.W. (1996) Tandem repeats 3′ of the IGHA genes in the human immunoglobulin heavy chain gene cluster. Genomics, 35, 189–195. [DOI] [PubMed] [Google Scholar]
- Kashiwagi H. et al. (2001) MUC1 and MUC2 expression in human gallbladder carcinoma: a clinicopathological study and relationship with prognosis. Oncol. Rep., 8, 485–489. [DOI] [PubMed] [Google Scholar]
- Katsanis N., Worley K.C. & Lupski J.R. (2001) An evaluation of the draft human genome sequence. Nature Genet., 29, 88–91. [DOI] [PubMed] [Google Scholar]
- Kouprina N. & Larionov V. (1999) in Current Protocols in Human Genetics (eds Dracopli, N. C. et al.) Vol. 1, 5.17.1–5.17.21. Wiley, New York, USA. [DOI] [PubMed] [Google Scholar]
- Kouprina N. et al. (1998) Construction of human chromosome 16- and 5specific circular YAC/BAC libraries by in vivo recombination in yeast (TAR cloning). Genomics, 53, 21–28. [DOI] [PubMed] [Google Scholar]
- Kouprina N. et al. (2003) Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res., 31, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem S.H. et al. (2002) The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene, 21, 769–777. [DOI] [PubMed] [Google Scholar]
- Polushin N. et al. (2001) 2′-modified oligonucleotides from methoxyoxalamido and succinimido precursors: synthesis, properties, and applications. Nucleosides Nucleotides Nucleic Acids, 4–7 507–511. [DOI] [PubMed] [Google Scholar]
- Ravin N.V. & Ravin V.K. (1999) Use of a linear multicopy vector based on the mini-replicon of temperate coliphage N15 for cloning DNA with abnormal secondary structures. Nucleic Acids Res., 27, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin S.V., Ioudinkova E.S., Trifonov E. & Scherrer K. (2001) Non-clonability correlates with genome instability: a case study of a unique DNA region. J. Mol. Biol., 307, 481–486. [DOI] [PubMed] [Google Scholar]
- Schroth G.P. & Ho P.S. (1995) Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res., 23, 1977–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple C.A., Morris S.W., Porteous D.J. & Evans K. (2002) Computational comparison of human genomic sequence assemblies for a region of chromosome 4. Genome Res., 12, 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J.N., Newlon C.S., Herskowitz I. & Hicks J.B. (1979) Isolation of a circular derivative of yeast chromosome III: implications for the mechanism of mating type interconversion. Cell, 2, 309–319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information


