Abstract
We report the occurrence of an isomerase with a putative (βα)8-barrel structure involved in both histidine and trypto-phan biosynthesis in Streptomyces coelicolor A3(2) and Mycobacterium tuberculosis HR37Rv. Deletion of a hisA homologue (SCO2050) putatively encoding N′-[(5′-phosphoribosyl)-formimino]-5 amino-imidazole-4-carboxamide ribonucleotide isomerase from the chromosome of S. coelicolor A3(2) generated a double auxotrophic mutant for histidine and tryptophan. The bifunctional gene SCO2050 and its orthologue Rv1603 from M. tuberculosis complemented both hisA and trpF mutants of Escherichia coli. Expression of the E. coli trpF gene in the S. coelicolor mutant only complemented the tryptophan auxo-trophy, and the hisA gene only complemented the histidine auxotrophy. The discovery of this enzyme, which has a broad-substrate specificity, has implications for the evolution of metabolic pathways and may prove to be important for understanding the evolution of the (βα)8-barrels.
Introduction
The evolution of enzymes and metabolic pathways are intrinsically related (Copley & Bork, 2000; Teichmann et al., 2001). Many enzymes involved in central metabolic pathways have (βα)8-barrel scaffolds, to which are attached different catalytic and substrate-binding folds (Gerlt & Babbitt, 2001; Todd et al., 2001). The enzymes N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carbox-amide ribonucleotide (ProFAR) isomerase (HisA, EC 5.3.1.16), which is involved in histidine biosynthesis (Lang et al., 2000), and N′-(5′-phosphoribosyl)-anthranilate (PRA) isomerase (TrpF, EC 5.3.1.24), which is involved in tryptophan biosynthesis (Priestle et al., 1987), are members of this structural family. It is generally accepted that the (βα)8-barrel enzymes involved in central metabolic pathways arose by divergent evolution (Copley & Bork, 2000; Hennsax et al., 2001; Gerlt & Babbitt, 2001; Nagano et al., 2002). Functional evidence for a common ancestry includes the directed evolution of the activity of TrpF from the HisA protein using random mutagenesis and selection in vivo (Jürgens et al., 2000). TrpF and HisA catalyse an Amadori rearrangement of their cognate aminoaldoses into the corresponding aminoketoses (Fig. 1; Hennsax et al., 2002).
Figure 1.
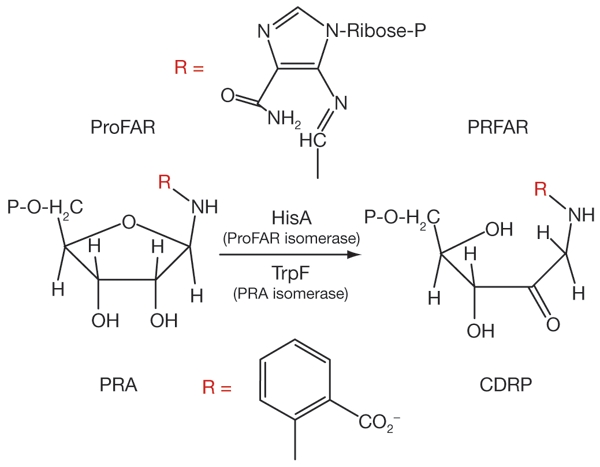
HisA and TrpF catalyse similar reactions. HisA and TrpF catalyse analogous Amadori rearrangements of N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (ProFAR) and N′-(5′-phosphoribosyl)anthranilate (PRA) into N′-[(5′-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide (PRFAR) and 1-[(2-carboxyphenyl)amino]-1-deoxyribulose 5-phosphate (CDRP), respectively. Red 'R's indicate the different side-chains in the two substrates.
The evolution of enzymes by divergence has a bearing on how biosynthetic pathways may have evolved to their current architecture. Among the hypotheses put forward on how metabolic pathways evolved the 'patchwork' hypothesis (Jensen, 1976) offers the most generally accepted explanation. This hypothesis states that metabolic pathways arose by the recruitment of enzymes with similar activity, and by subsequent modification of their substrate-binding ability. The model implies the existence of ancestral enzymes with broad substrate specificity, catalysing related reactions in different pathways. The recruitment of enzyme function, as inferred from predictions of the evolutionary relatedness of enzymes from different metabolic pathways (Parsot, 1987; Copley & Bork, 2000; Teichmann et al., 2001) and detection of promiscuous activities (O'Brien & Herschlag, 1999) is widely acknowledged.
In Escherichia coli and its relatives, the enzyme PRA isomerase (TrpF), and the next enzyme downstream in the tryptophan biosynthetic pathway, indole-3-glycerol-phosphate (IGP) synthase (TrpC; EC 4.1.1.48), are present on a single peptide chain encoded by trpC (Priestle et al., 1987). During our investigations into the regulation of tryptophan biosynthesis in Streptomyces coelicolor A3(2) (Hu et al., 1999), we failed to discover a trpF gene next to trpC. This paradox of a function without a gene was confirmed after completion of the sequencing of the complete genome of S. coelicolor (Bentley et al., 2002). A similar situation was encountered in the genomes of Mycobacterium tuberculosis HR37Rv (Cole et al., 1998) and M. leprae (Cole et al., 2001). Here, we report the occurrence of an isomerase, with a putative (βα)8-barrel structure predicted from its sequence, with a dual function in both histidine and aromatic amino-acid biosynthesis in S. coelicolor and M. tuberculosis. This report expands on the suggested physiological link between tryptophan and histidine biosynthesis observed in other organisms (Nester & Montoya, 1976). The discovery of a putative (βα)8-barrel enzyme, with a predicted ancient-like broad substrate specificity, may be relevant for understanding the evolution of this important structural family.
Results and Discussion
Functional genomics of trpF in silico
At present, the only actinomycete trpF sequence available in the database is that of Corynebacterium glutamicum (Matsui et al., 1986). Sequence analysis using this sequence as a probe, either using hidden Markov models (Karplus et al., 1998) or by carrying out BLAST searches (Altschul et al., 1997), failed to identify a homologue in S. coelicolor or the mycobacteria. It has been proposed in other organisms that TrpC and TrpF proteins share a common ancestry (Wilmanns et al., 1991; Gerlt & Babbitt, 2001). We tested whether the sequence of TrpC (open-reading frame (ORF) number SCO2039) of S. coelicolor could reveal a TrpF-like sequence from an actinomycete by performing PSI-BLAST searches (Altschul et al., 1997). Again, no TrpF-like sequence was identified. These observations indicate that the trpF genes of S. coelicolor and Mycobacterium species are of a different family to those found in other bacteria.
On the basis of the report by Jürgens et al. (2000) on the directed evolution of TrpF from HisA, we proposed that the HisA protein might have TrpF activity in S. coelicolor, which would account for the lack of trpF in this species. A hisA homologue was identified in S. coelicolor as part of a histidine biosynthetic cluster (Limauro et al., 1990). The completion of the genome sequencing project revealed that this cluster is localized on the chromosome upstream of a cluster of trp genes (Fig. 2A; Bentley et al., 2002). A similar bifunctionality of hisA is implied in M. tuberculosis by the synteny of the his and trp clusters in this species (Fig. 2A).
Figure 2.
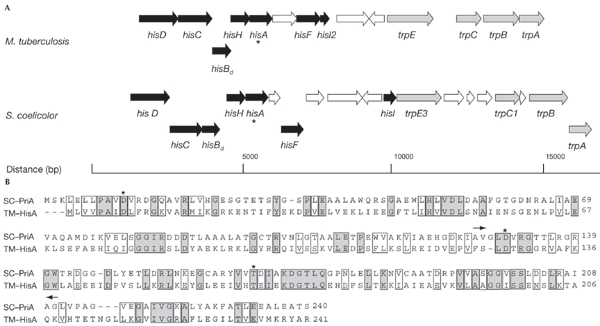
Synteny of the his/trp region and sequence similarity of PriA and HisA. (A) Organization of the his/trp cluster of Streptomyces coelicolor (Bentley et al., 2002) and Mycobacterium tuberculosis (Cole et al., 1998). The hisA homologues of S. coelicolor (SCO2050) and M. tuberculosis (Rv1603) are marked with asterisks. Genes involved in histidine (his; black) and tryptophan (trp; grey) biosynthesis are shown. (B) Sequence alignment of the HisA homologues of S. coelicolor (SC–PriA) and Thermotoga maritima (TM–HisA), which show 25% identity. Data from T. maritima (Jürgens et al., 2000; Hennsax et al., 2002) were used to identify the catalytically important residues in SCO2050 (Asp 11, Asp 130 and Thr 166 (asterisks)). The region of SCO2050 replaced in WH101 (this study) lies between Ala 126 and Gly 210 (indicated by arrows).
Tryptophan and histidine S. coelicolor auxotrophy
We disrupted the hisA homologue of S. coelicolor M145 (ORF number SCO2050, according to the annotation of Bentley et al. (2002)); this was done by deleting two of the three catalytically important amino-acid residues, Asp 130 and Thr 166, which were identified by aligning the sequences of SCO2050 and hisA from Thermotoga maritima (Fig. 2B; Jürgens et al., 2000; Hennsax et al., 2002). This in-frame deletion was constructed in a way that avoided possible polar effects. As hypothesized, the resulting mutant (WH101) could not grow on minimal medium unless supplemented with both histidine and tryptophan (Table 1). We propose that SCO2050 should be renamed priA (phosphoribosyl isomerase A) to reflect the common effects of TrpF and HisA on phosphoribosylated substrates.
Table 1.
Growth requirements of Streptomyces coelicolor WH101 and its transformants
| Strain | Genotype | Growth requirements | |
| WH101 | priA::scar | Histidine and tryptophan | |
| WH101(pIJ702) | priA::scar | Histidine and tryptophan | |
| WH101(pIJ702–PriASc) | priA::scar (priA+) | Prototrophic | |
| WH101(pIJ4123–TrpFEc) | priA::scar (trpF+) | Histidine | |
| WH101(pIJ4123–HisAEc) | priA::scar (hisA+) | Tryptophan |
Gene complementation studies
The priA gene was inserted into the plasmid pIJ702 to make pIJ702–PriASc, which was used to transform WH101 mutants. The resulting transformants were prototrophic, showing that the tryptophan and histidine auxotrophy of WH101 mutants is exclusively due to the loss of functional priA (Table 1). We tested whether the trpF and hisA genes of E. coli could complement the WH101 tryptophan and histidine auxotrophies. The E. coli genes were cloned into the Streptomyces expression vector pIJ4123 under the control of the thiostrepton-inducible promoter. Thiostrepton-dependent expression of trpF (using pIJ4123–TrpFEc) in WH101 mutants restored tryptophan independence only, whereas expression of hisA (using pIJ4123–HisAEc) restored histidine independence only (Table 1). We also investigated the ability of priA to complement independent hisA and trpF deletions in E. coli. For this purpose, priA was cloned into the expression vector pGEX-4T-1 to form pGEX–PriASc, which was used to transform E. coli auxotrophs with mutations in trpF (W3110 trpC (Fdel); see Darimont et al., 1998), and hisA (Hfr G6; see Matney et al., 1964). Complementation of both mutations was achieved by expression of priA (Table 2). The hisA and trpF genes of E. coli cloned into pGEX-4T-1, to create pGEX–HisAEc and pGEX–TrpFEc, respectively, were used as controls (Table 2).
Table 2.
Complementation of Escherichia coli strains W3110 trpC (Fdel) and Hfr G6
| Plasmid | Strain | ||
| W3110 trpC(Fdel) | Hfr G6 | ||
| pGEX-4T-1 | − | − | |
| pGEX–PriASc | + | + | |
| pGEX–PriAMt | + | + | |
| pGEX–HisAEc | − | + | |
| pGEX–TrpFEc | + | − |
Complementation detected (+) or not detected (−) by expression of the insert from the lacI-dependent promoter induced with 10 μM isopropylthiogalactoside.
The intergenic complementation of trpF and hisA by priA, and the partial complementation of priA by either trpF or hisA, confirms that the product of priA is involved in the biosynthesis of both histidine and tryptophan in S. coelicolor. This discovery places into a physiological context the remarkable observation that TrpF and HisA activities can co-exist in a single protein (Jürgens et al., 2000) despite the low identity (10%) between the sequences of these enzymes. Preliminary characterization of the in vitro activity of PriA has shown that this HisA-like enzyme has PRA isomerase activity (data not shown).
Mycobacterium tuberculosis has a priA gene
To test the possibility that mycobacteria were similar to streptomycetes in having a single gene that encodes both TrpF and HisA activity, we cloned the putative priA orthologue (ORF number Rv1603, according to Cole et al. (1998)) of M. tuberculosis H37Rv into the expression vector pGEX-4T-1, to produce pGEX–PriAMt. This plasmid was shown to complement hisA and trpF mutations in E. coli (Table 2). As there is no trpF gene in Mycobacterium species, these results suggest that the protein product of Rv1603 in M. tuberculosis has the same metabolic role as that of PriA in S. coelicolor. This indicates that the presence of PriA in S. coelicolor and mycobacteria is due to the retention of ancient characteristics, rather than the modification of HisA to provide TrpF activity and a subsequent loss of TrpF. The failure to detect any trpF-like sequence in S. coelicolor supports the former interpretation.
Why did the streptomycetes and mycobacteria not develop independent trpF genes? In general, the streptomycetes do not regulate amino-acid biosynthesis by feedback repression of gene expression (Hodgson, 2000). Therefore, as enzyme expression is not co-ordinately regulated on a pathwayspecific basis, enzymes with functions in multiple pathways are possible. The genome sequence of C. glutamicum shows that this actinobacterium has a trp operon that contains a fused trpCF gene. This strain does regulate amino acid biosynthesis by feedback regulation (Sano & Matsui, 1987), and a simple explanation is that its ancestor acquired a complete, regulated trp operon after it separated from the streptomycete and mycobacteria evolutionary lines. Crawford (1989) has previously proposed this possibility. It would be interesting to test the putative hisA product of C. glutamicum for PRA isomerase activity.
An evolutionary interpretation of the nature of PriA
An obvious mechanistic explanation for the bifunctionality of PriA would be that it shows a broad specificity for the substrates PRA and ProFAR (Fig. 1). Enzymes with broad substrate specificity are not rare, and examples of these have accumulated since the postulation of the patchwork hypothesis by Jensen (1976). Nevertheless, the existence of broad substrate specificity, which stands at the core of the patchwork hypothesis, is usually inferred from the promiscuous activities of enzymes in vitro (for a review, see O'Brien & Herschlag, 1999) and/or overlapping specificities in vivo. Examples of the latter include members of the superfamily of aminotransferases (Jensen & Gu, 1996). This family includes another example of an enzyme that functions in both histidine and aromatic amino acid biosynthesis, which was first identified by Nester & Montoya (1976).
A number of enzymes involved in the biosynthesis of branched-chain amino acids show specificity for multiple substrates, such that isoleucine and valine are produced in the same metabolic pathway (Umbarger, 1996). However, this situation is different to that of PriA, because the substrates of PriA show marked difference in size and shape (Fig. 1). Therefore, if the broad substrate specificity of a given enzyme is in fact an ancient feature, as suggested by Jensen (1976), this implies that this feature has been retained in PriA throughout the course of its evolution. Thus, we conclude that PriA is a 'molecular fossil'.
Speculation
We believe that priA did not evolve as a consequence of loss of trpF and subsequent broadening of the specificity of the HisA protein. On the basis of the broad substrate specificity of PriA, one possibility is that trpF and hisA could have evolved from priA after gene duplication and specialization, as suggested by the patchwork hypothesis (Jensen, 1976). This would be an example of divergent evolution, and implies that the ancestor of priA encoded one of the older members of the (βα)8-barrel protein family (Fani et al., 1994; Copley & Bork, 2000; Lang et al., 2000), although it is also possible that the priA ancestor evolved relatively recently (Nagano et al., 2002). An alternative possibility is that a TrpF function could have evolved from another enzyme, such as TrpC (Wilmanns et al., 1991; Gerlt & Babbitt, 2001), allowing PriA to lose its TrpF activity, an example of convergent evolution. Therefore, convergent or divergent evolution may have accounted for the evolution of the extant (βα)8-barrel proteins.
Methods
Computational sequence analysis and searches.
The sequences used for the searches were TrpCF (accession number E24723), the genome of C. glutamicum (accession number NC_003450), TrpC (accession number SCO2039) and the genome of S. coelicolor (accession number NC_003888). The program ClustalW was used for sequence alignment.
Growth requirements of strains.
Strains Hfr G6 and W3110 trpC (Fdel) were provided by the E. coli Genetic Stock Center. Minimal A medium (Miller, 1972) was used to test Hfr G6 and its derivatives, and modified Vogel–Bonner medium (Darimont et al., 1998) was used to test W3110 trpC (Fdel) and its derivatives. Tryptophan and histidine were added to a final concentration of 100 μg ml−1. Plasmids were selected for using ampicillin at 100 μg ml−1; induction of lacI-dependent promoters was carried out using 10 μM isopropylthiogalactoside. The minimal medium for streptomycetes (Kieser et al., 2000) was supplemented with tryptophan (37.5 μg ml−1) and histidine (50 μg ml−1) as appropriate. Thiostrepton (Sigma) was used at 50 μg ml−1 to select for plasmids and for induction of thiostrepton-inducible promoters. Apramycin (Sigma) selection was carried out using the antibiotic at 50 μg ml−1.
Deletion of SCO2050 from the chromosome of S. coelicolor.
The auxotrophic strain WH101 was constructed using REDIRECT® (Gust et al., 2003). The protocol, plasmids and strains were pro-vided by PBL Biomedical Laboratories. The oligonucleotides used for this were: 5′-TGGGTCGCCAAGGTCATCGCCGAGCACGGCGCAAGATCATTCCGGGGATCCGTCGACC-3′ and 5′-CTTCCCGACGATGGCCCCCTCGACACCGGCCGGACGAGTGTAGGCTGGAGCTGCTTC-3′ (the bases that are identical in the SCO2050 sequence are underlined). The disruption cassette was made by PCR using Expand high-fidelity DNA polymerase (Roche). SCO2050 was mutagenized in cosmid SC4G6 (Redenbach et al., 1996) by homologous recombination (double crossover) replacing 255 bp from the 5′ end of the gene. The disruption cassette was removed by the FLP-recombinase system, leaving behind a 'scar' of 81 nucleotides with no stop codons. The newly mutagenized cosmid (carrying only the original marker) was re-engineered, inserting the selectable marker aac(3)IV and an RP4 oriT. The resulting construct was introduced into S. coelicolor M145 by RP4-based conjugation (Kieser et al., 2000) and selected for using apramycin. Authentic double crossovers were obtained after two rounds of growth on fresh plates containing soya-flour mannitol medium (Kieser et al., 2000) without selection. The replacement of SCO2050 was identified in colonies that were apramycin sensitive and auxotrophic for tryptophan and histidine. The presence of the wild type or of the mutated form of SCO2050 was detected by PCR using the primers 5′-GGGCGAAACCGAAGGACTC-3′ and 5′-TCGTGGCCGCCGTGGAGAACG-3′, and sequencing was used to confirm that the desired replacement event had taken place.
Cloning of the hisA and trpF genes of E. coli, and priA of S. coelicolor and M. tuberculosis.
All DNA fragments were produced by PCR amplification, using primers with restriction sites engineered at their 5′ ends (see supplementary information online). PCR was performed using Pwo DNA polymerase (Roche). The PCR products were digested with the appropriate restriction enzymes and were ligated using T4 DNA ligase (Gibco) into the vectors pMTL22 (Chambers et al., 1988), pET3a, pET22a (Novagen) or pGEX-4T-1 (Amersham Pharmacia). The resulting plasmids were used to transform E. coli MC1061 using the calcium method (Sambrook et al., 1989). Fragments from restriction digests of the pMTL22 and pET constructs were ligated into plasmids pIJ702 and pIJ4123, and the resulting constructs were used to transform Streptomyces lividans TK24 protoplasts (Kieser et al., 2000). The desired constructs were isolated from TK24 and used to transform S. coelicolor WH101 protoplasts. The priA gene (SCO2050) from S. coelicolor was amplified by PCR from cosmid 4G6. The hisA and trpF genes of E. coli were amplified from chromosomal DNA. The trpF gene was also cut out of pMS401 (a gift from M. Samaddar and J. Blackburn) using NcoI and BamHI and sub-cloned into pET22a, from where trpF was subsequently removed using NdeI and BamHI and subcloned into pIJ4123. The priA orthologue (Rv1603) of M. tuberculosis H37Rv was amplified from chromosomal DNA, provided by D. Roper. Sequencing was used to confirm all constructs.
Supplementary data are available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/4-embor771-s1.mov).
Supplementary Material
Supplementary information
Acknowledgments
We thank B. Gust of the John Innes Centre for help in constructing the mutant strain. We are indebted to C. Orengo, D. Lee, A. Grant and S. Rison of University College, London, for help with the bioinformatic analysis and for useful discussions. We would also like to thank J. Blackburn and W. Patrick of Cambridge, UK, for providing us with pMS401 and for useful comments. We are grateful to D. Roper from our department for supplying us with mycobacterial DNA. F.B.-G is a recipient of a Conacyt scholarship, Mexico (number 111558).
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W. & Lipman D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley S.D. et al. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Chambers S.P., Prior S.E., Barstow D.A. & Minton N.P. (1988) The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene, 68, 139–149. [DOI] [PubMed] [Google Scholar]
- Cole S.T. et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393, 537–544. [DOI] [PubMed] [Google Scholar]
- Cole S.T. et al. (2001) Massive gene decay in the leprosy bacillus. Nature, 409, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Copley R.R. & Bork P. (2000) Homology among (βα)8 barrels: implications for the evolution of metabolic pathways. J. Mol. Biol., 303, 627–640. [DOI] [PubMed] [Google Scholar]
- Crawford I.P. (1989) Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu. Rev. Microbiol. 43, 567–600. [DOI] [PubMed] [Google Scholar]
- Darimont B., Stehlin C., Szadkowski H. & Kirschner K. (1998) Mutational analysis of the active site of indoleglycerol phosphate synthase from Escherichia coli. Protein Sci., 7, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R., Lio P., Chiarelli I. & Bazzicalupo M. (1994) The evolution of the histidine biosynthetic genes in prokaryotes: a common ancestor for the hisA and hisF genes. J. Mol. Evol., 38, 489–495. [DOI] [PubMed] [Google Scholar]
- Gerlt J.A. & Babbitt P.C. (2001) Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu. Rev. Biochem., 70, 209–246. [DOI] [PubMed] [Google Scholar]
- Gust B., Challis G.L., Fowler K., Kieser T. & Chater K.F. (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odour geosmin. Proc. Natl Acad. Sci. USA, 31 January 2003, 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennsax M., Hocker B., Wilmanns M. & Sterner R. (2001) Divergent evolution of (βα)8-barrel enzymes. Biol. Chem., 382, 1315–1320. [DOI] [PubMed] [Google Scholar]
- Hennsax M., Thoma R., Schmidt S., Hennig M., Kirschner K. & Sterner R. (2002) Two (βα)8-barrel enzymes of histidine and tryptophan biosynthesis have similar reaction mechanisms and common strategies for protecting their labile substrates. Biochemistry, 41, 12032–12042. [DOI] [PubMed] [Google Scholar]
- Hodgson D.A. (2000) Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol., 42, 47–238. [DOI] [PubMed] [Google Scholar]
- Hu D.S.-J., Hood D.W., Heidstra R. & Hodgson D.A. (1999) The expression of the trpD, trpC and trpBA genes of Streptomyces coelicolor A3(2) is regulated by growth rate and growth phase but not by feedback repression. Mol. Microbiol., 32, 869–880. [DOI] [PubMed] [Google Scholar]
- Jensen R.A. (1976) Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol., 30, 409–425. [DOI] [PubMed] [Google Scholar]
- Jensen R.A. & Gu W. (1996) Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol., 178, 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens C., Strom A., Wegener D., Hettwer S. & Wilmanns M. (2000) Directed evolution of a (βα)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc. Natl Acad. Sci. USA, 97, 9925–9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus K., Barrett C. & Hughey R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M.J., Buttner M.J., Chater K.F. & Hopwood D.A. (2000) Practical Streptomyces Genetics. The John Innes Foundation, Norwich, UK.
- Lang D., Thoma R., Hennsax M., Sterner R. & Wilmanns M. (2000) Structural evidence for evolution of the βα-barrel scaffold by gene duplication and fusion. Science, 289, 1546–1550. [DOI] [PubMed] [Google Scholar]
- Limauro D., Avitabile A., Capellano C., Puglia M.A. & Bruni C.B. (1990) Cloning and characterization of the histidine biosynthetic gene cluster of Streptomyces coelicolor A3(2). Gene, 90, 31–41. [DOI] [PubMed] [Google Scholar]
- Matney T.S., Goldschmidt E.P., Erwin N.S. & Scroggs R.A. (1964) A preliminary map of genomic sites for F-attachment in Escherichia coli K12. Biochem. Biophys. Res. Commun., 17, 278–281. [DOI] [PubMed] [Google Scholar]
- Matsui K., Sano K. & Ohtsubo E. (1986) Complete nucleotide and deduced amino acid sequences of the Brevibacterium lactofermentum tryptophan operon. Nucleic Acids Res., 14, 10113–10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972) in Experiments in Molecular Genetics, 431–432, Cold Spring Harbor Laboratory Press, New York, USA. [Google Scholar]
- Nagano N., Orengo C.A. & Thornton J.M. (2002) One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol., 321, 741–765. [DOI] [PubMed] [Google Scholar]
- Nester E.W. & Montoya A.L. (1976) An enzyme common to histidine and aromatic amino acid biosynthesis in Bacillus subtilis. J. Bacteriol., 126, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien P.J. & Herschlag D. (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol., 6, 91–105. [DOI] [PubMed] [Google Scholar]
- Parsot C. (1987) A common origin for enzymes involved in the terminal step of the threonine and tryptophan biosynthetic pathways. Proc. Natl Acad. Sci. USA, 84, 5207–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestle J.P., Grütter M.G., White J.L., Vincent M.G., Kania M., Wilson E., Jardetzky T.S., Kirschner K. & Jansonius J.N. (1987) Three-dimensional structure of the bifunctional enzyme N-(5′-phosphoribosyl)anthranilate isomerase-indole-3-glycerol-phosphate synthase from Escherichia coli. Proc. Natl Acad. Sci. USA, 84, 5690–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redenbach M., Kieser H.M., Denapaite D., Eichner A., Cullum J., Kinashi H. & Hopwood D.A. (1996) A set of ordered cosmids and a detailed genetic and physical map for the 8-Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol., 21, 77–96. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. & Maniatis T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, NY, USA. [Google Scholar]
- Sano K. & Matsui K. (1987) Structure and function of the trp operon control regions of Brevibacterium lactofermentum, a glutamic-acid-producing bacterium. Gene, 53, 191–200. [DOI] [PubMed] [Google Scholar]
- Teichmann S.A., Rison S.C.G., Thornton J.M., Riley M., Gough J. & Chothia C. (2001) The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J. Mol. Biol., 311, 693–708. [DOI] [PubMed] [Google Scholar]
- Todd A.E., Orengo C.A. & Thornton J.M. (2001) Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol., 307, 1113–1143. [DOI] [PubMed] [Google Scholar]
- Umbarger H.E. (1996) In Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Neidhardt, F.C.) 442–457. ASM, Washington, DC, USA. [Google Scholar]
- Wilmanns M., Hyde C.C., Davies D.R., Kirschner K. & Jansonius J.N. (1991) Structural conservation in parallel β/α-barrel enzymes that catalyze three sequential reactions in the pathway of tryptophan biosynthesis. Biochemistry, 30, 9161–9169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


