Abstract
The simian virus 40 large tumour-antigen (T antigen) DNA helicase is a hexameric structure; it has been proposed that, in viral DNA replication, two of these hexamers are combined to form a bipartite holoenzyme that acts concurrently at both forks of a replication bubble. In a search for structural components of this helicase complex, we have identified nucleolin as a specific binding protein for the T-antigen hexamer. We show that nucleolin, in co-operation with human topoisomerase I, mediates the cohesion of the T-antigen helicase holoenzyme during plasmid unwinding. Our results provide biochemical evidence for a direct role of nucleolin in DNA replication, in addition to its known function in ribosome biogenesis. The data presented here suggest that nucleolin enables the formation of a functional 'helicase-swivelase' complex at the replication fork.
Introduction
Simian virus 40 (SV40) DNA replication is initiated by a T-antigen double-hexamer complex. The two hexameric subunits of this complex bind in a head-to-head configuration to viral origins of DNA replication (ori). Because of this binding, the double-stranded structure of the ori DNA becomes distorted locally (for a review, see Bullock, 1997). Each hexameric subunit then becomes active as a processive DNA helicase at one of the two replcation forks, and daughterstrand synthesis is carried out by cellular replication factors. There are more than ten of these replication factors, most of which co-operate with each other and/or with the T antigen, resulting in the formation of a large complex, known as the replisome (for a review, see Herendeen & Kelly, 1996). Electron microscopic and biochemical studies of the DNA unwinding process have indicated that the T-antigen double-hexamer complex remains intact after replication initiation; it is then thought to function as a bipartite holoenzyme, catalysing bidirectional DNA unwinding simultaneously at both forks of a replication bubble (Wessel et al., 1992; Smelkova & Borowiec, 1997). A helicase that has two reactive centres, working in opposite directions, cannot move along DNA. Instead, the DNA must be translocated through the complex, which is consistent with the observation that replisomes are bound to the nuclear scaffold at replication centres (Cook, 1999; Falaschi, 2000). However, the studies described above have shown a requirement for additional cellular factors that stabilize the bipartite T-antigen helicase. These factors may also anchor the helicase to nuclear structures, in agreement with the finding that a subfraction of cellular T antigen is matrix-bound (Schirmbeck & Deppert, 1991).
On the basis of these functional requirements, we set out to find scaffold-associated proteins that interact specifically with T-antigen hexamers and, in this study, nucleolin was identified as the only candidate. However, nucleolin was able to stabilize the T-antigen double-hexamer complex in ori-dependent DNA unwinding only in co-operation with topoisomerase I (Topo I), which in turn binds to the T antigen and to nucleolin. The ternary complex formed by interactions between all three of these proteins seems to have a 'helicase-swivelase' activity, which is thought to be essential for DNA replication.
Results and Discussion
Nucleolin is a T-antigen hexamer-specific binding factor
We studied the binding of the hexameric T antigen (the only T-antigen form active in DNA replication) to nuclear-scaffold-associated cellular proteins by far-western overlay blot analysis. Protein preparations or cellular fractions from HeLa cells were separated by SDS–polyacrylamide gel electrophoresis and probed with purified T antigen (Fig. 1A). The binding buffer used contained Mg-ATP, which gives an efficient formation of T-antigen hexamers (Uhlmannschiffler et al., 2002). Two proteins, with relative molecular masses of approximately 35 kDa and 100 kDa, were identified as binding partners (Fig. 1A, lanes 1 and 5). The 35-kDa protein is probably a degradation product of the larger 100-kDa protein (p100). The p100 protein was purified to near homogeneity (Fig. 1A, lanes 2–4). p100 was bound by T antigen only in the presence of Mg-ATP (Fig. 1A, lanes 5–8), and not in the presence of EDTA in the binding buffer (data not shown). Therefore, the interaction between p100 and the T antigen seems to be specific for the hexameric form of the T antigen. Analysis of the amino-acid sequence of the amino-terminal region of p100 (the first 22 amino acids) suggested that p100 is human nucleolin (Srivastava et al., 1989). This was confirmed by western blot analysis of p100 with antibodies specific for the carboxyl terminus of nucleolin (amino acids 695–707; Fig. 1A, lane 9). The hexamerspecific association of the T antigen with nucleolin was confirmed by using native nucleolin purified from HeLa cells as a sample in a far-western analysis in which purified T-antigen monomers and hexamers (isolated by sucrose gradient centrifugation; Uhlmannschiffler et al., 2002) were used as probes (Fig. 1A, lanes 10 and 11).
Figure 1.
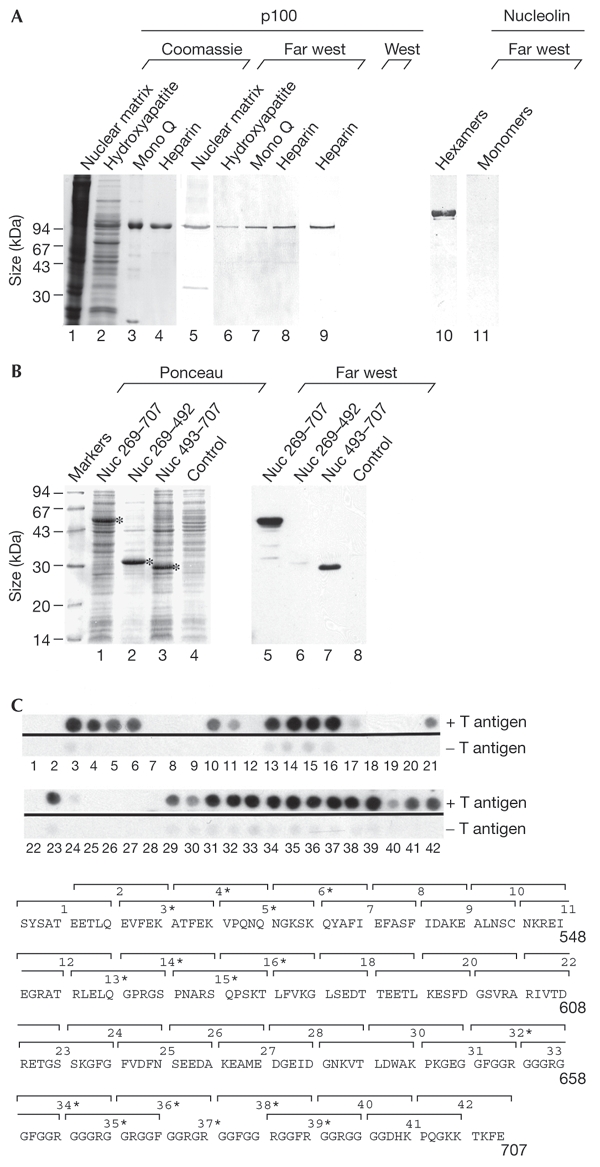
Interaction of nucleolin with T-antigen hexamers. (A) Extracts of nuclear matrix preparations from HeLa cells, and fractions thereof, which were obtained after each step of a conventional chromatographic purification program (lanes 2–4 and 6–9; the affinity resin used is indicated at the top of each lane), were separated by SDS–polyacrylamide gel electrophoresis. These samples were then analysed by Coomassie staining (lanes 1–4), far-western blotting, using T antigen as a probe in the presence of Mg-ATP (lanes 5–8), and by western blotting using antibodies raised against a peptide contaning the carboxy-terminal sequence of nucleolin (residues 695–707; lane 9). Far-western analysis of purified nucleolin (0.5 μg) probed with purified T-antigen hexamers (lane 10) and monomers (lane11) in the absence of Mg-ATP is also shown. (B) Crude extracts of Escherichia coli cells expressing human nucleolin deletion mutants and of control cells (expressing no recombinant protein) were analysed by far-western blotting using the T antigen as a probe in the presence of Mg-ATP (lanes 5–8), or by Ponceau staining of the blot (lanes 1–4). (C) Far-western analysis of overlapping oligopeptides (all were ten amino-acids in length) from the carboxy-terminal region of nucleolin using the T antigen as a probe in the presence of Mg-ATP (upper part). The oligopeptides are numbered, in ascending order, starting from that representing the sequence closest to the amino terminus of nucleolin. The sequence of the oligopeptides is indicated by the brackets drawn above the single-letter amino-acid sequence of nucleolin; T-antigen-interacting peptides are indicated by asterisks (lower part). Far west, far-western analysis; Nuc, nucleolin; West, western blot analysis.
An oligopeptide consisting of the T-antigen nuclear-localization sequence (NLS) has been reported to bind to the N terminus of nucleolin (Xue et al., 1993). However, we found that this oligopeptide was unable to inhibit the binding of T-antigen hexamers to nucleolin when added as a possible competitor for the binding of these proteins in far-western analyses (data not shown). Furthermore, histone H1 also failed to inhibit the T-antigen–nucleolin association (data not shown), although this histone is highly homologous to the N terminus of nucleolin (Hanakahi et al., 1997) and, interestingly, also interacts specifically with T-antigen hexamers (Ramsperger & Stahl, 1995). Thus, the N-terminal region of nucleolin does not seem to be involved in the interaction with T-antigen hexamers. In addition, nucleolin deletion mutants consisting of residues 269–707 and 493–707, which were expressed in Escherichia coli, were found to associate with the T antigen (Fig. 1B, lanes 5 and 7), whereas deletion mutant 269–492 was not (Fig. 1B, lane 6). Full-length nucleolin cannot be made in recombinant systems. A set of overlapping oligopeptides (ten amino acids each) consisting of regions of nucleolin were made in order to identify possible T-antigen-binding sites. Three such potential regions were identified in the nucleolin protein: in RNA binding sites three (amino acids 504–523) and four (residues 554–573) and in the glycine/alanine/arginine-rich domain (amino acids 644–693) (Ginisty et al., 1999) (Fig. 1C). According to their functions, these sites are likely to be exposed, at least in part, on the surface of native nucleolin, but further experiments must confirm these T-antigen contact sites in the context of larger nucleolin domains. Any future analysis of nucleolin binding sites on the T-antigen hexamer will depend on detailed knowledge of its structure.
The binding of the T antigen to nucleolin was also shown in vivo by co-immunopurification of the proteins from COS cell extracts (Fig. 2A and B). In these experiments, TC7 cells, which are the progenitors of COS cells and do not contain T antigen, were used in parallel as a negative control. The specificity of the binding of nucleolin to the hexameric form only of the T antigen was shown by the sensitivity of the interaction to EDTA in COS cell extracts, whereas binding of T antigen to p53, which is hexamer-independent, was insensitive (Fig. 2C). Interestingly, Topo I, which is reported to interact concurrently with both the T antigen and nucleolin (Simmons et al., 1996; Haluska et al., 1998), was not found to copurify with either (Fig. 2D), although it was present in the COS cell extract (data not shown). The ionic strength of the extraction buffer (300 mM NaCl, pH 9.0) probably prevented Topo I binding to T antigen and/or nucleolin. Indeed, a weaker binding of Topo I to both proteins was seen in co-immunoprecipitation experiments under different conditions. The T-antigen–Topo I interaction was also shown not to be hexamerspecific (data not shown); this is in contrast with the reported specific association of Topo I with complexes formed between the T-antigen double-hexamer and replication origins (Gai et al., 2002), but might be explained by the fact that our binding studies were done in the absence of DNA. Indeed, it has been reported that profound conformational changes in the N-terminal domain of the T antigen (the Topo I-binding site) take place on DNA binding (VanLoock et al., 2002).
Figure 2.
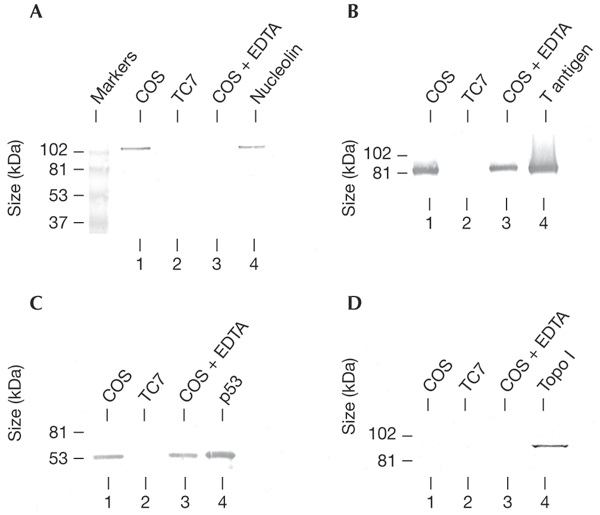
Analysis of the T-antigen–nucleolin interaction in vivo. T antigen was immunopurified from COS and TC7 cell extracts prepared in the presence of magnesium ions, and COS cell extracts were also prepared in the presence of EDTA. Aliquots of the immunopurified fractions were analysed by western blotting for the presence of nucleolin (A), T antigen (B), p53 (C), and topoisomerase I (Topo I) (D). As a positive control, lane 4 of each gel was loaded with the appropriate purified protein sample.
Role of nucleolin in bidirectional DNA unwinding
Both the T antigen (Scheffner et al., 1989; Uhlmannschiffler et al., 2002) and nucleolin (Ginisty et al., 1999) can bind RNA and alter RNA secondary structures. However, we could not detect alterations in these activities as a result of the T-antigen–nucleolin interaction in in vitro experiments using purified forms of these proteins (H.S., unpublished observations). Nucleolin has been suggested to function in DNA replication in several studies, although a specific role for nucleolin in this process has not been assigned (Daniely & Borowiec, 2000; Wang et al., 2001; Xu et al., 2001). A possible effect of nucleolin on the DNA replication function of T antigen hexamers has been tested in a DNA helicase assay using a plasmid containing SV40 ori DNA as a substrate, monitored by electron microscopy (EM) (Wessel et al., 1992; Ramsperger & Stahl, 1995; Uhlmannschiffler et al., 2002). The plasmid was linearized at a site opposite the ori sequence. Unwound single-stranded DNA (ssDNA) was stabilized by the binding of the E. coli ssDNA binding protein (SSB), which was used instead of replication protein A (RPA), the human homologue of SSB, because the 14-kDa subunit of RPA is known to interact with nucleolin, thereby inhibiting DNA unwinding. However, this only happens in vivo in conditions of cellular stress, when most nucleolin is released from the nucleolus into the nucleus (Daniely & Borowiec, 2000). Under normal conditions, the small subfraction of nucleolin that is located outside the nucleolus, and which is probably bound to the nuclear matrix, may not interact with RPA, but instead with the T-antigen helicase or with both T antigen and RPA in a non-inhibitory manner. In the presence of SSB we did not observe an inhibitory influence of nucleolin on the DNA unwinding activity of T-antigen hexamers. In a typical experiment (see Methods), up to 40% of the DNA molecules were involved in unwinding reactions (and showed unwinding bubbles of different sizes) after short incubation times under orispecific conditions, irrespective of the absence (data not shown) or presence of nucleolin (used in equimolar amounts to T antigen; Fig. 3A, lower panels). Most of the completely doublestranded DNA (dsDNA) molecules (not yet unwound) contained T-antigen double-hexamers bound to the ori, and T-antigen–nucleolin interactions were clearly visible, as shown by an increase in the size of most ori-bound protein complexes on electron micrographs (compare upper panel of Fig. 3A with the inset in the lower panel). In addition, nucleolin did not affect the unwinding efficiency of the T antigen in helicase reactions carried out using a small ori-containing dsDNA fragment as a T-antigen substrate, as monitored by gelshift electrophoresis (data not shown; for experimental details, see Uhlmannschiffler et al., 2002). We speculated as to whether nucleolin binding to the T antigen stabilizes the double-hexamer complex, formed during initiation, in a subsequent unwinding step. Resultant structures of unwinding intermediates, in which T-antigen double-hexamers act simultaneously at both forks of replication bubbles, contain two loops of ssDNA that appear to emerge from, and return to, the helicase (described as 'rabbit-ears-containing structures') (Wessel et al., 1992). These structures were seen occasionally in this study in unwinding reactions performed in the absence of nucleolin (less than 3% of unwinding intermediates; data not shown). However, these structures were not found to be induced by nucleolin.
Figure 3.
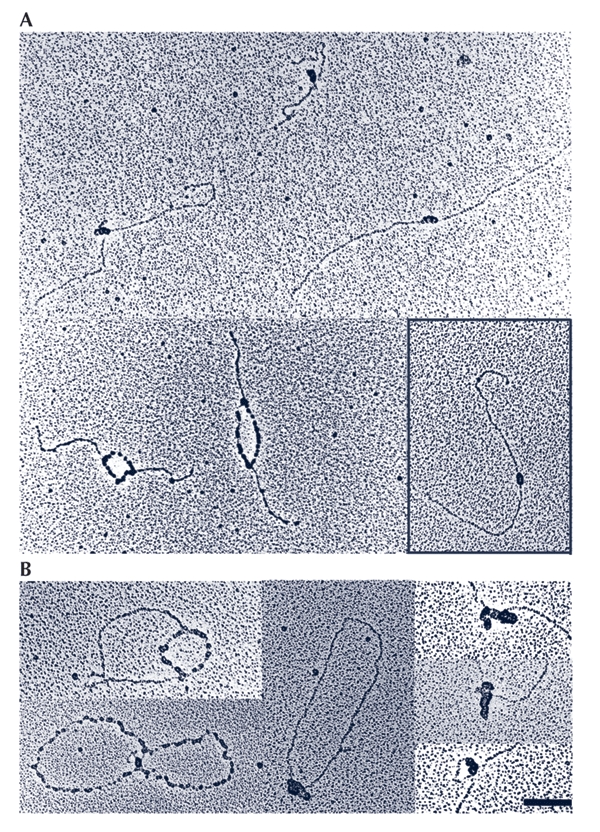
Analysis of nucleoprotein complexes from T-antigen helicase reactions by electron microscopy. (A) Nucleoprotein complexes from a reaction carried out in the presence of nucleolin with an origin of replication (ori)-containing linearized plasmid as a substrate. Top panel: three characteristic double-stranded (ds) DNA molecules (not unwound) with their ori-bound protein complexes are shown. Bottom panel: two partially unwound DNA molecules with their single-stranded (ss) DNA regions coated with Escherichia coli ssDNA binding protein are shown. The inset shows a typical dsDNA molecule with T antigen bound to the origin from a control reaction carried out in the absence of nucleolin. (B) Nucleoprotein complexes from reactions performed in the presence of topoisomerase I (Topo I) with ori-containing circular covalently-closed DNA. The left panels show two partially unwound DNA molecules, and the middle panel a dsDNA molecule, which has not yet been unwound, with an ori-bound T-antigen–Topo I complex. In the right panel, three characteristic sections of dsDNA molecules, which have not been unwound, with their ori-bound protein complexes are shown. The ori position was determined by measuring the lengths of the DNA molecules. Scale bar represents 100 nm.
Topo I has been reported to stimulate T-antigen-mediated DNA replication in vitro (Gai et al., 2000), but it is not known how this protein is recruited to the replication complex, despite its known physical interaction with the T antigen (Simmons et al., 1996; Haluska et al., 1998). Furthermore, unwinding studies using SV40 minichromosomes as a substrate have revealed that additional cellular factors are essential for the co-operation of T antigen and Topo I in performing a helicaseswivelase function (Ramsperger & Stahl, 1995). By binding simultaneously to Topo I and to T-antigen hexamers, nucleolin could act as a mediator in the association of these two proteins. The mutual association of all three proteins could therefore hold together the T-antigen double-hexamer, leading to a bipartite helicaseswivelase complex. In the absence of nucleolin, rabbit-ears-containing structures, used as an indication for the formation of such a complex, were not seen when T antigen and Topo I were used in plasmid-unwinding assays with either the linearized (data not shown; see also Ramsperger & Stahl, 1995) or the circular covalently closed (ccc) form of the ori-containing plasmid (Fig. 3B). However, ori-bound protein complexes on dsDNA obtained under these conditions were much larger in size than those obtained with T antigen alone or with T antigen and nucleolin. This suggests that there is an interaction of Topo I with ori-bound T antigen, as previously reported (Gai et al., 2000), whereby more molecules of Topo I than of nucleolin were found to be bound to each double hexamer.
Surprisingly, when nucleolin and Topo I were both added to the unwinding reactions, rabbit-ears-containing structures were formed efficiently, suggesting that the interaction of nucleolin, Topo I and T antigen leads to the formation of bipartite helicase complexes that are involved in the unwinding of ccc DNA. The formation of these bipartite T-antigen–helicase complexes was most efficient when there was approximately one molecule each of Topo I and nucleolin per T-antigen hexamer; in these conditions, more than 50% of all unwinding intermediates showed the rabbit-ears-containing structures (a typical field view from these unwinding experiments is shown in Fig. 4).
Figure 4.

Simultaneous action of protein complexes at both forks of replication bubbles in bidirectional circular covalently closed (ccc) DNA unwinding. A characteristic field view of an unwinding experiment performed with T antigen, nucleolin, topoisomerase I and cccDNA-containing origins of replication is shown. More than 50% of the unwinding intermediates showed what are know n as 'rabbit-ears-containing structures' (Wessel et al., 1992). The inset shows a two-fold enlargement of one of these structures. Scale bar represents 100 nm.
As neither nucleolin nor Topo I on their own can stabilize two T-antigen hexamers during DNA unwinding, the most obvious interpretation of our experiments, which were carried out using highly purified proteins, is that both nucleolin and Topo I are present in the helicase complexes, although this needs to be demonstrated directly in future experiments. Without knowing the exact stoicheiometric composition, we propose that each hexameric subunit of a T-antigen dodecamer complex interacts with the C terminus of a nucleolin molecule, the N terminus of which is bound to Topo I, which itself is associated with the other T-antigen hexamer (Fig. 5). Thus, nucleolin may act as a clamp to assist the proper orientation of T-antigen hexamers and Topo I. As Topo I interacts with the T antigen and nucleolin through its N terminus (Haluska et al., 1998), its catalytically active C-terminal domain should have unrestricted access to the DNA in front of the fork. In this model, the movement of torsional strain across replicated sections, leading to a catenation of daughter strands, would be prevented. Topo I would also be able to continually relieve the superhelical tension generated during the unwinding of chromatin, in which DNA is otherwise partially shielded against any relaxation activity (Ramsperger & Stahl, 1995). In SV40 DNA replication, this complex is most likely to form at the ori, and the ability of nucleolin to recognize DNA regions with high base-unpairing potential (Dickinson & Kohwishigemtsu, 1995) could contribute to the initiation process.
Figure 5.
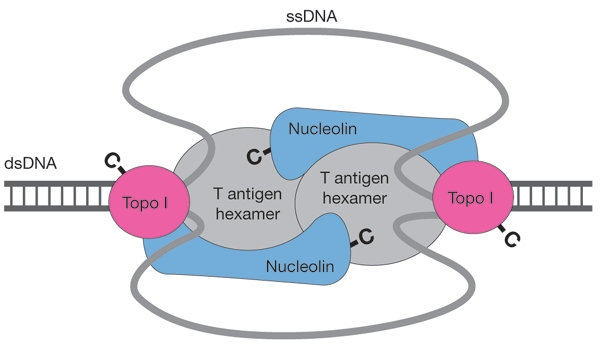
Model of the structure–function relationships in a T-antigen double-hexamer–nucleolin–topoisomerase I (Topo I) complex in bidirectional DNA unwinding. The orientation of the proteins within the complex is indicated by the topology of their carboxyl termini (-C). ssDNA, single-stranded DNA; dsDNA, double-stranded DNA.
A direct role of nucleolin in DNA replication has been suggested before, as specific inhibition of this process by G-rich oligonucleotides correlates positively with their ability to bind nucleolin (Xu et al., 2001). A detailed analysis of the function of nucleolin in the SV40 DNA replication system has led to the suggestion that nucleolin may modulate the helicase activity of the SV40 replication complex (Xu et al., 2001), and also possibly that of the minichromosome-maintenance complex, which is thought to resemble the cellular DNA-replication helicase (Labib & Diffly, 2001).
Nucleolin is a structural component of the nucleoskeleton (Dickinson & Kohwishigemtsu, 1995; Gotzmann et al., 1997), and as part of the DNA unwinding complex it could function in binding replisomes to the nuclear scaffold. Furthermore, as it is cell-cycle regulated (Kel et al., 2001), nucleolin, like Yph1 (Du & Stillman, 2002), might link ribosome biogenesis and DNA replication, which both require high levels of energy, to a common regulatory mechanism.
Methods
Protein purification.
Topo I (Strausfeld & Richter, 1989) and nucleolin (Belenguer et al., 1990) were purified from HeLa cells, and T antigen from overexpressing SF9 cells as described in Ramsperger & Stahl (1995). T-antigen monomers and hexamers were prepared as described in Uhlmannschiffler et al. (2002). T antigen was immunopurified from COS cells as described in Ramsperger & Stahl (1995), except that cell extracts were prepared in 300 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 0.5% Igepal, 1 mM dithiothreitol, 100 mM Tris-HCl, pH 9.0. Where indicated, the buffer contained 5 mM EDTA instead of MgCl2.
Nucleolin deletion constructs.
Nucleolin deletion constructs (nuc269–707, nuc269–492 and nuc493–707) were expressed in E. coli from pQE30-based plasmids, made by insertion of nucleolin cDNA sequences generated by PCR from the pNFor4 plasmid (Hanakahi et al., 1997).
Nucleolin oligopeptides.
Overlapping oligopeptides (of ten amino acids) of the C-terminal region of nucleolin were synthesized by the SPOTs method using Auto-spot Robot ASP-222 equipment (ABIMED). Peptides (including those representing the T-antigen NLS, CYPDEVKRKKKP, and the reverse sequence, PKKKRKVEDPYC) were synthesized using an Applied Biosystems 433A Peptide Synthesizer. Polyclonal antibodies 121 and 421 were purified as described in Ramsperger & Stahl (1995).
Antibodies.
Anti-nucleolin antibodies were raised in rabbits against a peptide (GDHKPQGKKTKFE) corresponding to the C-terminal sequence of nucleolin, and were coupled to haemocyanine. Anti-Topo I antibodies were purchased from TopoGen, Inc.
Purification of p100.
Proteins associated with the nuclear scaffold of HeLa cells were stabilized by sodium tetrathionate (0.5 mM) treatment during matrix preparation, and were extracted from 5 × 109 cells after DTT (50 mM) treatment, which was carried out essentially as described in Belgrader et al. (1991). The S100 supernatant was adsorbed to hydroxyapatite. Proteins were eluted with potassium phosphate (pH 8.0, 300 mM) and precipitated with ammonium sulphate (50% saturation). The protein precipitates were dissolved in A buffer (25 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, 10 mM β-mercaptoethanol, 7.5% glycerol) and fractionated by chromatography using a 3-ml MonoQ column with a Pharmacia fast performance liquid chromoatography system. After elution with an NaCl gradient (0.05–1.00 M) in A buffer, p100 was identified in the fractions containing 300–400 mM NaCl. Pooled fractions were diluted in A buffer (1:1 v/v), separated by chromatography on a 1-ml heparin column (HiTrap; Pharmacia) and washed twice with A buffer, containing 175 mM (first wash), and 400 mM NaCl (second wash). Proteins were eluted with 500 mM NaCl and were then concentrated by a second ammonium sulphate precipitation and dissolved in 0.5 ml of A buffer plus 150 mM NaCl.
Other methods.
The interaction of p100/nucleolin with the T antigen was analysed in far-western assays with T antigen (30–40 nM) in the presence of 2 mM ATP and 2.5 mM MgCl2 (or alternatively 2 mM EDTA instead of Mg-ATP) as described in Ramsperger & Stahl (1995). When isolated T-antigen hexamers and monomers were used as a probe, neither Mg-ATP nor EDTA were used.
Ori-dependent helicase reactions (30 μl) were carried out as described previously (Wessel et al., 1992; Ramsperger & Stahl, 1995; Uhlmannschiffler et al., 2002) and contained 12 pmol of T antigen and 55 fmol of the ori-containing plasmid pSV-08 (Uhlmannschiffler et al., 2002) either in its ccc form or linearized by BsaI digestion. Where indicated, 2.5 pmol of nucleolin and/or Topo I were added. After incubation at 37 °C for 30 min, nucleoprotein complexes were fixed using 0.1% glutaraldehyde at 37 °C for 20 min, spread using benzylalkyldimethylammonium chloride (BAC) and rotary-shadowed with tungsten as described in Wessel et al. (1992), Ramsperger & Stahl (1995) and Uhlmannschiffler et al. (2002). Micrographs were taken with a Zeiss 10 transmission electron microscope.
Acknowledgments
We thank S. Jungbluth for excellent technical assistance, N. Schuster for the kind gift of anti-nucleolin antibodies, and W.Nastainczyk and P. Scholtes, Universität des Saarlandes, Homburg, for peptide synthesis. This work was supported during its initial phase by the Deutsche Forschungsgemeinschaft.
References
- Belenguer P., Caizergues-Ferrer M., Labbe J.C., Doree M. & Amalric F. (1990) Mitosisspecific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol. Cell. Biol., 10, 3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgrader P., Siegel A.J. & Berezney R. (1991) A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. J. Cell Sci., 98, 281–291. [DOI] [PubMed] [Google Scholar]
- Bullock P.A. (1997) The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol., 32, 503–568. [DOI] [PubMed] [Google Scholar]
- Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. [DOI] [PubMed] [Google Scholar]
- Daniely Y. & Borowiec J.A. (2000) Formation of a complex between nucleolin and replication protein A after cell stress prevents initiation of DNA replication. J. Cell Biol., 149, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson L.A. & Kohwishigematsu T. (1995) Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol. Cell. Biol., 15, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y.C. & Stillman B. (2002) Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell, 109, 835–848. [DOI] [PubMed] [Google Scholar]
- Falaschi A. (2000) Eukaryotic DNA replication: a model for a fixed double replisome. Trends Genet., 16, 88–92. [DOI] [PubMed] [Google Scholar]
- Gai D., Roy R., Wu C. & Simmons D.T. (2000) Topoisomerase I associates specifically with simian virus 40 large-T-antigen double hexamer-origin complexes. J. Virol., 74, 5224–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B. & Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci., 112, 761–772. [DOI] [PubMed] [Google Scholar]
- Gotzmann J., Eger A., Meissner M., Grimm R., Gerner C., Sauermann G. & Foisner R. (1997) Two-dimensional electrophoresis reveals a nuclear matrix-associated nucleolin complex of basic isoelectric point. Electrophoresis, 18, 2645–2653. [DOI] [PubMed] [Google Scholar]
- Haluska P.J., Saleem A., Edwards T.K. & Rubin E.H. (1998) Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res., 26, 1841–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi L.A., Dempsey L.A., Li M.J. & Maizels N. (1997) Nucleolin is one component of the B cellspecific transcription factor and switch region binding protein, LR1. Proc. Natl Acad. Sci. USA, 94, 3605–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herendeen D. & Kelly T.J. (1996) in SV40 DNA Replication (ed. Blow, J. J.) 29–65. Oxford University Press, New York, USA. [Google Scholar]
- Kel A.E., Kel-Margoulis O.V., Farnham P.J., Bartley S.M., Wingender E. & Zhang M.Q. (2001) Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol., 309, 99–120. [DOI] [PubMed] [Google Scholar]
- Labib K. & Diffley J.F. (2001) Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev., 11, 64–70. [DOI] [PubMed] [Google Scholar]
- Ramsperger U. & Stahl H. (1995) Unwinding of chromatin by the SV40 large T antigen DNA helicase. EMBO J., 14, 3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Knippers R. & Stahl H. (1989) RNA unwinding activity of the SV40 large T antigen. Cell, 57, 955–963. [DOI] [PubMed] [Google Scholar]
- Schirmbeck R. & Deppert W. (1991) Structural topography of simian virus 40 DNA replication. J. Virol., 65, 2578–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D.T., Melendy T., Usher D. & Stillman B. (1996) Simian virus 40 large T antigen binds to topoisomerase I. Virology, 222, 365–374. [DOI] [PubMed] [Google Scholar]
- Smelkova N.V. & Borowiec J.A. (1997) Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol., 71, 8766–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Fleming P.J., Pollard H.B. & Burns A.L. (1989) Cloning and sequencing of the human nucleolin cDNA. FEBS Lett., 250, 99–105. [DOI] [PubMed] [Google Scholar]
- Strausfeld U. & Richter A. (1989) Simultaneous purification of DNA topoisomerase I and II from eukaryotic cells. Prep. Biochem., 19, 37–48. [DOI] [PubMed] [Google Scholar]
- Uhlmannschiffler H., Seinsoth S. & Stahl H. (2002) Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res., 30, 3192–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLoock M.S., Alexandrov A., Yu X., Cozzarelli N.R. & Egelman E.H. (2002) SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr. Biol., 12, 472–476. [DOI] [PubMed] [Google Scholar]
- Wang Y., Guan J., Wang H., Wang Y., Leeper D. & Iliakis G. (2001) Regulation of DNA replication after heat shock by replication protein a–nucleolin interactions. J. Biol. Chem., 276, 20579–20588. [DOI] [PubMed] [Google Scholar]
- Wessel R., Schweizer J. & Stahl H. (1992) Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bi-directional unwinding from the viral origin of DNA replication. J. Virol., 66, 804–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Hamhouyia F., Thomas S.D., Burke T.J., Girvan A.C., McGregor W.G., Trent J.O., Miller D.M. & Bates P.J. (2001). Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem., 276, 43221–43230. [DOI] [PubMed] [Google Scholar]
- Xue Z., Shan X., Lapeyre B. & Melese T. (1993) The amino terminus of mammalian nucleolin specifically recognizes SV40 T-antigen type nuclear localization sequences. Eur. J. Cell Biol., 62, 13–21. [PubMed] [Google Scholar]


