Abstract
Metronidazole and related 5-nitroimidazoles are the only available drugs in the treatment of human urogenital trichomoniasis caused by the protozoan parasite Trichomonas vaginalis. The drugs are activated to cytotoxic anion radicals by their reduction within the hydrogenosomes. It has been established that electrons required for metronidazole activation are released from pyruvate by the activity of pyruvate:ferredoxin oxidoreductase and transferred to the drug by a low-redox-potential carrier, ferredoxin. Here we describe a novel pathway involved in the drug activation within the hydrogenosome. The source of electrons is malate, another major hydrogenosomal substrate, which is oxidatively decarboxylated to pyruvate and CO2 by NAD-dependent malic enzyme. The electrons released during this reaction are transferred from NADH to ferredoxin by NADH dehydrogenase homologous to the catalytic module of mitochondrial complex I, which uses ferredoxin as electron acceptor. Trichomonads acquire high-level metronidazole resistance only after both pyruvate- and malate-dependent pathways of metronidazole activation are eliminated from the hydrogenosomes.
For more than 40 years, metronidazole and related derivatives of 5-nitroimidazole have been the drugs of choice in the treatment of infections caused by anaerobic or microaerophilic microbes, both prokaryotic and eukaryotic. The susceptible organisms are characterized by the presence of low-redox-potential electron-transporting systems that are absent in aerobes. These pathways involve ferredoxin-like electron carriers that use the nitroimidazole prodrugs as electron acceptors, generating short-lived reactive anion radicals that inflict multiple types of cellular damage and subsequent cell death (5, 14).
In Trichomonas vaginalis, an amitochondriate flagellate causing human urogenital trichomoniasis, metronidazole is activated within hydrogenosome. This double-membrane-bound organelle harbors a catabolic pathway in which pyruvate or malate is oxidatively decarboxylated with concomitant generation of electrons. Pyruvate is converted to acetyl coenzyme A (acetyl-CoA) and CO2 by a pyruvate:ferredoxin oxidoreductase (PFOR). While acetyl-CoA is utilized in a substrate-level synthesis of ATP, the released electrons are transferred via ferredoxin to hydrogenase that produces molecular hydrogen (15). Malate is converted to pyruvate and CO2 by malic enzyme. The electrons released from malate reduce NAD+, from which they are transferred to ferredoxin by NADH:ferredoxin oxidoreductase activity (17) of the NADH dehydrogenase (NDH) module of complex I (6). Metronidazole enters the hydrogenosomes by diffusion and acts as a high-affinity electron acceptor, which is reduced by ferredoxin to its cytotoxic form (7). PFOR-dependent generation of electrons is considered to be the key pathway responsible for the metronidazole activation, although a low-level NADH-dependent activity-reducing metronidazole, which was not studied in more detail, was observed in the bovine parasite Tritrichomonas foetus (13). Whether the recently described malate-dependent electron transport (17) contributes to the metronidazole activation in T. vaginalis was not known.
Here we provide the direct evidence that the malate-dependent pathway is capable of metronidazole reduction in the wild-type as well as in PFOR-deficient T. vaginalis strains developed in vitro. Based on monitoring the formation of metronidazole anion radicals by electron paramagnetic resonance (EPR) spectroscopy, we show that malic enzyme, NDH, and ferredoxin are required for the PFOR-independent metronidazole reduction, thus constituting an alternative electron-transporting chain which participates in metronidazole activation in hydrogenosomes.
MATERIALS AND METHODS
Organisms.
Trichomonas vaginalis strain TV 10-02, which is susceptible to metronidazole (9), and its laboratory-induced derivatives lacking PFOR activity, which express low (TV 10-02 MR 5) or high (TV 10-02 MR 100) levels of metronidazole resistance (8, 17), were used in the experiments monitoring the formation of metronidazole anion radicals by isolated hydrogenosomes. T. vaginalis strain T1 (provided by Patricia Johnson, University of California, Los Angeles, Calif.) was used for the purification of NDH. The cells were grown in trypticase-yeast extract-maltose (TYM) medium (3) supplemented with 10% heat-inactivated horse serum and 0.05% agar (wt/vol) at 37°C. Cultures of the resistant strains were maintained in a TYM medium with metronidazole (5 μg ml−1 for the MR 5 strain and 50 μg ml−1 for the MR 100 strain). Large-volume cultures for isolation of hydrogenosomes were grown without agar and metronidazole.
Cell fractionation.
The cells (approximately 2 liters of culture) were harvested by centrifugation, washed with ST buffer (250 mM sucrose, 0.5 mM KCl, 10 mM Tris-HCl, pH 7.2) and suspended in ST buffer with 50 μg of N-α-tosyl-l-lysine chloromethyl ketone (TLCK) per ml and 10 μg of leupeptin per ml. The cells were disrupted by sonication and subjected to differential centrifugation as described previously (4). The resulting hydrogenosome-enriched fraction was further purified by isopycnic centrifugation on 45% Percoll (19). These highly purified hydrogenosomes were used throughout this study.
Purification of NDH.
NDH was purified from the hydrogenosomes by liquid chromatography on cation exchange and hydroxyapatite columns as described in an earlier publication (6). Aliquots from purification steps were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the proteins were visualized by Coomassie staining.
Preparation of T. vaginalis recombinant ferredoxin.
Recombinant T. vaginalis [2Fe-2S] ferredoxin without a hydrogenosomal targeting sequence was expressed in Escherichia coli and isolated by two steps of liquid chromatography as described previously (19).
Enzyme activity determinations.
Activity of malic enzyme was determined spectrophotometrically at 340 nm as the rate of malate-dependent reduction of NAD+ as described previously (4). Activities of PFOR and NDH were determined under anaerobic conditions in 100 mM potassium phosphate buffer, pH 7.4, containing 25 mM mercaptoethanol and 10 mM methyl viologen. The PFOR activity was determined with 6 mM pyruvate and 0.5 mM CoA; 8.5 mM NADH was the substrate for NDH. Activities of both enzymes were determined spectrophotometrically at 600 nm using a molar extinction coefficient, ε600, of 6,300 M−1 cm−1. Activity of NDH in the course of purification was monitored using 2,6-dichloroindophenol (DCIP) as the electron acceptor. The reaction mixture contained 50 mM KCl, 100 mM Tris-HCl, pH 8 (assay buffer), 3.3 mM NADH, and 100 μM DCIP. The reduction of acceptor was monitored at 600 nm, and the ε600 of DCIP was taken as 21000 M−1 cm−1. All spectrophotometric determinations were done at 25°C. One unit of enzyme activity was defined as amount of protein catalyzing the consumption of 1 micromole of substrate or the formation of 1 micromole of product per minute. Protein concentrations were determined by the method of Lowry.
The activities of PFOR, malic enzyme, and NDH were determined in isolated hydrogenosomes of all strains prior to the EPR experiments. All the activities were in the same range as those determined previously for the same strains (17), except for NDH (called NADH:ferredoxin oxidoreductase in the reference), for which the activity was found in this work to be about twofold higher in wild-type and low-resistance strains.
EPR spectroscopy.
EPR spectroscopy was used to detect the metronidazole nitro anion radicals formed by isolated hydrogenosomes and also by purified NDH. Assay mixtures used to detect the metronidazole radical formation catalyzed by PFOR, malic enzyme, and NDH within isolated organelles were similar to those used for spectrophotometric determination of the respective enzymatic activities, except for the omission of the methyl viologen electron acceptor in the PFOR and NDH reactions. The assay mixture (0.8 ml) contained 43 mM metronidazole, approximately 50 μg/ml T. vaginalis ferredoxin, 25 mM mercaptoethanol, and 1.2 mg/ml Triton X-100, plus either 7.5 mM pyruvate plus 0.27 mM CoA, 14 mM malate plus 1.1 mM NAD+, or 2.2 mM NADH. The solution was degassed by argon flow for 10 min, and the reaction was started by the addition of approximately 300 μg of hydrogenosomal protein of a wild-type, low-resistance, or highly resistant strain. Upon addition of the protein sample, each mixture was immediately drawn into a 150-μl flat detection cell. EPR spectra of the hydrogenosomal preparations were recorded at 25°C on a Bruker ESP 300 spectrometer (Bruker BioSpin). Formation of metronidazole nitro anion radicals by purified NDH was monitored in the assay buffer containing 2.2 mM NADH and 12 mM metronidazole. The mixture (2 ml) was flushed with oxygen-free nitrogen for 10 min, and then recombinant T. vaginalis ferredoxin (approximately 700 μg) was added and the reaction was started by the addition of approximately 4 μg of purified NDH. Spectra were recorded on a Bruker ELEXSYS E580 spectrometer at 25°C. Hyperfine coupling constants for the metronidazole anion radicals were determined using the SimFonia program.
RESULTS
Formation of metronidazole anion radicals in isolated hydrogenosomes.
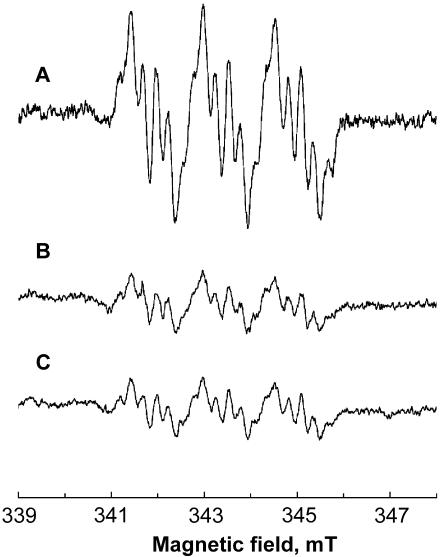
In order to test whether electrons generated by malic enzyme are used in metronidazole activation, we monitored the formation of metronidazole anion radicals by EPR spectroscopy in the reaction mixture containing hydrogenosomes, T. vaginalis ferredoxin, malate, NAD+, and metronidazole. The signal corresponding to reduced metronidazole was detectable within 1 to 2 min after the reaction was started by the addition of hydrogenosomes isolated from the wild-type, metronidazole-sensitive T. vaginalis (Fig. 1B). The activity of PFOR, which is a well-known catalyst of metronidazole-reductive activation in trichomonads (12, 13), was used as a positive control. Formation of metronidazole anion radicals was observed as in the reaction with malic enzyme (Fig. 1A). The amplitude of the signal generated by malate-dependent activity was lower than that generated by PFOR, indicating a lower activity of metronidazole reduction catalyzed by the alternative system (Fig. 1).
FIG. 1.
EPR spectra demonstrating formation of metronidazole anion radicals by the hydrogenosomes of wild-type, drug-sensitive T. vaginalis in the presence of 43 mM metronidazole. (A) Signal generated in the presence of pyruvate, CoA, and ferredoxin (PFOR activity). (B) Signal generated in the presence of malate, ferredoxin, and NAD+ (malic enzyme activity). (C) Signal generated in the presence of NADH and ferredoxin (NDH activity). The spectra were recorded at 25°C with 20 mW of microwave power, a frequency of 9.64 GHz, and a modulation amplitude of 0.19 mT. The hyperfine coupling constants were aNO2N = 1.565 mT, a4H = 0.542 mT, and aCH3H = 0.229 mT.
In the next experiment, NADH only was used as an electron donor for metronidazole reduction by the hydrogenosomes of drug-sensitive trichomonads; this sole substrate should have been sufficient if NDH was involved in metronidazole reduction. Indeed, the metronidazole anion radicals were formed and detected with the same amplitude as that in the case of the malic enzyme-catalyzed reaction (Fig. 1C), indicating that NDH activity is the limiting factor in malate-dependent reduction of metronidazole. Reduction of ferredoxin by NDH is relatively slow (approximately 10 μmol/min per mg of purified protein [unpublished data]), as opposed to the rapid reoxidation of reduced ferredoxin by metronidazole (21). Addition of external ferredoxin in all reactions had only a marginal effect; omitting ferredoxin resulted in somewhat lower amplitudes of the signals, showing that the ferredoxin concentration in the hydrogenosomal preparations was sufficient to support the drug reduction.
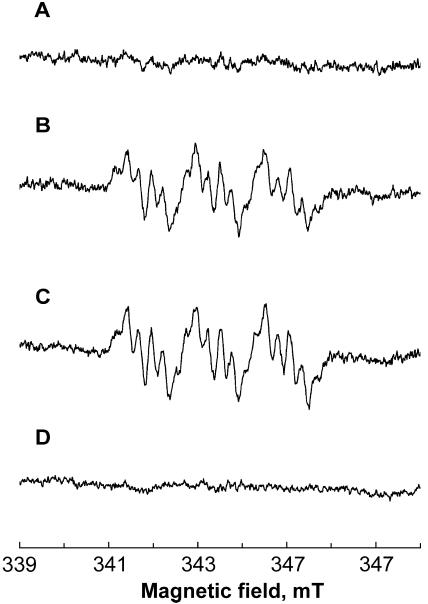
Using the same experimental setup as described above, the hydrogenosomes from the PFOR-deficient strain (TV 10-02 MR 5), displaying a low level of resistance, were tested for their capacity to catalyze the reductive activation of metronidazole. Trace A in Fig. 2 shows that metronidazole was not reduced when pyruvate and CoA were added into the reaction, consistent with the absence of PFOR activity in this strain. However, metronidazole anion radicals were formed by activities of both malic enzyme and NDH (Fig. 2B and C, respectively). As described above, the amplitudes of the signals were comparable in malic enzyme- and NDH-catalyzed reactions.
FIG. 2.
EPR spectra demonstrating formation of metronidazole anion radicals by the hydrogenosomes of low-resistance (strain TV 10-02 MR 5; traces A to C) and highly resistant (strain TV 10-02 MR 100; trace D) T. vaginalis strains in the presence of 43 mM metronidazole. (A) Absence of signal in the presence of pyruvate, ferredoxin, and CoA. (B) Signal generated in the presence of malate, ferredoxin, and NAD+ (malic enzyme activity). (C) Signal generated in the presence of NADH and ferredoxin (NDH activity). (D) Absence of signal in the presence of hydrogenosomes of highly resistant T. vaginalis, NADH, and ferredoxin (NDH activity). Instrument settings were as described for Fig. 1.
Finally, we tested the formation of metronidazole anion radicals using the hydrogenosomes from the T. vaginalis strain displaying a high level of metronidazole resistance (TV 10-02 MR 100). This strain lacks detectable activities of PFOR, malic enzyme, and NDH and does not express ferredoxin (17). Consistent with the absence of all these proteins, metronidazole was reduced neither by the PFOR-dependent reaction nor by the alternative pathway involving malic enzyme and NDH (Fig. 2D; only the trace corresponding to NDH activity is shown).
Reduction of metronidazole by NDH.
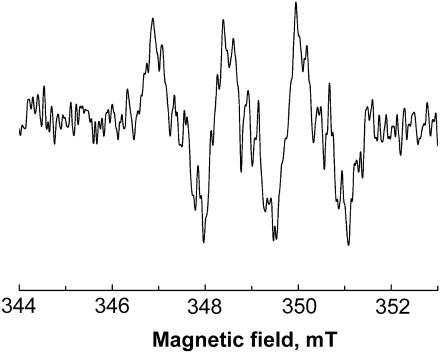
To provide the direct evidence that NDH provides electrons for metronidazole activation, we tested the formation of metronidazole anion radicals by use of the near-homogeneous enzyme isolated from T. vaginalis hydrogenosomes (Fig. 3). EPR spectroscopy detected metronidazole anion radicals in the assay mixture containing NADH, purified NDH, and recombinant T. vaginalis ferredoxin (Fig. 4). When ferredoxin was omitted from the reaction, metronidazole was not reduced, confirming that electrons from NADH must be transferred by a low-redox-potential carrier with appropriate specificity to reduce the drug.
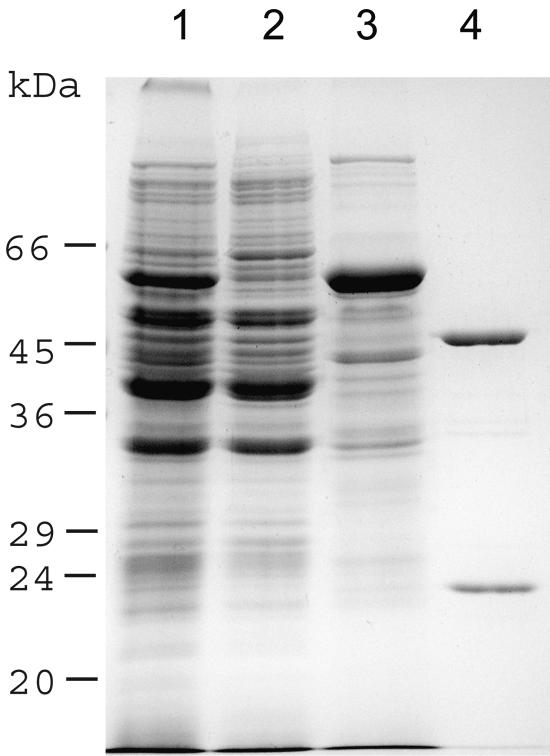
FIG. 3.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of Coomassie-stained subcellular fractions of T. vaginalis and isolated NDH. Lanes: 1, total homogenate; 2, cytosol; 3, hydrogenosomes; 4, two subunits of NDH purified from hydrogenosomes.
FIG. 4.
EPR spectrum of metronidazole anion radical formed by purified NDH in the presence of NADH and Trichomonas ferredoxin. The spectrum was recorded with microwave power of 0.505 mW, a frequency of 9.7584 GHz, and a modulation amplitude of 0.4 mT.
DISCUSSION
In this study, we report on a novel PFOR-independent metronidazole-activating pathway present in T. vaginalis hydrogenosomes that consists of malic enzyme, NDH, and ferredoxin. Until recently, the principal hydrogenosomal activity responsible for the reductive activation of metronidazole had been ascribed to PFOR (2, 12, 16), based on the findings (i) that PFOR catalyzed generation of nitro anion radicals in Tritrichomonas foetus hydrogenosomal fraction when pyruvate, CoA, and metronidazole were present (13) and (ii) that PFOR activity was lost in in vitro-derived strains of T. foetus (1) and T. vaginalis (8) resistant to high concentrations of metronidazole. These strains were able to grow in vitro in the presence of >100 μg/ml metronidazole. However, more-recent studies of metabolic changes accompanying the in vitro induction of a high-level metronidazole resistance in T. vaginalis revealed that PFOR activity is among the first enzymes that disappear from the hydrogenosomes in the process of resistance development. The early-stage parasites, which were already PFOR deficient, were still quite susceptible to the drug, growing in the presence of no more than 5 μg/ml metronidazole (17). These observations suggested that other hydrogenosomal enzymes participating in redox reactions account for metronidazole activation. Hydrogenosomes contain a great abundance of malic enzyme, which by oxidative decarboxylation of malate provides reducing power in the form of NADH (4), and a NADH:ferredoxin (methyl viologen) oxidoreductase, elusive until recently, that could recycle NADH by reducing ferredoxin (6, 18, 20). In T. vaginalis, the activities of these enzymes gradually decrease in the course of resistance development, and the highly resistant phenotype is acquired only after malic enzyme and NADH:ferredoxin oxidoreductase activities are markedly reduced or completely disappear from the hydrogenosome together with ferredoxin (7, 17). Based on these observations, it has been speculated that malic enzyme may provide electrons for the drug activation in the absence of PFOR (7, 17). Indeed, using EPR spectroscopy, we show here that in vitro formation of nitro anion radicals is catalyzed by isolated hydrogenosomes supplied with NAD+, malate, and metronidazole, demonstrating that malic enzyme is able to generate electrons required for the drug reduction. NADH resulting from oxidative decarboxylation of malate is then the only substrate that is sufficient to support metronidazole reduction in purified hydrogenosomes, both wild type and those lacking PFOR, indicating that the distal part of the alternative metronidazole-activating electron transport pathway is catalyzed by an NADH-utilizing enzyme. This enzyme is apparently an NDH, a homolog of the NADH dehydrogenase module of mitochondrial respiratory complex I (6), as the near-homogeneous preparation of this protein from trichomonad hydrogenosomes reduced metronidazole at the expense of NADH. However, NDH cannot reduce the drug directly. The terminal electron carrier donating electrons to metronidazole appears to be ferredoxin, since metronidazole anion radicals could be detected only when Trichomonas ferredoxin was added into the reaction mixture containing purified NDH and NADH. This observation is consistent with the results of several studies that implicate ferredoxins as proximal electron donors in metronidazole reduction (11, 16, 21).
The widely accepted concept of an indispensable role of ferredoxin in metronidazole activation was questioned recently by Land and coworkers (10), who found that (presumably single-gene) ferredoxin knockout does not confer the metronidazole resistance in transformed T. vaginalis. As one possible explanation of this phenomenon, the authors suggested the presence of other ferredoxins or flavodoxins in T. vaginalis that would be divergent enough not to be targeted by the gene replacement machinery (10). Indeed, several other ferredoxin and flavodoxin genes have subsequently been annotated during the Trichomonas vaginalis genome-sequencing project (http://www.tigr.org/tdb/e2k1/tvg/). Our results presented in this study do not rule out the existence of other electron-generating pathways in trichomonad hydrogenosomes that could account for metronidazole activation; however, the presence of such systems awaits experimental verification.
In conclusion, we have demonstrated the presence of a novel pathway of metronidazole reduction in T. vaginalis hydrogenosomes. Unlike the PFOR-dependent activity, where pyruvate is the source of electrons, the alternative pathway uses electrons released from malate in the form of NADH plus H+ by the action of malic enzyme. NADH dehydrogenase then recycles NADH by reducing ferredoxin, which provides electrons to metronidazole. Thus, ferredoxin plays a pivotal role in both pyruvate- and malate-dependent activations of metronidazole.
Acknowledgments
This work was performed in the framework of the COST B-22 program and was supported by the OC.B22.001 grant provided by the Ministry of Education of the Czech Republic. Further support was obtained from the IGA grant agency of the Ministry of Health of the Czech Republic (grant NB/7377-3 to P.S.), from the Grant Agency of the Czech Republic (grant 204/04/0435 to J.T.), and from the United Kingdom Biochemistry and Biotechnology Research Council (to R.C.).
REFERENCES
- 1.Cerkasovova, A., J. Cerkasov, and J. Kulda. 1984. Metabolic differences between metronidazole resistant and susceptible strains of Tritrichomonas foetus. Mol. Biochem. Parasitol. 11:105-118. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, A., R. Cammack, D. Linstead, and D. Lloyd. 1985. The generation of metronidazole radicals in hydrogenosomes isolated from Trichomonas vaginalis. J. Gen. Microbiol. 131:2141-2144. [DOI] [PubMed] [Google Scholar]
- 3.Clark, C. G., and L. S. Diamond. 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin. Microbiol. Rev. 15:329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drmota, T., P. Proost, R. M. Van, F. Weyda, J. Kulda, and J. Tachezy. 1996. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol. Biochem. Parasitol. 83:221-234. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, D. I. 1993. Nitroimidazole drugs—action and resistance mechanisms. II. Mechanisms of resistance. J. Antimicrob. Chemother. 31:201-210. [DOI] [PubMed] [Google Scholar]
- 6.Hrdy, I., R. P. Hirt, P. Dolezal, L. Bardonova, P. G. Foster, J. Tachezy, and T. M. Embley. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618-622. [DOI] [PubMed] [Google Scholar]
- 7.Kulda, J. 1999. Trichomonads, hydrogenosomes and drug resistance. Int. J. Parasitol. 29:199-212. [DOI] [PubMed] [Google Scholar]
- 8.Kulda, J., J. Tachezy, and A. Cerkasovova. 1993. In vitro induced anaerobic resistance to metronidazole in Trichomonas vaginalis. J. Eukaryot. Microbiol. 40:262-269. [DOI] [PubMed] [Google Scholar]
- 9.Kulda, J., M. Vojtechovska, J. Tachezy, P. Demes, and E. Kunzova. 1982. Metronidazole resistance of Trichomonas vaginalis as a cause of treatment failure in trichomoniasis—a case report. Br. J. Vener. Dis. 58:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Land, K. M., M. G. Gadillo-Correa, J. Tachezy, S. Vanacova, C. L. Hsieh, R. Sutak, and P. J. Johnson. 2004. Targeted gene replacement of a ferredoxin gene in Trichomonas vaginalis does not lead to metronidazole resistance. Mol. Microbiol. 51:115-122. [DOI] [PubMed] [Google Scholar]
- 11.Lindmark, D. G., and M. Muller. 1976. Antitrichomonad action, mutagenicity, and reduction of metronidazole and other nitroimidazoles. Antimicrob. Agents Chemother. 10:476-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marczak, R., T. E. Gorrell, and M. Muller. 1983. Hydrogenosomal ferredoxin of the anaerobic protozoon, Tritrichomonas foetus. J. Biol. Chem. 258:12427-12433. [PubMed] [Google Scholar]
- 13.Moreno, S. N., R. P. Mason, and R. Docampo. 1984. Distinct reduction of nitrofurans and metronidazole to free radical metabolites by Tritrichomonas foetus hydrogenosomal and cytosolic enzymes. J. Biol. Chem. 259:8252-8259. [PubMed] [Google Scholar]
- 14.Muller, M. 1986. Reductive activation of nitroimidazoles in anaerobic microorganisms. Biochem. Pharmacol. 35:37-41. [DOI] [PubMed] [Google Scholar]
- 15.Muller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879-2889. [DOI] [PubMed] [Google Scholar]
- 16.Quon, D. V., C. E. d'Oliveira, and P. J. Johnson. 1992. Reduced transcription of the ferredoxin gene in metronidazole-resistant Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 89:4402-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasoloson, D., S. Vanacova, E. Tomkova, J. Razga, I. Hrdy, J. Tachezy, and J. Kulda. 2002. Mechanisms of in vitro development of resistance to metronidazole in Trichomonas vaginalis. Microbiology 148:2467-2477. [DOI] [PubMed] [Google Scholar]
- 18.Steinbuchel, A., and M. Muller. 1986. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 20:57-65. [DOI] [PubMed] [Google Scholar]
- 19.Sutak, R., P. Dolezal, H. L. Fiumera, I. Hrdy, A. Dancis, M. Gadillo-Correa, P. J. Johnson, M. Muller, and J. Tachezy. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 101:10368-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thong, K. W., and G. H. Coombs. 1987. Comparative study of ferredoxin-linked and oxygen-metabolizing enzymes of trichomonads. Comp. Biochem. Physiol. B 87:637-641. [DOI] [PubMed] [Google Scholar]
- 21.Vidakovic, M., C. R. Crossnoe, C. Neidre, K. Kim, K. L. Krause, and J. P. Germanas. 2003. Reactivity of reduced [2Fe-2S] ferredoxins parallels host susceptibility to nitroimidazoles. Antimicrob. Agents Chemother. 47:302-308. [DOI] [PMC free article] [PubMed] [Google Scholar]