Abstract
The soybean SIRE1 family of Ty1/copia retrotransposons encodes an envelope-like gene (env-like). We analysed the DNA sequences of nine SIRE1 insertions and observed that the gag/pol and env-like genes are in the same reading frame and separated by a single UAG stop codon. The six nucleotides immediately downstream of the stop codon conform to a degenerate nucleotide motif, CARYYA, which is sufficient to facilitate stop codon suppression in tobacco mosaic virus. In vivo stop codon suppression assays indicate that SIRE1 sequences confer leakiness to the UAG stop codon at an efficiency of 5%. These data suggest that SIRE1 retro-elements use translational suppression to express their envelope-like protein; this is in contrast with all characterized retroviruses, which express the envelope protein from a spliced genomic messenger RNA.
Introduction
Retrotransposons are repetitive genomic elements that share a close relationship with retroviruses. Both retrotransposons and retroviruses express gag and pol genes, which are necessary for their replication via an RNA intermediate. One distinction between the two types of retroelements is the presence of an envelope (env) gene after pol, which enables a retrovirus to be infectious. However, some retrotransposons have an open reading frame (ORF) after pol (Eickbush & Malik, 2002; Malik et al., 2000), which is often referred to as an env-like gene. The env-like ORF in the gypsy element of Drosophila is known to mediate infection (Kim et al., 1994; Song et al., 1994). Two different lineages of retroelements with an ORF after pol have been described in plant genomes: the Athila group of Ty3/gypsy elements, which were first characterized in Arabidopsis (Wright & Voytas, 1998; Wright & Voytas, 2002), and the SIRE1 family of Ty1/copia elements from soybean (Laten et al., 1998). Because the cell wall is thought to present a barrier to retroviral infection, the function of the env-like ORF in these plant retroelements remains controversial.
Viral protein expression can be regulated in a number of ways, including splicing, frameshifting, and stop codon suppression (Farabaugh, 1996; Gesteland & Atkins, 1996). However, in all characterized retroviruses, expression of the env gene is achieved through splicing of the genomic RNA (Coffin et al., 1997). Some retrotransposons with the env-like ORF use the same method for env expression, illustrating another similarity between these elements and characterized retroviruses. For example, the gypsy elements and BAGY2, an Athila homologue from barley, make a spliced env-like mRNA (Avedisov & Ilyin, 1994; Pelisson et al., 1994; Vicient et al., 2001). The Athila elements from Arabidopsis all have predicted splice acceptors at the beginning of their env-like genes, suggesting that, as for BAGY2, their env-like genes are translated from a spliced product (Wright & Voytas, 2002).
SIRE1 and its homologues are the only known Ty1/copia elements with an env-like ORF after pol (Kapitonov & Jurka, 1999; Laten et al., 1998; Peterson-Burch et al., 2000). In this study, we characterized multiple SIRE1 insertions and found that the env-like ORF is separated from pol by a single stop codon. We also showed that expression of the SIRE1 env-like ORF can result from stop codon suppression. Interestingly, this type of regulation is very similar to another known example of stop codon suppression in tobacco mosaic virus (TMV), and could potentially yield similar ratios of Gag to Env protein as are generated by the splicing of retroviral messenger RNAs.
Results and Discussion
The SIRE1 family is estimated to be present at between 500 and 800 copies in the soybean genome (Laten & Morris, 1993). The first such element reported, SIRE1-1, has a stop codon 24 nucleotides after the pol termination codon, and this was presumed to begin the env-like ORF (Laten et al., 1998). We sequenced several additional members of the SIRE1 family and found that, in contrast to SIRE1-1, all have gag/pol and env-like ORFs separated from one another by a single UAG stop codon (Fig. 1A). In addition, we observed that all had the hexanucleotides CAAUUA, CAGCUA or CAACUA directly after the stop codon UAG, giving a consensus of CARYUA (where R represents purines and Y represents pyrimidines; Fig. 1B). These hexanucleotides conform to nucleotide contexts previously shown to facilitate stop codon suppression. For example, in yeast, the degenerate sequence CA(A/G)N(U/C/G)A (where N is any nucleotide) was found to promote greater than 5% readthrough when located downstream of a UAG stop (Namy et al., 2001). Skuzeski et al. (1991), using in vivo assays to study stop codon suppression in TMV, showed that the presence of the degenerate nucleotide motif CARYYA immediately after any stop codon was sufficient for stop codon suppression to occur. In fact, several plant viruses, not only TMV, use the CARYYA motif to allow translational readthrough of stop codons (Beier & Grimm, 2001; Harrell et al., 2002). As the three SIRE1 sequence variants all conformed to the TMV CARYYA motif, as well as the CA(A/G)N(U/C/G)A sequence found in the yeast study, this suggested that the expression of the Env-like protein of SIRE1 is regulated by stop codon suppression.
Figure 1.
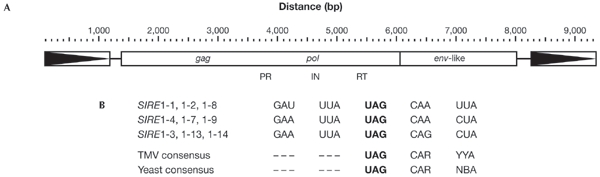
Organization of the SIRE1 open reading frames. (A) gag and pol are in one open reading frame (ORF) separated from the envelope-like (env-like) gene by a single stop codon. Boxed arrows represent the long terminal repeats. (B) The three SIRE1 sequence variations after the UAG stop codon conform to the CARYYA consensus of tobacco mosaic virus (TMV; Skuzeski et al., 1991) and the CARNBA consensus sufficient for stop codon suppression in yeast (Namy et al., 2001) (R, purines; Y, pyrimidines; N, any nucleotide; B represents U, C or G). The numbers after SIRE1 define distinct insertions. PR, protease; IN, integrase; RT, reverse transcriptase.
To test the SIRE1 translational readthrough hypothesis, we used a stop codon suppression assay similar to the one used by Skuzeski et al. (1991). In this assay, a β-glucuronidase (GUS) reporter gene was placed in-frame and downstream from a UAG stop codon that was surrounded by different SIRE1 nucleotide contexts. The three SIRE1 sequence variants all conferred leakiness to the stop codon UAG (Fig. 2A). Although the SIRE1 hexanucleotide sequence CAGCUA conforms to the CAR YYA consensus sequence, it was not specifically tested by Skuzeski et al. (1991). Our test of the CAGCUA sequence further supports the suggestion that the CARYYA motif is able to mediate translational readthrough. In our experiments, and in agreement with estimations made for TMV (Skuzeski et al., 1991), the levels of translation through the stop codon averaged 5% of those achieved for a GUS control lacking the stop codon.
Figure 2.
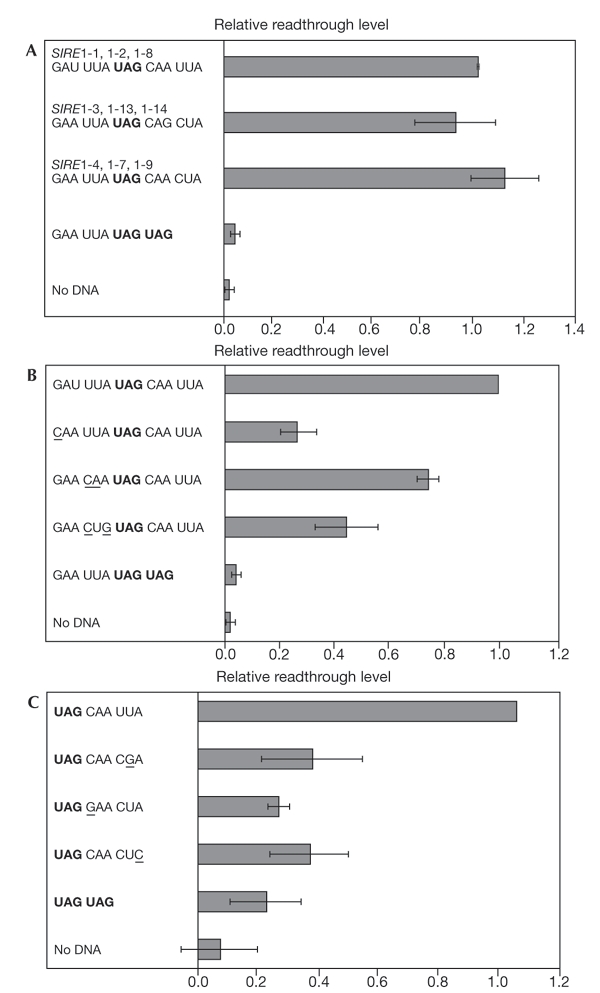
Translational readthrough of SIRE1. For these experiments, levels of readthrough are relative to the initial readthrough level of the CAAUUA SIRE1 variant. UAG stop codons are in bold. Mutations in the SIRE1 sequences are underlined. Error bars represent one standard deviation above and below the average. (A) The three SIRE1 sequence variations result in the same level of readthrough, averaging 5% of the normal activity of a β-glucuronidase reporter gene (data not shown). (B) Readthrough levels of clones with mutations in sequences upstream of the stop codon. (C) Readthrough levels of clones with mutations in the CARYYA consensus. R, any purine. Y, any pyrimidine.
To test the effect of mutations in the SIRE1 sequences, specific changes were made to the nucleotides either upstream or downstream of the UAG stop codon. As found for TMV (Skuzeski et al., 1991), mutations made before the SIRE1 stop codon reduced the readthrough efficiency, as shown by lower levels of GUS expression. GUS expression in the mutants, however, was still significantly greater than that in the negative controls (Fig. 2B). One of the mutations made changed the SIRE1 UUA codon immediately upstream of the stop codon to CAA. This mutation mimics the TMV sequence (CAAUAGCAAUUA), and conferred slightly lower levels of readthrough than the SIRE1 wild-type sequence (UUAUAGCAAUUA). These data further indicate that the level of readthrough in SIRE1 compares well with that of TMV.
For mutations after the UAG stop codon, we changed three of the four bases that are conserved in the SIRE1 CARYUA consensus—the first cytosine and the second and sixth adenines. Although wild-type SIRE1 sequences still allowed readthrough, mutations deviating from the SIRE1 CARYUA consensus reduced expression to the level of the negative controls (Fig. 2C). In addition, one of the mutations of the SIRE1 consensus, CAACGA, still conformed to the yeast degenerate readthroughsequence CA(A/G)N(U/C/G)A. However, we did not detect significant GUS activity in tobacco protoplasts for this stop codon context. This result may represent a difference between the translational suppression apparatus in yeast and plants.
The nucleotide sequences we tested may not be the only signal that affects the leakiness of the stop codon between pol and env in the wild-type SIRE1 retroelement. Another example of a stop codon suppression signal, which is not immediately adjacent to the stop codon, occurs in murine leukaemia virus. In this retrovirus, a pseudoknot structure after the stop codon separating gag and pol facilitates recoding of the stop codon by a suppressor transfer RNA (Honigman et al., 1991; Wills et al., 1991). In barley yellow dwarf virus (an RNA plant virus), a sequence that lies 700 bp downstream of the stop codon is required for suppression of the coat protein stop codon (Brown et al., 1996). However, our data show clearly that the CARYYA motif is crucial for SIRE1 stop codon suppression.
All viral readthrough products that have been studied are either polymerase proteins or coat protein extensions (Beier & Grimm, 2001). The Env-like protein does not show similarity to polymerases, and, because it follows reverse transcriptase, it cannot be a coat protein extension. Reverse transcriptase extensions have not been previously observed.
One other possibility is that the SIRE1 Env-like protein functions analogously to the Env protein in mammalian retroviruses—in viral infection. Assuming that the nucleotides tested represent all the nucleotides influencing stop codon suppression, we would predict that for approximately 5% of the times that a SIRE1 mRNA is translated, a very large (approximately 250 kDa) Gag–Pol–Env fusion protein would be produced. If the Env-like protein is used for infection, it would most likely be cleaved from the Gag–Pol–Env polyprotein by an element-encoded or host-encoded protease to release a functional protein. In mammalian retroviruses, such as simian immunodeficiency virus and human immunodeficiency virus 1, Gag to Env ratios have been estimated at anywhere from 6:1 to 60:1 in different infective strains (Chertova et al., 2002, and references therein). A 5% readthrough level of the SIRE1 stop codon corresponds to a 20:1 Gag to Env-like protein ratio, which is consistent with the ratios seen in other characterized retroviruses. If the Env-like protein does mediate infection, stop codon suppression would be a novel means of regulating the production of this retroelement protein.
Methods
SIRE1 sequences.
Sequences for the SIRE1 elements have the following GENBANK accession numbers: SIRE1-1 (AF053008), SIRE1-2 (AY205606), SIRE1-3 (AY205607), SIRE1-4 (AY205608), SIRE1-7 (AY205609), SIRE1-8 (AY205610), SIRE1-9 (AY205611), SIRE1-13 (AY205612) and SIRE1-14 (AY205613). A detailed analysis of these SIRE1 sequences will appear elsewhere (Laten, H., E.H., Farmer, L., Lin, E. & D.V., unpublished data).
DNA constructs.
All DNA constructs used for stop codon suppression used a cauliflower mosaic virus 35S promoter fused to the GUS coding sequence (pDW919). PCR mutagenesis was used to incorporate the SIRE1 stop codon and flanking nucleotides into the GUS coding sequence. The forward primer for PCR was 5′-TAAAGGCGCCCAGTCCCTTATGNNNNNNUAGNNNNNNTTACGTCCTGTAGAAACCCCAACC-3′, which contains a NarI site at the 5′ end. The positions where nucleotides varied in the different constructs made are shown by Ns. The start codon (ATG) for GUS and the SIRE1 stop codon (UAG) are in bold. The sequence used for the reverse primer (5′-TACGTACACTTTTCCCGGCAATAAC-3′) corresponds to the region in the GUS coding sequence after a BclI site. After amplification, PCR products were digested with NarI and BclI and cloned into sites for these enzymes in pDW919. All constructs were sequenced to confirm the presence of the nucleotide changes.
Readthrough assays.
For transient assays, Nicotiana tabacum cv. SR1 protoplasts were isolated from sterile leaf tissue (Van den Elzen et al., 1985) in K3 medium (Kao & Michayluk, 1975) with 0.4 M sucrose. The protoplasts were collected in Babcock bottles by centrifugation at 500 r.p.m. for 10 min. After two washes in this medium, the protoplasts were resuspended in K3 medium with 0.4 M glucose (K3/G1). Protoplasts were electroporated with approximately 70 μg of plasmid DNA at 437 V cm−1 and 1200 μF. For electroporation, the K3/G1 medium was supplemented with 2 M KCl to produce a 20 ms pulse using a BioRad Gene Pulser II electroporater. Transiently transformed protoplasts were diluted five-fold in K3/G1 media and incubated at 30 °C for 24 h. GUS activity assays were carried out to measure expression of the GUS reporter gene using the substrate MUG (4-methylumbelliferyl-β-D-glucuronide (Sigma) (Jefferson, 1987). The total protein content was determined for each sample (Bradford, 1976). Multiple MUG readings for each sample were averaged and normalized to total protein. Within each experiment, tested samples were expressed as relative levels of the wild-type SIRE1 sample (containing the sequence CAAUUA after the stop codon). Each experiment was repeated three times, relative levels of readthrough were averaged and standard deviations were determined.
Acknowledgments
We gratefully acknowledge Jeffrey Townsend for preparation and electroporation of the tobacco protoplasts. We also acknowledge Howard Laten for the sharing of clones and sequences. We thank John Atkins and Pasha Baranov for helpful comments and suggestions on the readthrough assays. We thank Patricia Lonosky and Elizabeth Pettit for a critical reading of this manuscript.
References
- Avedisov S.N. & Ilyin Y.V. (1994) Identification of spliced RNA species of Drosophila melanogaster gypsy retrotransposon. New evidence for retroviral nature of the gypsy element. FEBS Lett., 350, 147–150. [DOI] [PubMed] [Google Scholar]
- Beier H. & Grimm M. (2001) Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res., 29, 4767–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown C.M., Dinesh-Kumar S.P. & Miller W.A. (1996) Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV protein gene stop codon. J. Virol., 70, 5884–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E. et al. (2002) Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol., 76, 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin J., Hughes S.H. & Varmus H.E. (1997) Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, USA. [PubMed] [Google Scholar]
- Eickbush T. & Malik H. (2002) In Mobile DNA II (eds Craig, N.L., Craigie, R., Gellert, M. & Lambowitz, A.M.) 1111–1144. ASM, Washington, DC, USA. [Google Scholar]
- Farabaugh P.J. (1996) Programmed translational frameshifting. Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- Gesteland R.F. & Atkins J.F. (1996) Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem., 65, 741–768. [DOI] [PubMed] [Google Scholar]
- Harrell L., Melcher U. & Atkins J.F. (2002) Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res., 30, 2011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigman A., Wolf D., Yaish S., Falk H. & Panet A. (1991) cis acting RNA sequences control the gag-pol translation readthrough in murine leukemia virus. Virology, 183, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Reporter, 5, 387–405. [Google Scholar]
- Kao K.N. & Michayluk M.R. (1975) Nutrient requirements for growth of Vicia hajastana cells and protoplasts at very low population density in liquid media. Planta, 126, 105–110. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V. & Jurka J. (1999) Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica, 107, 27–37. [PubMed] [Google Scholar]
- Kim A., Terzian C., Santamaria P., Pelisson A., Purd'homme N. & Bucheton A. (1994) Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 91, 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten H.M., Majumdar A. & Gaucher E.A. (1998) SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc. Natl Acad. Sci. USA, 95, 6897–6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten H.M. & Morris R.O. (1993) SIRE-1, a long interspersed repetitive DNA element from soybean with weak sequence similarity to retrotransposons: initial characterization and partial sequence. Gene, 134, 153–159. [DOI] [PubMed] [Google Scholar]
- Malik H., Henikoff S. & Eickbush T. (2000) Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res., 10, 1307–1318. [DOI] [PubMed] [Google Scholar]
- Namy O., Hatin I. & Rousset J.P. (2001) Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep., 2, 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A., Song S.U., Prud'homme N., Smith P.A., Bucheton A. & Corces V.G. (1994) gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissuespecific control of the Drosophila flamenco gene. EMBO J., 13, 4401–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson-Burch B.D., Wright D.A., Laten H.M. & Voytas D.F. (2000) Retroviruses in plants? Trends Genet., 16, 151–152. [DOI] [PubMed] [Google Scholar]
- Skuzeski J.M., Nichols L.M., Gesteland R.F. & Atkins J.F. (1991) The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol., 218, 365–373. [DOI] [PubMed] [Google Scholar]
- Song S.U., Gerasimova T., Kurkulos M., Boeke J.D. & Corces V.G. (1994) An Env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev., 8, 2046–2057. [DOI] [PubMed] [Google Scholar]
- Van den Elzen P., Lee K.Y., Townsend J. & Bedbrook J. (1985) Simple binary vectors for DNA transfer to plant cells. Plant Mol. Biol., 5, 149–154. [DOI] [PubMed] [Google Scholar]
- Vicient C.M., Kalendar R. & Schulman A.H. (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res., 11, 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills N.M., Gesteland R.F. & Atkins J.F. (1991) Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA, 88, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.A. & Voytas D.F. (1998) Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics, 149, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D.A. & Voytas D.F. (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res., 12, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]


