Abstract
Tn7::In2-8 contains sat2-aadB-catB2(ΔattC)-dfrA1-sat2-aadA1-orfX in the variable region of a class 2 integron embedded in the Tn7-like transposon. This novel transposon was inserted in its preferred site downstream of the glms gene in Acinetobacter baumannii. Acquisition of the pseudocassette catB2 could have arisen by a secondary-site integrase-mediated intermolecular recombination event.
Integrons are elements that contain the genetic determinants of the components of a site-specific recombination system that recognizes and captures mobile gene cassettes (8).
The different integrons are classified according to their respective integrase genes. Class 2 integrons are embedded in the Tn7 family of transposons. Tn7 includes a defective integron consisting of the gene cassettes dfrA1-sat2-aadA1-orfX in its variable region (9, 16). In the GenBank, there are only six class 2 integrons described so far, Tn1825 (20), Tn4132 (24), Tn7::ISI-ere-A (2), a class 2 integron with GenBank accession no. AB161461 (1), a recently described class 2 integron in Burkholderia cenocepacia (DQ082896), and Tn7 (9). The class 2 integron integrase, which is less than 50% homologous to the IntI1 integrase, is not functional due to the presence of an internal stop codon (9). Class 2 integrons have been reported in Acinetobacter spp. isolates throughout the world (7, 14, 19).
Here, we describe a novel rearrangement of a class 2 integron, Tn7::In2-8, in three isolates of Acinetobacter baumannii, with new cassettes in the variable region of a class 2 integron.
A. baumannii AB28 was isolated in 2003 in a public hospital (H1) of Buenos Aires from the tracheal aspiration of a patient in the coronary unit (intensive care unit). Susceptibility tests were performed using a disk diffusion method, NCCLS, 2003 (13). AB28 was resistant to gentamicin, amikacin, ceftazidime, and ciprofloxacin, and it had an intermediate susceptibility to imipenem and meropenem according to the breakpoint of the NCCLS method (13). The pulsed-field gel electrophoresis analysis performed with the AB28 isolate showed that it belonged to clone VI (data not shown).
Total DNA of AB28 was extracted as previously described (18) and subjected to PCR amplifications with specific primers for the class 2 integrase gene (15). To detect the inserted gene cassettes, the class 2 integron variable region was amplified with primers 125′CS and 23′CS (Table 1), which annealed in the flanking sequences of this region (intI2 and tnsE genes). An amplicon product of approximately 6,400 bp was obtained, which was bigger than expected for Tn7 (4,383 bp). In order to characterize this integron, PCR cartography (Table 1) and sequence analysis were performed using an Applied Biosystems 373 sequencer. The sequences were analyzed with Genetics Computer Group software (Wisconsin package, version 10.3).
TABLE 1.
Oligonucleotide sequences used in this study
| Primer name | Sequence (5′-3′) | Reference or source |
|---|---|---|
| 125′CS | TTTTTGTGCTGCCATATCCGTG | This work |
| 23′CS | TGGGCTGAGAGAGTGGT | This work |
| satF | TGAGCAGGTGGCGGAAAC | This work |
| satR | TCATCCTGTGCTCCCGAG | This work |
| dhfR1 | AGCTGTTCACCTTTGGC | 10 |
| dhfR1R | CCTGAAATCCCCAGCAA | This work |
| aadA1 | TCGATGACGCCAACTAC | 10 |
| catB2R | AACAAAGAAAAGGCATTGC | This work |
| catB2F | CTGACTGAGCAGGTGAAG | This work |
| aadBF | GTAACACGCAAGCACGATGA | This work |
| aadBR | GCCTGTAGGACTCTATGTGC | This work |
| glmsF | GGCGGTCAGTTGTATGTCTT | 6 |
| Tn75′R | GACTCGTCCCGTCTTATGAG | 6 |
| tnsER | TCGATTTGCTGCTTTTGATG | This work |
| tnsEFF | TTGCTCTCTAACCACTCT | This work |
| tnsDR | CCGTCTAATTTGATAATCTTC | This work |
| tnsDF | GGGATTGTTAGTCCTAAGC | This work |
| tnsCR | GCTATCCCAGTCGCTGGG | This work |
| tnsCF | GTTTATCGTGATACGGGGG | This work |
| tnsBR | GAGCAAGCATTTACAAAAGC | This work |
| tnsBF | CATGTGGTCCAAGAACATAAG | This work |
| tnsAR | GCTAACAGTACAAGAAGTTCC | This work |
| tnsAF | CTCCATATTCACTACTTGGCT | This work |
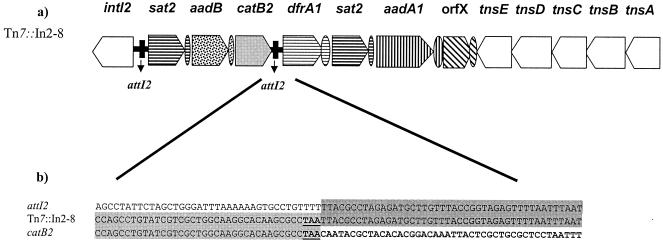
The analysis revealed the presence of a duplication of the sat2 cassette, which confers resistance to streptothricin, and the presence of the following two new resistant determinants for a class 2 integron: the aadB gene cassette (conferring resistance to gentamicin, kanamycin, and tobramycin) and the catB2(ΔattC) pseudocassette (conferring resistance to chloramphenicol). The sequence analysis established that downstream of the stop codon of the catB2 gene there is a 259-bp region with 100% homology to the attI2 class 2 integron recombination site replacing the typical attC site. Subsequent to this region, the same cassettes of the variable region of Tn7 were found, resulting in the novel integron structure sat2-aadB-catB2(ΔattC)-dfrA1-sat2-aadA1-orfX (Fig. 1).
FIG. 1.
Schematic representation of the structure of Tn7::In2-8. a) The attI2 site is indicated as shown, and the ovals represent the attC sites of the gene cassettes. All intI2 genes sequenced so far reveal an internal stop codon. b) Nucleotide sequence of catB2 pseudocassette and comparison of this sequence with the attC site of the catB2 cassette (accession number AJ487034) and the 5′CS region of the class 2 integron (accession number NC_002525).
Sequence analysis of this region revealed that a nonelucidated mechanism might be involved in the acquisition of the pseudocassette catB2 in the variable region of Tn7::In2-8. On one hand, the site where the excision in the attI2 was done, TT′TTA (the prime denotes the recombination point), does not belong either to the core sites GTTRRRY of attI1 and attC, which are the sites that IntI1 preferentially recognizes, or to the secondary sites conformed by the consensus GWTMW (5). Previously, this consensus sequence (GWTMW) has been found in the fused ant(3")-Ia(ΔattC)-blaOXA-9 cassette in Tn1331 (21). On the other hand, the site where the cleavage in the attC of the catB2 occurred, GCCTAA′C, corresponds to the inverse core site of the attC, complementary to GTTRRRY. In fact, cassettes are excised and then integrated at the preferential core site GTTRRRY, with the crossing over between the G and the first T (17) as different from the secondary site. An illegitimate integrase-mediated intermolecular recombination event involving similar sequences has been suggested for the creation of the fused ant(3")-Ih-(ΔattC)-aac(6′)-IId in SCH909 (3). Therefore, the acquisition of the pseudocassette catB2 in Tn7::In2-8 could have been caused by an analogous event involving the inverted core site of the attC site of catB2 (CTAA′CAAT) and the end of an attI2 site (GTTTT′TTACG) preceding the dfrA1 cassette of Tn7.
Using different primers designed for the 3′CS region where the transposition genes reside (Table 1), we carried out PCR and sequence analysis. We found the five transposition genes described before (6, 22) with 100% homology to the transposition genes of accession number X17693 (4).
We have also confirmed that this new rearrangement was inserted in the chromosome of A. baumannii using the specific primers described by Gay et al. (6). This set of primers amplifies the attTn7 site of Escherichia coli and a region of the Tn7 transposon. By PCR and sequence analysis, we found that Tn7::In2-8 was inserted downstream of the glms housekeeping gene (11, 12, 23).
We also proceeded to test if this new rearrangement could be found in our collection of multiresistant A. baumannii isolates. For that purpose, we performed PCRs with the primers Inti2R and CatB2R in 65 clinical isolates of our collection, and only 2 isolates (AB29 and AB1) were positive for this reaction. Moreover, PCR cartography showed the same novel rearrangement observed in AB28. One of the isolates (AB29) corresponds to the same hospital (H1), the same year (2003), and the same clone (data not shown). It came from a patient's blood from the clinical medical area (not an intensive care unit). Possibly, clonal spread between these two different patients may have occurred since it is the same clone with the same resistance pattern and both isolates carried this new class 2 integron. The other isolate (AB1) came from another hospital (H2) and was collected from a patient's blood in 1994, and its pulsed-field gel electrophoresis analysis showed that it belonged to clone IV (data not shown). Susceptibility tests revealed that this isolate (AB1) was resistant to only ceftazidime, aztreonam, and piperacillin.
AB29 and AB1 were also localized in the chromosome downstream of the glms housekeeping gene, as previously described for the transposon Tn7 (12).
This study reports a new rearrangement, Tn7::In2-8, found in two non-epidemiologically related multiresistant A. baumannii clones containing aadB and catB2, never described in a class 2 integron context before. The acquisition of new determinants of resistance in the variable region of integrons mediated by an illegitimate integrase intermolecular recombination event is likely to play an important role not only in the evolution of bacterial and plasmid genomes but also in the creation of novel rearrangements in the variable region of integrons that frequently contribute to the multiresistance challenge.
Nucleotide sequence accession number.
The sequence of In2-8 has been submitted to GenBank under accession number DQ176450.
Acknowledgments
We are very grateful to Nancy Messier for helpful comments in the manuscript. We thank Nancy Craig for fruitful discussions about the nomenclature of novel Tn7-like transposons. We thank Paul H. Roy for collaborating in the sequencing of the PCR products.
M.S.R and C.Q. are the recipients of a C.O.N.I.C.E.T. fellowship. D.C. is a member of the Carrera del Investigador Científico, CONICET-Argentina. This study was supported by a grant from UBACYT M403, Buenos Aires, Argentina, to D.C.
REFERENCES
- 1.Ahmed, A. M., H. Nakano, and T. Shimamoto. 2005. Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: description of an unusual class 2 integron. J. Antimicrob. Chemother. 55:371-374. [DOI] [PubMed] [Google Scholar]
- 2.Biskri, L., and D. Mazel. 2003. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 47:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centron, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores, C., M. I. Qadri, and C. Lichtenstein. 1990. DNA sequence analysis of five genes, tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 18:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Francia, M. V., F. de la Cruz, and J. M. Garcia Lobo. 1993. Secondary-sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 6.Gay, N. J., V. L. Tybulewicz, and J. E. Walker. 1986. Insertion of transposon Tn7 into the Escherichia coli glmS transcriptional terminator. Biochem. J. 234:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez, G., K. Sossa, S. Mella, R. Zemelman, H. Bello, and M. Domiguez. 1998. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol. Lett. 161:125-128. [DOI] [PubMed] [Google Scholar]
- 8.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integron: capture and spread of gene by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 9.Hansson, K., L. Sundstrom, A. Pelletier, and P. Roy 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lichtenstein, C., and S. Brenner. 1982. Unique insertion site of Tn7 transposition into the E. coli chromosome. Nature 297:601-603. [DOI] [PubMed] [Google Scholar]
- 12.McKown, R. L., K. A. Orle, T. Chen, and N. Craig. 1988. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol. 170:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NCCLS. 2003. Methods for diffusion antimicrobial susceptibility test for bacteria that grow aerobically. M2-A8. NCCLS, Wayne, Pa.
- 14.Oh, J. Y., K. S. Kim, Y. W. Jeong, J. W. Cho, J. C. Park, and J. C. Lee. 2002. Epidemiological typing and prevalence of integrons in multiresistant Acinetobacter strains. APMIS 110:247-252. [DOI] [PubMed] [Google Scholar]
- 15.Orman, B. E., S. A. Piñeiro, S. Arduino, M. Galas, R. Melano, M. I. Caffer, D. O. Sordelli, and D. Centrón. 2002. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radstrom, P., O. Skold, G. Swedberg, J. Flensburg, P. H. Roy, and L. Sundstrom. 1994. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J. Bacteriol. 176:3257-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Recchia, G. D., H. W. Stokes, and R. M. Hall. 1994. Characterization of specific and secondary recombination sites recognized by the integron DNA integrase. Nucleic Acids Res. 22:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Seward, R. J., and K. J. Towner. 2002. Detection of integrons in world-wide nosocomial isolates of Acinetobacter spp. Clin. Microbiol. Infect. 5:308-318. [DOI] [PubMed] [Google Scholar]
- 20.Tietze, E., J. Brevet, H. Tschape, and W. Voigt. 1988. Cloning and preliminary characterization of the streptothricin resistance determinants of the transposons Tn1825 and Tn1826. J. Basic Microbiol. 28:129-136. [DOI] [PubMed] [Google Scholar]
- 21.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 22.Waddell, C. S., and N. L. Craig. 1988. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 2:137-149. [DOI] [PubMed] [Google Scholar]
- 23.Walker, J., N. Gay, M. Saraste, and A. Eberle. 1984. DNA sequence around the Escherichia coli unc operon. Biochem. J. 224:799-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young, H., M. J. Qumsich, and M. L. Mcintosh. 1994. Nucleotide sequence and genetic analysis of the type Ib trimethoprim-resistant, Tn4132-encoded dihydrofolate reductase. J. Antimicrob. Chemother. 34:715-725. [DOI] [PubMed] [Google Scholar]