Abstract
Human immunodeficiency virus type I reverse transcriptase (RT) possesses distinct DNA polymerase and RNase H sites, whereas integrase (IN) uses the same active site to perform 3′-end processing and strand transfer of the proviral DNA. These four enzymatic activities are essential for viral replication and require metal ions. Two Mg2+ ions are present in the RT polymerase site, and one or two Mg2+ ions are required for the catalytic activities of RNase H and IN. We tested the possibility of inhibition of the RT polymerase and RNase H as well as the IN 3′-end processing and transfer activities of purified enzymes by a series of 3,7-dihydroxytropolones designed to target two Mg2+ ions separated by ∼3.7 Å. The RT polymerase and IN 3′ processing and strand transfer activities were inhibited at submicromolar concentrations, while the RNase H activity was inhibited in the low micromolar range. In all cases, the lack of inhibition by tropolones and O-methylated 3,7-dihydroxytropolones was consistent with the active molecules binding the metal ions in the active site. In addition, inhibition of the DNA polymerase activity was shown to depend on the Mg2+ concentration. Furthermore, selective inhibitors were identified for several of the activities tested, leaving some potential for design of improved inhibitors. However, all tested compounds exhibited cellular toxicity that presently limits their applications.
Human immunodeficiency virus type 1 (HIV-1) is the causative agent of AIDS. Synthesis and integration of the HIV-1 proviral DNA into the host cell genome are the result of four catalytic activities. The virally encoded reverse transcriptase (RT) contains polymerase and RNase H active sites (1, 15, 22), whereas the integrase (IN) possesses 3′-end processing and strand transfer activities achieved by the same catalytic site (6, 18). All four activities are essential for HIV-1 replication, but only the polymerase is currently targeted by FDA-approved inhibitors. Several drugs inhibiting the IN strand transfer activity have proceeded to clinical trials (10).
Combinations of RT and protease inhibitors offer highly effective, durable treatment options (10). However, the high incidence of resistance in therapy-experienced and newly infected patients underscores the need for new antiretroviral agents (11, 17, 32, 51). Increasing the number of anti-AIDS drugs would also facilitate the management of their side effects (37).
One strategy to minimize the emergence of drug resistance may be the design of compounds that would interact with amino acids or cofactors that are essential for catalysis. A common feature of the RT and IN catalytic activities is the requirement of a metal cofactor, which is most likely Mg2+ in vivo. Two Mg2+ ions separated by 3.57 Å have been observed in the polymerase active site of RT in complex with a primer/template DNA duplex and an incoming nucleotide (27). The three aspartate residues that bind Mg2+ and the metal ions themselves are essential for DNA synthesis (28, 40).
Divalent metal ions, such as Mg2+ or Mn2+, are also essential for HIV-1 RNase H activity (7), but the number of ions involved in the RNA cleavage reaction is still unclear. Two Mn2+ ions separated by 4 Å and coordinated to D443, E478, D498, and D549 have been observed in the isolated HIV-1 RNase H domain (8), whereas only one Mg2+ ion, bound to D443 and D549, was seen in the RNase H domain of RT bound to a DNA duplex, in the presence of an incoming deoxynucleoside triphosphate (dNTP) (27).
Similarly, IN activities require divalent metal ions (4, 30, 49), but the number of metal ions involved in catalysis is still subject to debate (23). Structural studies of IN revealed a single binding site for Mg2+ and Mn2+ (20, 35), while two Cd2+ or Zn2+ ions separated by 3.6 to 3.7 Å were observed (4, 5, 49). Indeed, it is believed that a second Mg2+ ion is carried into the integrase active site by the substrate (31).
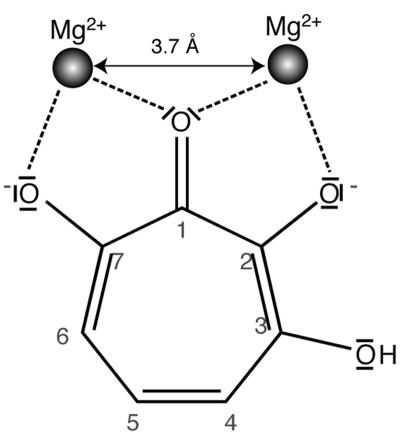
In this study, we tested the possibility of inhibition of the RT polymerase, RNase H, and the IN 3′-end processing and transfer activities by compounds designed to target two Mg2+ ions separated by ∼3.7 Å. To this aim, we used 3,7-dihydroxytropolones (Fig. 1), which have been shown to inhibit inositol monophosphatase by binding the Mg2+ ions of the catalytic site, thus preventing binding of the substrate (41, 42). To our knowledge, this is the first time that this strategy has been used to inhibit the polymerase activity of HIV-1 RT, and there has been only one published rational attempt to simultaneously target two metal ions in the RNase H active site (29). While this work was in progress, it became increasingly likely that several IN inhibitor families identified by “blind” or focused screening bind magnesium in the IN active site (12, 23, 39). However, it is not completely clear whether these inhibitors bind one or two metal ions (19, 23). Recently, a rationally designed IN inhibitor that can bind two Mg2+ ions has been described (33).
FIG. 1.
Structure of 3,7-dihydroxytropolone and schematic drawing of its interaction with two Mg2+ ions separated by 3.7 Å (modified from reference 42 with permission of the publisher).
In this study, we showed that the RT polymerase and IN 3′ processing and strand transfer activities were inhibited at submicromolar concentrations, while 3,7-dihydroxytropolones inhibited the RNase H activity in the low micromolar range. In all cases, the lack of inhibition by modified tropolones and O-methylated 3,7-dihydroxytropolones is consistent with the active molecules binding the metal ions in the active site. In line with this mechanism, inhibition of the RT polymerase by 3,7-dihydroxytropolones and acetylated derivatives was very sensitive to the magnesium concentration. Furthermore, selective inhibitors were identified for several of the activities tested. Even though all tested compounds were toxic to cells, the numerous possibilities for derivatization of the dihydroxytropolone core potentially offer the prospect for increase in selectivity and decrease in toxicity of these inhibitors.
MATERIALS AND METHODS
Oligodeoxyribonucleotides.
The sequences of the oligodeoxyribonucleotides used in this study are as follows: for ODN, 5′-GTC CCT GTT CGG GCG GGA-3′; ODN35, 5′-TTA GCC AGA GAG GCT CCC AGG CTC AGA TCT GGT CT-3′; LE18, 5′-TAC GAA TTC TAA TAC GA CGA CTC ACT ATA GGT CTC TCT GGT TAG ACC AGA TCT GAG C-3′; U5B, 5′-GTG TGG AAA ATC TCT AGC AGT-3′; U5B-2, 5′-GTG TGG AAA ATC TCT AGC A-3′; and U5A, 5′-ACT GCT AGA GAT TTT CCA CAC-3′. All oligodeoxyribonucleotides were purchased from Eurogentec (Belgium).
Templates, primers, and enzymes.
RNA1-311, encompassing nucleotides (nt) 1 to 311 of HIV genomic RNA (MAL isolate), was used as the template for the RT assays. It was synthesized by in vitro transcription and purified as previously described (36).
The template used for the RNase H assays was obtained by PCR with two partially complementary oligodeoxyribonucleotides generating a double-stranded DNA containing the T7 RNA polymerase promoter upstream of the region coding for nucleotides 1 to 47 of HIV genomic RNA (RNA1-47) (HXB2 isolate). Ten amplification cycles (30 s at 94°C, 30 s at 38°C, and 30 s at 72°C) were performed with 100 pmol of primers ODN35 and LE18 and 4 U DyNAzyme. The PCR product was purified on a 3% low-melting-point agarose gel containing ethidium bromide. The band containing the transcription template was cut out under UV light and heated for 20 min at 50°C in the presence of phenol. The extracted DNA was ethanol precipitated and resuspended in water. RNA1-47 was in vitro transcribed and purified as described previously (36). RNA1-47 (2 μg) was dephosphorylated with 2 U calf intestine phosphatase for 1 h at 37°C in 100 mM NaCl, 50 mM Tris-HCl (pH 7.9), 10 mM MgCl2, and 1 mM dithioerythritol. After a phenol-chloroform extraction, RNA1-47 was ethanol precipitated and resuspended in water.
RNA1-47 and oligodeoxyribonucleotides ODN, U5B, and U5B-2 were 5′ end labeled with [γ-32P]ATP (3,000 Ci/mmol) and phage T4 polynucleotide kinase. ODN and RNA1-47 were purified on 8% denaturing polyacrylamide gels; U5B and U5B-2 were purified with a MicroSpin G-25 column (Amersham Bioscience).
In order to form the primer/template complexes, RNA1-311 and ODN or RNA1-47 and ODN35 were first denatured in water for 2 min at 90°C and chilled on ice. Nucleic acids were annealed at 70°C for 20 min in 100 mM NaCl and cooled on ice for 30 min. To prepare the substrates for integrase, U5A was annealed to either U5B or U5B-2 in 100 mM NaCl by being heated at 80°C, followed by a slow cooling.
An RNA1-47 ladder was prepared by incubating RNA1-47 (15,000 cpm) and 1 μg Saccharomyces cerevisiae tRNA for 15 min at 90°C in 0.1 M NaHCO3-Na2CO3. The RNA1-47 fragments were precipitated by addition of 0.2 M LiClO4, and the pellet was resuspended in water.
Plasmid expressing wild-type HIV-1 RT was kindly provided to us by Torsten Unge (Uppsala, Sweden), together with the protocols for protein overexpression and purification. RNase H− RT was obtained by introducing the E478Q mutation in this plasmid (44). Wild-type IN was expressed and purified as described previously (30).
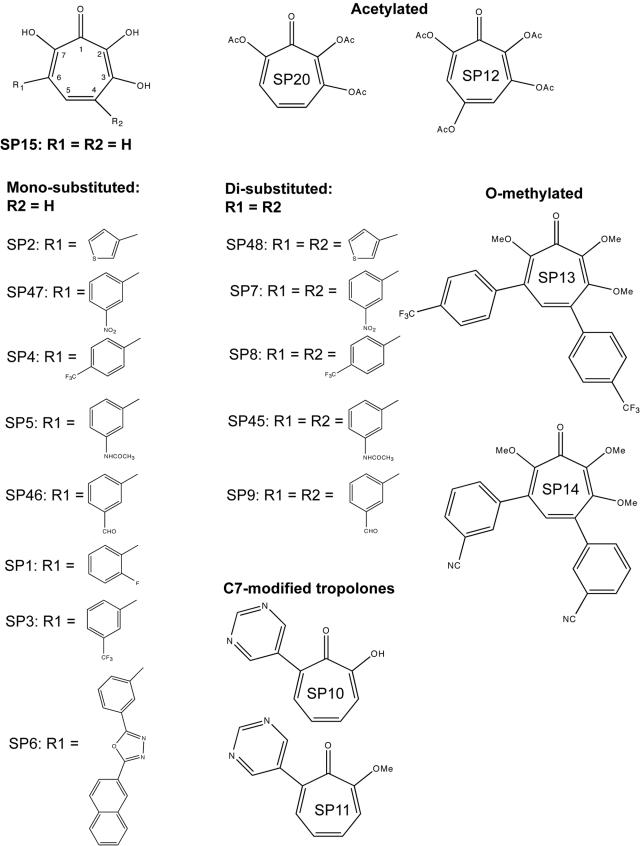
The 3,7-dihydroxytropolones, mono- or disubstituted at positions 4 or 4,6 (referred to as the SP compounds [Fig. 2]), were synthesized as previously described (41). Stock solutions (20 mM) were prepared in dimethyl sulfoxide. We checked that this solvent did not affect any of the tested enzymatic activities at the maximum concentration used in this study.
FIG. 2.
Structures of the tropolones and dihydroxytropolones used in this study. OAc, acetate.
RT assays. (i) Minus-strand “strong-stop” DNA synthesis.
ODN/RNA1-311 (primer/template) complex (10 nM) was preincubated at 37°C for 4 min with 30nM HIV-1 RNase H− RT in 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 6 mM MgCl2, and 1 mM dithioerythritol. Reverse transcription was initiated by addition of dNTPs (50 μM each) in the presence or absence of SP compounds. Reactions were stopped at times ranging from 30 s to 30 min by addition of 1 volume of formamide containing 50 mM EDTA, and the reaction products were analyzed on 8% denaturing polyacrylamide gels. Radioactivity was quantified with a BioImager BAS 2000 (Fuji), using the MacBAS 2.5 (Fuji) software.
(ii) Polymerase-dependent RNase H cleavage.
ODN35/RNA1-47 complex (10 nM) was added to 10 nM wild-type HIV-1 RT in the presence or absence of SP compounds. RNase H assays were carried out for 15 s to 30 min, and reactions were stopped by mixing an equal volume of formamide containing 50 mM EDTA. Cleavage products were resolved using denaturing 15% polyacrylamide gels and analyzed as described above.
3′-end processing and strand transfer for IN assays.
Processing and strand transfer reactions were performed using 1.25 nM U5A/U5B and 6.25 nM U5A/U5B-2 DNA/DNA complexes, respectively, in a buffer containing 20 mM HEPES (pH 7.2), 1 mM dithiothreitol, and 10 mM MgCl2 or MnCl2. The reactions were initiated by addition of 100 nM IN, and the mixtures were incubated at 37°C for up to 1 h in the absence or presence of SP compounds. Reactions were stopped by phenol-chloroform extraction. The DNA products were precipitated with ethanol, resuspended in water, and separated on denaturing 16% polyacrylamide gels. Gels were analyzed with a STORM 840 PhosphorImager (Molecular Dynamics) and quantified with Image Quant software.
Inhibition of viral replication and cytotoxicity assays.
The origin of viruses and the techniques used for measuring inhibition of virus replication have been previously described (43). Briefly, with MT-4 cells, determination of antiviral activity of the SP compounds was based on a reduction of HIV-1 strain IIIB-induced cytopathogenicity, the metabolic activity of the cell being measured by the property of mitochondrial dehydrogenase to reduce yellow 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) to blue formazan. For CEM-SS cells, the production of HIV-1 strain LAI was measured by quantification of the RT activity associated with the virus particles released in the culture supernatant (38). Infected MT-4 and CEM-SS cells were cultured in the presence of different concentrations of SP compounds for 5 days before virus production determination. The 50% inhibitory concentration (IC50) was derived from the computer-generated median effect plot of the dose/effect data. In parallel experiments, cytotoxicity of the SP compounds was measured by the MTT test with uninfected cells after a 5-day incubation. The 50% cytotoxic concentration (CC50) is the concentration at which the optical density at 540 nm was reduced by 50%.
RESULTS
Polymerase activity.
The effect of the SP compounds (Fig. 2) on HIV-1 RT activity was tested with in vitro minus-strand “strong-stop” DNA synthesis. This DNA is the “runoff” reverse transcription product of a primer annealed to the primer binding site. In this study, we used a labeled 18-mer DNA primer and RNA1-311, which corresponds to nt 1 to 311 of HIV-1 MAL genomic RNA, as the template. In order to avoid complications due to concomitant inhibition of RNase H by some of the SP compounds (see below), an RNase H− RT was used for this assay. Twenty molecules (Fig. 2) were tested at different concentrations, and minus-strand “strong-stop” DNA, which corresponds to a 178-nucleotide primer extension in the MAL isolate, was quantified after separation of the products by denaturing polyacrylamide gel electrophoresis.
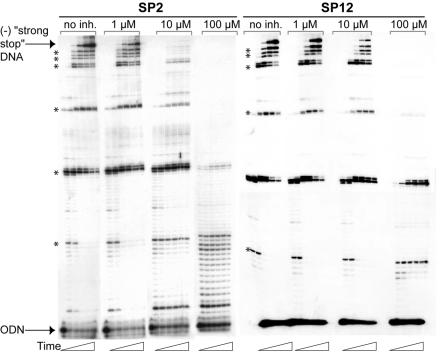
Figure 3 shows an example of gels obtained with the SP2 and SP12 compounds. In the absence of SP compounds, minus-strand “strong-stop” DNA synthesis was efficient. When adding SP2, minus-strand “strong-stop” DNA synthesis was already inhibited by >50% at 1 μM, drastically reduced at 10 μM, and undetectable at 100 μM (Fig. 3). These experiments revealed that inhibition of DNA synthesis was sequence independent. Synthesis of intermediate products was also inhibited, with the longer products disappearing first, at the expense of the shorter ones. At 100 μM SP2, only very short products were synthesized, and the product pattern was reminiscent of distributive polymerization.
FIG. 3.
Inhibition of minus-strand “strong-stop” DNA synthesis by SP2 and SP12. Radiolabeled ODN/RNA1-311 complex (10 nM) was extended with 30 nM HIV-1 RNase H− RT. DNA synthesis was performed in the absence (no inhibitor [inh.]) or presence of 1 μM, 10 μM, or 100 μM of SP 2 or SP12 for 0 or 30 s or 1, 5, 10, 20, or 30 min. The minus-strand (−) strong-stop product is labeled, and the pausing sites are indicated by asterisks.
The IC50 for each compound was determined by using the 30-min data set (Table 1). The parental SP15 compound was not the best inhibitor of the polymerase activity. Four monosubstituted compounds, SP2, SP3, SP5, and SP46, inhibited minus-strand “strong-stop” DNA synthesis at low micromolar or submicromolar concentrations, indicating that some R1 groups favor interaction of 3,7-dihydroxytropolones with RT. The monosubstituted compounds were generally more potent inhibitors of minus-strand “strong-stop” DNA synthesis than the disubstituted compounds (Table 1). Five pairs of mono- and disubstituted compounds with identical substituents could be directly compared. In three pairs (SP47/SP7, SP4/SP8, and SP5/SP45), the monosubstituted compound was 20- to 60-fold more efficient than its disubstituted counterpart, while for the other pairs (SP46/SP9 and SP2/SP48) the difference was only two- to fivefold.
TABLE 1.
IC50s of the SP compounds on the polymerase and RNase H activities of HIV-1 RT and on the 3′ processing and transfer activities of HIV-1 IN
| Inhibitor | IC50 (μM [95% confidence interval])a
|
|||
|---|---|---|---|---|
| RT
|
IN
|
|||
| Polymerase | RNase H | 3′ Processing | Transfer | |
| Parent compound | ||||
| SP15 | 3.2 (2.6-3.9) | 20 (5.2-76) | 40 (11-140) | 11 (6.9-17) |
| Monosubstituted | ||||
| SP2 | 0.66 (0.34-1.3) | 14 (3.5-59) | 13 (9.6-18) | 2.6 (0.62-11) |
| SP47 | 2.3 (0.89-6.2) | 1.3 (0.6-3.1) | 0.71 (0.37-1.3) | 0.78 (0.30-2.0) |
| 0.36 (0.29-0.46)* | ||||
| SP4 | 2.7 (0.76-9.6) | 58 (38-90) | 13 (7.1-24) | 120 (7.1-210) |
| SP5 | 1.1 (0.47-2.8) | 18 (7.1-46) | 11 (7.6-16) | 5.3 (0.91-31) |
| SP46 | 0.57 (0.36-0.92) | 12 (5.6-28) | 0.57 (0.39-0.84) | 1.2 (0.44-3.5) |
| 0.25 (0.21-0.28)* | ||||
| SP1 | 20 (7.4-56) | 330 (91-1,200) | 200 (98-410) | 17 (6.4-46) |
| SP3 | 1.6 (0.39-6.9) | 59 (18-190) | 29 (4.1-210) | 7.1 (1.0-49) |
| SP6 | 25 (4.7-130) | 32 (25-43) | 54 (40-72) | 18 (6.2-50) |
| Disubstituted | ||||
| SP48 | 3.2 (0.53-18.7) | 46 (25-83) | 2.3 (1.7-3.2) | 3.1 (1.1-8.6) |
| SP7 | 51 (5.6-470) | 15 (6.2-35) | 3.2 (1.8-5.5) | 3.7 (0.74-18) |
| 1.2 (0.56-2.5)* | ||||
| SP8 | 170 (43-640) | 550 (58-5,200) | 8.8 (5.0-15) | 3.8 (1.2-12) |
| SP45 | 52 (27-99) | 900 (470-1,700) | 0.15 (0.075-0.28) | 1.7 (0.60-4.7) |
| SP9 | 1.2 (0.42-3.6) | 4.7 (1.2-18) | 3.1 (2.2-4.5) | 1.1 (0.23-5.4) |
| 1.4 (0.90-2.0)* | ||||
| C-7 modified | ||||
| SP10 | >1,000 | >1,000 | >1,000 | >1,000 |
| SP11 | >1,000 | >1,000 | >1,000 | >1,000 |
| Acetylated | ||||
| SP12 | 7.6 (6.9-8.3) | 65 (45-93) | 90 (24-340) | 62 (15-270) |
| SP20 | 30 (8.0-110) | 1,000 (980-1,100) | >1,000 | NDb |
| O-methylated | ||||
| SP13 | 190 (52-710) | 730 (200-2,600) | >1,000 | >1,000 |
| SP14 | 360 (220-590) | >1,000 | 700 (110-4,400) | >1,000 |
IC50s and 95% confidence intervals were determined from a single fit when the IC50 was >10 μM or from at least three measurements when the IC50 was <10 μM. All data were obtained with MgCl2, except those indicated by an asterisk, which were obtained with 10 mM MnCl2.
ND, not determined.
Several compounds were inactive or weakly active against the polymerase activity of HIV-1 RT. Notably, the C-7-modified tropolones (SP10 and SP11), which have only two adjacent oxygens on the seven-membered ring, had no activity at 1 mM, and the O-methylated 3,7-dihydroxytropolones (SP13 and SP14) were only weakly active. These results were in keeping with our hypothesis that hydroxytropolones might inhibit the polymerase activity by interfering with the Mg2+ ions in the active site.
SP12 and SP20 inhibited minus-strand “strong-stop” DNA synthesis, even though they were less potent inhibitors than SP15 (Fig. 3 and Table 1). For instance, SP20 was 10-fold less efficient than the corresponding nonacetylated compound, SP15. As for SP2, a sequence-independent inhibition pattern was observed (Fig. 3). Inhibition of the RT polymerase activity by SP12 and SP20 was unexpected, as the hydroxyl groups that are supposed to chelate the magnesium ions are acetylated in these compounds (Fig. 2). The first pKa of SP15 is 6.7, and deprotonation of a hydroxyl group results in increased chelating potency (42). Accordingly, acetylated hydroxytropolones were inactive against inositol monophosphatase (41, 42). Thus, our results raised the possibility that some or all SP compounds inhibit the RT polymerase activity by an unpredicted mechanism.
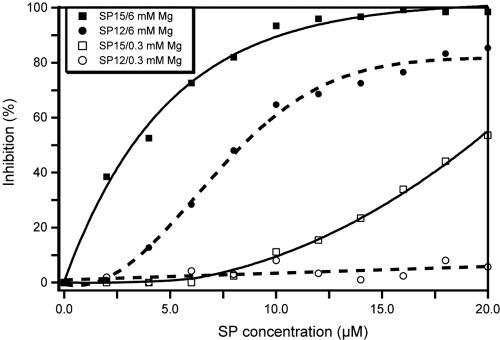
To address this question and to gain insight into the role of Mg2+ chelation in the inhibition process, we studied the inhibition of minus-strand “strong-stop” DNA synthesis by SP12 and SP15 at two different magnesium concentrations (Fig. 4). At 6 mM Mg2+, the IC50 of SP15 was 3.2 μM, while SP12 was about twofold less active (IC50 of 7.6 μM). Interestingly, inhibition potency of these compounds dramatically decreased at low magnesium concentration: at 0.3 mM Mg2+, the IC50 of SP15 was 19 ± 1 μM, while inhibition of minus-strand “strong-stop” DNA synthesis by SP12 was hardly detectable (Fig. 4). These results suggest that the SP compounds can bind the RT polymerase active site only in the presence of Mg2+ ions, in agreement with an inhibition mechanism involving chelation of the catalytic ions.
FIG. 4.
Effect of Mg2+ concentration on inhibition of minus-strand “strong-stop” DNA synthesis by SP15 and SP12. Minus-strand “strong-stop” DNA synthesis was performed for 30 min in the presence of 6 mM or 0.3 mM Mg2+, in the absence or presence of increasing concentrations of SP12 or SP15.
RNase H activity.
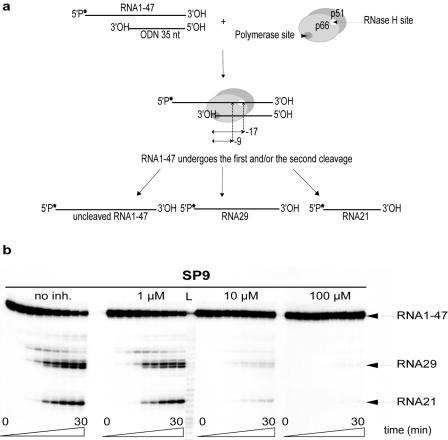
Two modes of RNase H cleavage have been described: a polymerase-dependent cleavage (21), directed by the 3′ end of the DNA strand, and a polymerase-independent mechanism guided by the 5′ end of the RNA template (48). The first mode of cleavage is thought to occur in concert with DNA polymerization to degrade the genomic RNA during minus-strand DNA synthesis, while the second may contribute to the degradation of larger genomic RNA fragments left after DNA 3′-end-directed cleavage and may also play a role in the formation and removal of the polypurine tract. During polymerase-dependent cleavage, the polymerase active site of the enzyme is positioned at the recessed 3′ end of the DNA strand (Fig. 5a). The RNase H active site is positioned approximately 17 nt from the polymerase active site, and its position from the 3′ end of the DNA determines the position of the primary cleavage. RT then repositions and makes a secondary cleavage 7 to 9 nt from the recessed 3′ end of the DNA (Fig. 5a). We tested this mode of cleavage using RNA1-47 as viral RNA and ODN35 as DNA.
FIG. 5.
Inhibition of RNase H activity by 3,7-dihydroxytropolones. (a) Experimental design and expected products of polymerase-dependent RNase H activity. In this mode of cleavage, the polymerase active site is positioned at the 3′ end of the DNA primer (ODN35) and determines the position of the cleavage 17 nt downstream, yielding RNA29. RT is also able to cut 9 nt from the 3′ end of the DNA strand, producing RNA21. The 32P-labeled 5′ end of RNA1-47 is indicated by an asterisk. (b) Inhibition of RNase H activity by SP9. The reaction was initiated by the addition of 10 nM HIV-1 RT to 10 nM of RNA1-47/ODN35 complex, in the absence (no inhibitor [inh.]) or presence of 1 μM, 10 μM, or 100 μM of SP9. The reaction was stopped after 0, 15, or 30 s or 1, 3, 5, 10, 20, or 30 min. Lane L corresponds to a RNA ladder that was used to determine the sizes of the products.
As shown in Fig. 5b, both the primary (RNA29) and the secondary (RNA21) cleavage products accumulated over time in the absence of SP compounds. In addition, weaker primary cuts generated fragments 28 and 32 nt in length (Fig. 5b). However, all degradation products progressively disappeared as the SP9 concentration was increased. Formation of the two major products was slightly slower at 1 μM, strongly inhibited at 10 μM, and undetectable at 100 μM (Fig. 5b). Interestingly, the two cleavages were inhibited with the same efficiency. The SP compounds were tested for inhibition of the polymerase-dependent RNase H activity, and the results are summarized in Table 1.
The best inhibitors of RNase H were the monosubstituted 3,7-dihydroxytropolone SP47 and the disubstituted compound SP9. When comparing mono- and disubstituted 3,7-dihydroxytropolones with identical substituents, the monosubstituted compounds were 3.3- to 50-fold-more-potent RNase H inhibitors, with the exception of the SP46/SP9 pair, in which the disubstituted compound was slightly more active. In addition, all SP compounds were more potent against polymerase than against RNase H activity, except SP47 and SP7, suggesting that the meta-nitrophenyl group may form specific interactions in the RNase H active site. Several molecules had a rather good specificity for the polymerase activity: SP2, SP3, SP4, and SP46 were at least 20-fold more efficient against polymerase than against RNase H (Table 1). Interestingly, these compounds are all monosubstituted 3,7-dihydroxytropolones. By contrast, none of the tested compounds displayed a high specificity for the RNase H activity, and only SP47 and SP9 were significantly more efficient than the parent compound.
The C-7-modified tropolones SP10 and SP11 and the methylated 3,7-dihydroxytropolones SP13 and SP14, which did not significantly inhibit the RT polymerase activity, displayed little or no activity against RNase H at 1 mM. Interestingly, the acetylated dihydroxytropolones SP12 and SP20, which were moderate polymerase inhibitors, were weaker RNase H inhibitors (Table 1). In view of the results obtained previously with inositol monophosphatase (41, 42), these data suggest that hydroxytropolones inhibit the RT RNase H activity by binding the Mg2+ ions in the active site.
Integrase activities.
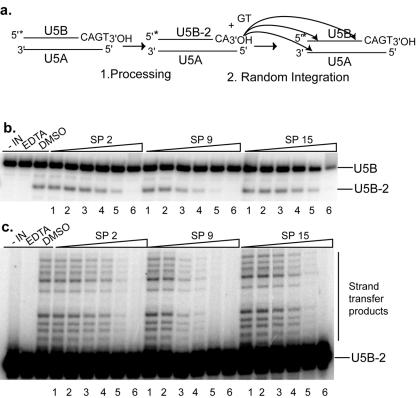
The structures of the HIV-1 RNase H domain and the IN core present strong similitude, and their enzymatic mechanisms are probably also closely related (16, 50). Thus, it was interesting to test the effects of the SP compounds on the 3′ processing and strand transfer activities of HIV-1 IN. To this end, we constructed a DNA duplex corresponding to the U5 region of the 3′ long terminal repeat of the proviral DNA (Fig. 6a). Both reactions consist in a nucleophilic attack of a phosphodiester bond by a hydroxyl group and require either Mg2+ or Mn2+ as a cofactor. During the processing reaction, the enzyme removes two 3′ nucleotides at each terminus of the proviral DNA, resulting in overhanging CA ends (Fig. 6a). In the following strand transfer reaction, the 3′ processed end performs a nucleophilic attack on a phosphodiester bond of the target DNA (6). In the in vitro assay with purified IN, the preprocessed 3′ end is randomly integrated in the target duplex (Fig. 6a).
FIG. 6.
Inhibition of the 3′-end processing and strand transfer activities of HIV-1 IN by 3,7-dihydroxytropolones. (a) Experimental design and expected products of the 3′-end processing and strand transfer reactions. The 3′-end processing removes 2 nt at the 3′ end of U5B, yielding U5B-2. During strand transfer, U5B-2 is randomly integrated into a homologous strand or into U5A, yielding high-molecular-weight products. The 5′-radiolabeled end of U5B is indicated by an asterisk. (b) Dose-response effect of SP2, SP9, and SP15 on IN 3′ processing activity. The 5′-end-labeled oligonucleotide U5B annealed to U5A was incubated for 1 h at 37°C with 100 nM IN. (c) Inhibition of IN transfer activity by SP2, SP9, and SP15. The 5′-end-labeled oligonucleotide U5B-2 annealed to U5A was incubated for 1 h at 37°C with 100 nM IN. In panels b and c, lane “−IN” is a control without integrase, lane “EDTA” is a negative control containing 20 mM EDTA, and lane “DMSO” is a control with dimethyl sulfoxide but without SP compounds. Lanes 1 to 6 correspond, respectively, to 0.3 μM, 1 μM, 3 μM, 10 μM, 30 μM, and 100 μM SP2, SP9, or SP15.
3′-end processing.
Figure 6b shows the results of 3′-end processing inhibition by SP2, SP9, and SP15. Processing of the 21-mer duplex liberates the 3′-terminal dinucleotide GT and the processed U5B oligonucleotide (U5B-2). IN activity was assayed in the presence of increasing SP concentrations. As shown by 15% polyacrylamide gel autoradiography, the addition of increasing concentrations of SP2, SP9, and SP15 led to significant inhibition of 3′-end processing (Fig. 6b), yielding IC50 values of 13 μM, 3.1 μM, and 40 μM, respectively. The IC50 values for inhibition of the 3′-end processing are summarized in Table 1.
There was no clear-cut difference between mono- and disubstituted 3,7-dihydroxytropolones regarding the 3′-end processing activity. Two monosubstituted (SP47 and SP46) and one disubstituted (SP45) compound inhibited 3′-end processing at submicromolar concentrations. However, all disubstituted 3,7-dihydroxytropolones had IC50s lower than 10 μM, while the IC50s of most monosubstituted SP compounds were greater than 10 μM (Table 1). The methylated dihydroxytropolones SP13 and SP14 and the C-7-modified tropolone SP10 were completely inactive, whereas SP11 displayed a very low activity. The acetylated compound SP12 was a rather poor inhibitor (IC50 of 90 μM) of the 3′ processing activity, while SP20 was totally inactive.
In vitro, IN can use either Mg2+ or Mn2+ ions as a cofactor, even though Mg2+ is most likely the cation of biological relevance. Thus, we compared the levels of inhibition of 3′-end processing by two mono- and two disubstituted SP compounds in buffers containing 10 mM of either MgCl2 or MnCl2. In all cases, the SP compounds were 2- to 2.6-fold more potent with Mn2+ than with Mg2+ (Table 1).
Strand transfer.
The inhibitory effect of SP compounds on the strand transfer reaction was assayed using a preprocessed duplex (U5B-2/U5A) as the IN substrate. Monosubstituted SP47 and SP46 and disubstituted SP45 and SP9 were potent inhibitors of the strand transfer reaction (IC50 of <2 μM) (Fig. 6c and Table 1). Mono- and disubstituted 3,7-dihydroxytropolones with identical substituents had similar IC50s, with the exception of the SP4/SP8 pair, in which the monosubstituted SP4 compound was poorly active (Table 1). SP4 was an exception, as all other 3,7-dihydroxytropolones with free hydroxyl groups had IC50s smaller than 20 μM. On the other hand, the C-7-modified tropolones (SP10 and SP11) and the methylated 3,7-dihydroxytropolones (SP13 and SP14) did not inhibit strand transfer (Table 1). The acetylated SP12 was sixfold less active than the parent compound, SP15.
Biochemical data and modeling indicated that the IN active site adopts different conformations during the two catalytic steps of the integration process (6, 18) and that it is possible to find step-specific IN inhibitors (12, 25). Interestingly, our data showed that SP1 is a rather good inhibitor of the transfer step (IC50 of 17 μM) but a very poor inhibitor of 3′-end processing (Table 1). Conversely, SP45 preferentially inhibited the 3′-end processing step, indicating that substituted 3,7-dihydroxytropolones can be selective inhibitors of the 3′-end processing or of the transfer reaction. Nevertheless, most of the SP compounds were equally efficient with both IN activities (SP5, SP7, SP8, SP9, SP12, SP46, SP47, and SP48).
Cytotoxicity assays and inhibition of viral replication.
The inhibitory and cytotoxic effects of SP molecules were studied with human lymphoblastoid CEM-SS and MT-4 cells infected with HIV-1 LAI and HIV-1 IIIB strains, respectively. In the HIV-1 IIIB/MT-4 cell culture assay, SP compounds were toxic in the micromolar-concentration range, displaying CC50 values from 3 to 30 μM (Table 2). In this assay, no SP compound displayed antiviral activity at concentrations below its CC50 (IC50 > CC50). As a control, zidovudine was active in the low nanomolar range, without significant cytotoxicity (Table 2).
TABLE 2.
Inhibition of HIV-1 replication (IC50) and cytotoxicity (CC50) of selected SP compounds on CEM-SS and MT-4 cell lines
| Inhibitor | HIV-1 LAI/CEM-SS
|
HIV-1 IIIB/MT-4
|
||
|---|---|---|---|---|
| IC50 (μM) | CC50 (μM) | IC50 (μM) | CC50 (μM) | |
| SP1 | 39 | 64 | >CC50 | 30 |
| SP2 | 3 | 7 | >CC50 | 3 |
| SP3 | 3 | 11 | >CC50 | 3 |
| SP4 | 5 | 9 | >CC50 | 5 |
| SP5 | 10 | 20 | >CC50 | 8 |
| SP7 | 6 | 7.5 | >CC50 | 43 |
| SP9 | 3 | 6 | >CC50 | 5 |
| SP10 | 40 | 48 | >CC50 | 29 |
| SP12 | 5 | 7 | >CC50 | 6 |
| SP15 | 0.3 | 9 | >CC50 | 3 |
| AZTa | 0.004 | 0.01 | ||
AZT, zidovudine.
In the HIV-1 LAI/CEM-SS cell culture system, the CC50 values of the SP compounds were slightly lower than with the MT-4 cells and ranged from 3 to 43 μM. In addition, in this cell line, SP compounds inhibited replication of HIV-1 LAI with IC50s ranging from 0.3 to 40 μM. However, for most compounds the IC50 was only two- to threefold lower than the CC50 (Table 2). Thus, we cannot exclude that the antiviral effects we observed were indeed a direct consequence of the cytotoxicity of the SP compounds. For instance, the inhibition of HIV LAI replication by SP10 (IC50 of 40 μM) was likely due to its toxicity (CC50), since this C-7-modified tropolone was inactive against all RT and IN enzymatic activities. However, it remains possible that some of the SP compounds have a real antiviral activity, especially SP15 and SP3, which have IC50 values 30- and 3.7-fold lower, respectively, than their CC50s.
DISCUSSION
The high incidence of resistance in therapy-experienced and newly infected patients and the adverse effects of the existing treatments underscore the need for new HIV-1 inhibitors (11, 17, 32, 51). The RT polymerase and RNase H activities as well as the IN 3′-end processing and strand transfer activities all require divalent cations and are essential for viral replication. While several inhibitors of RT polymerase activity are in the clinic and inhibitors of IN strand transfer activity have been tested in clinical trials (9), RNase H and 3′-end processing inhibitors are only at the earlier steps of the drug discovery pipeline.
In this study, we tested the inhibitory properties of 3,7-dihydroxytropolones on four enzymatic activities. Our choice was motivated by the observation that these compounds inhibit inositol monophosphatase by binding the two Mg2+ ions in the catalytic site (41, 42). Two Mg2+ ions separated by 3.57 Å are present in the polymerase active site of RT in complex with a primer/template DNA duplex and an incoming nucleotide (27), and a pair of metal ions has also been observed in some crystal structures of the RNase H domain (8) and IN (4, 5, 49).
Inhibitors, including some with marked specificity, were identified for each of the four enzymatic activities tested. Interestingly, the parental compound, SP15, was never the best inhibitor, indicating that the efficiency of 3,7-dihydroxytro- polones can be improved by aromatic groups at positions 4 and/or 6 of the tropolone ring. Some of the 3,7-dihydroxytropolones we tested inhibited the RT polymerase and the IN 3′-end processing and strand transfer activities at submicromolar concentrations, while these compounds were generally less potent against RNase H. To our knowledge, this is the first rational attempt to inhibit HIV-1 RT by simultaneously binding the two catalytic ions. α,γ-Diketo acids (47) and dihydroxypyrimidinecarboxylic acid (46) were recently shown to inhibit the RNA-dependent RNA polymerase from hepatitis C virus, but whether these compounds interact with one or both metal ions in the polymerase active site is not clear.
The 4-monosubstituted 3,7-dihydroxytropolone SP47 and the 4,6-disubstituted 3,7-dihydroxytropolone SP9 inhibited HIV-1 RT RNase H activity at low micromolar concentrations. This compares favorably with previously identified RNase H inhibitors such as ilimaquinone (IC50 of ∼20 μM) (34), N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone (BBNH) (IC50 of 3.5 μM) (2), and the new RNase H-specific thiophene diketo acid {4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid} (IC50 of 3.2 μM) (45). Interestingly, both BBNH and the thiophene diketo acid potentially inhibit RNase H activity by chelating one metal ion in this active site. Finally, a paper submitted at the same time as this paper showed that β-thujaplicinol, a natural monohydroxytropolone derivative, strongly inhibits HIV-1 and HIV-2 RNase H, while it is a weak inhibitor of Escherichia coli and human RNase H (3). This result further stresses the potential of hydroxytropolones as HIV inhibitors.
Several of the 3,7-dihydroxytropolones we tested were potent inhibitors of the 3′-end processing and strand transfer activities of HIV-1 IN (IC50 of <2 μM). By comparison, diketo acids inhibit IN at submicromolar concentrations (25), and styrylquinoline derivatives have IC50 values between 0.5 and 5 μM (12). However, while diketo acids preferentially inhibit strand transfer, most of the tested 3,7-dihydroxytropolones equally inhibited the two IN activities, with the noticeable exception of SP1, which selectively inhibits strand transfer, and SP45, which preferentially inhibits 3′-end processing.
While some of the 3,7-dihydroxytropolones preferentially inhibited one of the four enzymatic activities, some had little or no specificity (e.g., SP6, SP9, and SP47). It seems, however, unlikely that these compounds inhibit RT and IN by binding to their nucleic acid substrates. Indeed, 3,7-dihydroxytropolones are negatively charged at physiological pH (∼1.5 net negative charge at pH 7), and the uncharged O-methylated 3,7-dihydroxytropolones, which are more likely to bind nucleic acids, are completely inactive. In addition, by using surface plasmon resonance, Budihas et al. (3) showed that natural tropolone derivatives do not bind nucleic acids. In line with these results, we found no evidence of 3,7-dihydroxytropolone binding to DNA/DNA and RNA/DNA duplexes by band-shift and fluorescence competition assays (J. Didierjean et al., unpublished data).
The data presented here suggest that 3,7-dihydroxytropolones inhibit DNA polymerase, RNase H, and IN activities by chelating the Mg2+ ions of these active sites, even though more detailed studies will be required to prove this point. The fact that the C-7-modified tropolones (SP10 and SP11) and the O-methylated 3,7-dihydroxytropolones (SP13 and SP14) were inactive against these four enzymatic activities suggests that the inhibitors simultaneously bind two metal ions. Acetylated dihydroxytropolones have lower affinity for Mg2+ than do their unmodified counterparts (42). Accordingly, acetylated compounds (SP12 and SP20) had no or limited activity against RNase H and IN activities and were also inactive against inositol monophosphatase (41, 42). These compounds were fairly active against the RT polymerase activity, but SP20 was 10-fold less active than the corresponding parent compound (SP15). In the RNase H site, the 4-Å distance between the metal ions (8) is not ideal for interaction with tropolones, in contrast with the polymerase active site (27). Therefore, acetylated dihydroxytropolones might be able to interact with the metal ions only in the polymerase active site. Alternatively, only one Mg2+ ion might be present in the RNase H site (27), preventing binding of the weaker ligands.
Inhibition of DNA synthesis by acetylated and unmodified 3,7-dihydroxytropolones (SP12 and SP15) was dramatically reduced at low Mg2+ concentrations, in keeping with the chelation mechanism. We propose that binding of Mg2+ in the catalytic site is required to bind 3,7-dihydroxytropolones and that these compounds compete with one or several oxygens of the catalytic amino acid side chains for Mg2+ chelation. Thus, 3,7-dihydroxytropolones should be more efficient at high (saturating) Mg2+ concentrations. Crystallographic studies showed that binding of two Mg2+ ions in the polymerase site requires binding of the incoming dNTP (14, 27). One ion is bound to the β and γ phosphates of dNTP and enters with it in the catalytic site; its KD is about 0.1 mM. The second ion is bound in a subsequent step, and recent studies showed that its KD is in the millimolar range (13). Thus, high-millimolar Mg2+ concentrations are required to saturate the polymerase active site and to reach maximal inhibition by 3,7-dihydroxytropolones.
When we compared levels of inhibition of the IN 3′-end processing activity between MgCl2 and MnCl2, we observed that the SP compounds were slightly more efficient in the presence of Mn2+. At first sight, one could expect strongly decreased inhibition with MnCl2, as the oxygen atoms and hence 3,7-dihydroxytropolones have much weaker affinity for Mn2+ than for Mg2+. However, as mentioned above, we think there is competition between 3,7-dihydroxytropolones and the side chains of the catalytic amino acids of IN (D64, D116, and E152) for binding of the metal ions. As these amino acids and the SP inhibitors both bind the metal ions via oxygen atoms, they should bind Mn2+ ions less tightly than Mg2+. However, their relative affinity for Mn2+ and Mg2+ should be the same and we expected little difference in inhibition of the SP compounds with MgCl2 and MnCl2. In fact, our results are consistent with those obtained with diketo acids on the IN strand transfer activity. The diketo acids that bind metal ions solely via oxygen atoms have similar activities in the presence of Mg2+ and Mn2+, while those binding metal ions via nitrogen atoms were at least 1 order of magnitude more active with MnCl2 (23). Obviously, even though our results suggest that SP compounds inhibit HIV-1 RT and IN by chelation of the metal ions in their catalytic sites, additional studies will be required to demonstrate this mechanism.
Whereas the tests with purified enzymes showed that 3,7-dihydroxytropolones have interesting potential as HIV-1 inhibitors, cell-based assays indicated that their cytotoxicity represents a major challenge. This toxicity is most likely due to the inhibition of cellular bimetallic enzymes. Given the number of enzymes that use metal ions for catalysis, it could be argued that it will be impossible to find 3,7-dihydroxytropolones that specifically inhibit HIV-1 enzymes. In this respect, the diketo acid derivatives constitute an interesting example. These compounds were first identified by high-throughput screening for IN inhibitors (25). Some of these compounds block HIV-1 replication by specifically inhibiting the IN strand transfer activity and exhibit low toxicity (24-26), even though they act by binding one of the metal ions in the catalytic site (23) and could potentially interfere with a number of cellular enzymes. Recently, other members of the diketo acid family have been shown to specifically inhibit HIV-1 RNase H (45) and hepatitis C virus RNA polymerase (47). These examples show that inhibitors that interfere with metal ions in the enzyme catalytic sites can be specific. Along these lines, 3,7-dihydroxytropolone constitutes a very interesting platform allowing a number of very different derivatives to be synthesized. In addition, modeling and structural studies of the enzyme/3,7-dihydroxytropolone complexes should allow rational design of more-potent inhibitors that might also be more specific.
Acknowledgments
We thank Chantal and Bernard Ehresmann for fruitful discussions and for their constant interest during this work and Philippe Walter for the gift of RT.
This work was supported by grants from the Agence Nationale de Recherches sur le SIDA (ANRS) to R.M. and from Sidaction to S.R.P. F.Q. is a predoctoral fellow of the ANRS.
REFERENCES
- 1.Arts, E. J., and S. F. Le Grice. 1998. Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Prog. Nucleic Acid Res. Mol. Biol. 58:339-393. [DOI] [PubMed] [Google Scholar]
- 2.Borkow, G., R. S. Fletcher, J. Barnard, D. Arion, D. Motakis, G. I. Dmitrienko, and M. A. Parniak. 1997. Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert- butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 36:3179-3185. [DOI] [PubMed] [Google Scholar]
- 3.Budihas, S. R., I. Gorshkova, S. Gaidamakov, A. Wamiru, M. K. Bona, M. A. Parniak, R. J. Crouch, J. B. McMahon, J. A. Beutler, and S. F. Le Grice. 2005. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33:1249-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujacz, G., J. Alexandratos, A. Wlodawer, G. Merkel, M. Andrake, R. A. Katz, and A. M. Skalka. 1997. Binding of different divalent cations to the active site of avian sarcoma virus integrase and their effects on enzymatic activity. J. Biol. Chem. 272:18161-18168. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. C., J. Krucinski, L. J. Miercke, J. S. Finer-Moore, A. H. Tang, A. D. Leavitt, and R. M. Stroud. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. USA 97:8233-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, T. K., and D. R. Davies. 2004. Structure and function of HIV-1 integrase. Curr. Top. Med. Chem. 4:965-977. [DOI] [PubMed] [Google Scholar]
- 7.Cirino, N. M., C. E. Cameron, J. S. Smith, J. W. Rausch, M. J. Roth, S. J. Benkovic, and S. F. Le Grice. 1995. Divalent cation modulation of the ribonuclease functions of human immunodeficiency virus reverse transcriptase. Biochemistry 34:9936-9943. [DOI] [PubMed] [Google Scholar]
- 8.Davies, J. F., II, Z. Hostomska, Z. Hostomsky, S. R. Jordan, and D. A. Matthews. 1991. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252:88-95. [DOI] [PubMed] [Google Scholar]
- 9.De Clercq, E. 2004. Antiviral drugs in current clinical use. J. Clin. Virol. 30:115-133. [DOI] [PubMed] [Google Scholar]
- 10.De Clercq, E. 2004. HIV-chemotherapy and -prophylaxis: new drugs, leads and approaches. Int. J. Biochem. Cell Biol. 36:1800-1822. [DOI] [PubMed] [Google Scholar]
- 11.Demeter, L. M., H. J. Ribaudo, A. Erice, S. H. Eshleman, S. M. Hammer, N. S. Hellmann, and M. A. Fischl. 2004. HIV-1 drug resistance in subjects with advanced HIV-1 infection in whom antiretroviral combination therapy is failing: a substudy of AIDS Clinical Trials Group Protocol 388. Clin. Infect. Dis. 39:552-558. [DOI] [PubMed] [Google Scholar]
- 12.Deprez, E., S. Barbe, M. Kolaski, H. Leh, F. Zouhiri, C. Auclair, J. C. Brochon, M. Le Bret, and J. F. Mouscadet. 2004. Mechanism of HIV-1 integrase inhibition by styrylquinoline derivatives in vitro. Mol. Pharmacol. 65:85-98. [DOI] [PubMed] [Google Scholar]
- 13.Deval, J., K. Alvarez, B. Selmi, M. Bermond, J. Boretto, C. Guerreiro, L. Mulard, and B. Canard. 2005. Mechanistic insights into the suppression of drug resistance by human immunodeficiency virus type 1 reverse transcriptase using alpha-boranophosphate nucleoside analogs. J. Biol. Chem. 280:3838-3846. [DOI] [PubMed] [Google Scholar]
- 14.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, Jr., A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284:1095-1111. [DOI] [PubMed] [Google Scholar]
- 15.Ding, J., S. H. Hughes, and E. Arnold. 1997. Protein-nucleic acid interactions and DNA conformation in a complex of human immunodeficiency virus type 1 reverse transcriptase with a double-stranded DNA template-primer. Biopolymers 44:125-138. [DOI] [PubMed] [Google Scholar]
- 16.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981-1986. [DOI] [PubMed] [Google Scholar]
- 17.Dykes, C., J. Najjar, R. J. Bosch, M. Wantman, M. Furtado, S. Hart, S. M. Hammer, and L. M. Demeter. 2004. Detection of drug-resistant minority variants of HIV-1 during virologic failure of indinavir, lamivudine, and zidovudine. J. Infect. Dis. 189:1091-1096. [DOI] [PubMed] [Google Scholar]
- 18.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52:319-333. [DOI] [PubMed] [Google Scholar]
- 19.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 96:13040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 95:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89:10763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotte, M., X. Li, and M. A. Wainberg. 1999. HIV-1 reverse transcription: a brief overview focused on structure-function relationships among molecules involved in initiation of the reaction. Arch. Biochem. Biophys. 365:199-210. [DOI] [PubMed] [Google Scholar]
- 23.Grobler, J. A., K. Stillmock, B. Hu, M. Witmer, P. Felock, A. S. Espeseth, A. Wolfe, M. Egbertson, M. Bourgeois, J. Melamed, J. S. Wai, S. Young, J. Vacca, and D. J. Hazuda. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. USA 99:6661-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, Jr., M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. Jin, I.-W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 101:11233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 26.Hazuda, D. J., S. D. Young, J. P. Guare, N. J. Anthony, R. P. Gomez, J. S. Wai, J. P. Vacca, L. Handt, S. L. Motzel, H. J. Klein, G. Dornadula, R. M. Danovich, M. V. Witmer, K. A. Wilson, L. Tussey, W. A. Schleif, L. S. Gabryelski, L. Jin, M. D. Miller, D. R. Casimiro, E. A. Emini, and J. W. Shiver. 2004. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science 305:528-532. [DOI] [PubMed] [Google Scholar]
- 27.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 28.Kaushik, N., N. Rege, P. N. Yadav, S. G. Sarafianos, M. J. Modak, and V. N. Pandey. 1996. Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Biochemistry 35:11536-11546. [DOI] [PubMed] [Google Scholar]
- 29.Klumpp, K., J. Q. Hang, S. Rajendran, Y. Yang, A. Derosier, P. Wong Kai In, H. Overton, K. E. Parkes, N. Cammack, and J. A. Martin. 2003. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 31:6852-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leh, H., P. Brodin, J. Bischerour, E. Deprez, P. Tauc, J. C. Brochon, E. LeCam, D. Coulaud, C. Auclair, and J. F. Mouscadet. 2000. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry 39:9285-9294. [DOI] [PubMed] [Google Scholar]
- 31.Lins, R. D., T. P. Straatsma, and J. M. Briggs. 2000. Similarities in the HIV-1 and ASV integrase active sites upon metal cofactor binding. Biopolymers 53:308-315. [DOI] [PubMed] [Google Scholar]
- 32.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 33.Long, Y. Q., X. H. Jiang, R. Dayam, T. Sanchez, R. Shoemaker, S. Sei, and N. Neamati. 2004. Rational design and synthesis of novel dimeric diketoacid-containing inhibitors of HIV-1 integrase: implication for binding to two metal ions on the active site of integrase. J. Med. Chem. 47:2561-2573. [DOI] [PubMed] [Google Scholar]
- 34.Loya, S., and A. Hizi. 1993. The interaction of illimaquinone, a selective inhibitor of the RNase H activity, with the reverse transcriptases of human immunodeficiency and murine leukemia retroviruses. J. Biol. Chem. 268:9323-9328. [PubMed] [Google Scholar]
- 35.Maignan, S., J. P. Guilloteau, Q. Zhou-Liu, C. Clement-Mella, and V. Mikol. 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 282:359-368. [DOI] [PubMed] [Google Scholar]
- 36.Marquet, R., F. Baudin, C. Gabus, J. L. Darlix, M. Mougel, C. Ehresmann, and B. Ehresmann. 1991. Dimerization of human immunodeficiency virus (type 1) RNA: stimulation by cations and possible mechanism. Nucleic Acids Res. 19:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monier, P. L., and R. Wilcox. 2004. Metabolic complications associated with the use of highly active antiretroviral therapy in HIV-1-infected adults. Am. J. Med. Sci. 328:48-56. [DOI] [PubMed] [Google Scholar]
- 38.Moog, C., A. Wick, P. Le Ber, A. Kirn, and A. M. Aubertin. 1994. Bicyclic imidazo derivatives, a new class of highly selective inhibitors for the human immunodeficiency virus type 1. Antivir. Res. 24:275-288. [DOI] [PubMed] [Google Scholar]
- 39.Neamati, N., Z. Lin, R. G. Karki, A. Orr, K. Cowansage, D. Strumberg, G. C. G. Pais, J. H. Voigt, M. C. Nicklaus, H. E. Winslow, H. Zhao, J. A. Turpin, J. Yi, A. M. Skalka, T. R. Burke, Jr., and Y. Pommier. 2002. Metal-dependent inhibition of HIV-1 integrase. J. Med. Chem. 45:5661-5670. [DOI] [PubMed] [Google Scholar]
- 40.Patel, H. P., A. Jacobo-Molina, J. Ding, C. Tantillo, A. D. Clark, Jr., R. Raag, R. G. Nanni, S. H. Hughes, and E. Arnold. 1995. Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry 34:5351-5363. [DOI] [PubMed] [Google Scholar]
- 41.Piettre, S. R., C. Andre, M. C. Chanal, J. B. Ducep, B. Lesur, F. Piriou, P. Raboisson, J. M. Rondeau, C. Schelcher, P. Zimmermann, and A. J. Ganzhorn. 1997. Monoaryl- and bisaryldihydroxytropolones as potent inhibitors of inositol monophosphatase. J. Med. Chem. 40:4208-4221. [DOI] [PubMed] [Google Scholar]
- 42.Piettre, S. R., A. Ganzhorn, J. Hoflack, K. Islam, and J. M. Hornsperger. 1997. alpha-Hydroxytropolones: a new class of potent inhibitors of inositol monophosphatase and other bimetallic enzymes. J. Am. Chem. Soc. 119:3201-3204. [Google Scholar]
- 43.Puech, F., G. Gosselin, I. Lefebvre, A. Pompon, A. M. Aubertin, A. Kirn, and J. L. Imbach. 1993. Intracellular delivery of nucleoside monophosphates through a reductase-mediated activation process. Antivir. Res. 22:155-174. [DOI] [PubMed] [Google Scholar]
- 44.Schatz, O., F. V. Cromme, F. Gruninger-Leitch, and S. F. Le Grice. 1989. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 257:311-314. [DOI] [PubMed] [Google Scholar]
- 45.Shaw-Reid, C. A., V. Munshi, P. Graham, A. Wolfe, M. Witmer, R. Danzeisen, D. B. Olsen, S. S. Carroll, M. Embrey, J. S. Wai, M. D. Miller, J. L. Cole, and D. J. Hazuda. 2003. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 278:2777-2780. [DOI] [PubMed] [Google Scholar]
- 46.Summa, V., A. Petrocchi, V. G. Matassa, M. Taliani, R. Laufer, R. De Francesco, S. Altamura, and P. Pace. 2004. HCV NS5b RNA-dependent RNA polymerase inhibitors: from alpha,gamma-diketoacids to 4,5-dihydroxypyrimidine- or 3-methyl-5-hydroxypyrimidinonecarboxylic acids. Design and synthesis. J. Med. Chem. 47:5336-5339. [DOI] [PubMed] [Google Scholar]
- 47.Summa, V., A. Petrocchi, P. Pace, V. G. Matassa, R. De Francesco, S. Altamura, L. Tomei, U. Koch, and P. Neuner. 2004. Discovery of alpha, gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J. Med. Chem. 47:14-17. [DOI] [PubMed] [Google Scholar]
- 48.Wisniewski, M., M. Balakrishnan, C. Palaniappan, P. J. Fay, and R. A. Bambara. 2000. Unique progressive cleavage mechanism of HIV reverse transcriptase RNase H. Proc. Natl. Acad. Sci. USA 97:11978-11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wlodawer, A. 1999. Crystal structures of catalytic core domains of retroviral integrases and role of divalent cations in enzymatic activity. Adv. Virus Res. 52:335-350. [DOI] [PubMed] [Google Scholar]
- 50.Yang, W., and T. A. Steitz. 1995. Recombining the structures of HIV integrase, RuvC and RNase H. Structure 3:131-134. [DOI] [PubMed] [Google Scholar]
- 51.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]