Abstract
Many flaviviruses cause significant human disease worldwide. The development of flavivirus chemotherapy requires reliable high-throughput screening (HTS) assays. Although genetic systems have been developed for many flaviviruses, their usage in antiviral HTS assays has not been well explored. Here we compare three cell-based HTS assays for West Nile virus (WNV) drug discovery: (i) an assay that uses a cell line harboring a persistently replicating subgenomic replicon (containing a deletion of viral structural genes), (ii) an assay that uses packaged virus-like particles containing replicon RNA, and (iii) an assay that uses a full-length reporting virus. A Renilla luciferase gene was engineered into the replicon or into the full-length viral genome to monitor viral replication. Potential inhibitors could be identified through suppression of luciferase signals upon compound incubation. The antiviral assays were optimized in a 96-well format, validated with known WNV inhibitors, and proved useful in identifying a new inhibitor(s) through HTS of a compound library. In addition, because each assay encompasses multiple but discrete steps of the viral life cycle, the three systems could potentially be used to discriminate the mode of action of any inhibitor among viral entry (detected by assays ii and iii but not by assay i), replication (including viral translation and RNA synthesis; detected by assays i to iii), and virion assembly (detected by assay iii but not by assays i and ii). The approaches described in this study should be applicable to the development of cell-based assays for other flaviviruses.
West Nile virus (WNV) belongs to the genus Flavivirus in the family Flaviviridae. Besides WNV, many flaviviruses are significant human pathogens, including dengue, yellow fever, Japanese encephalitis, Murray Valley encephalitis, and tick-borne encephalitis viruses (5). Flavivirus virions are approximately 50 nm in diameter (18, 29) and contain a single-stranded, plus-sense RNA genome of about 11 kb in length (6). The genomic RNA contains a single long open reading frame (ORF) that is flanked by a 5′ untranslated region (5′-UTR) and a 3′-UTR (see Fig. 1A). The ORF encodes a polyprotein that is co- and posttranslationally processed by viral and cellular proteases into 10 mature proteins: 3 structural proteins (the capsid [C], premembrane [prM] or membrane [M], and envelope [E] proteins) and 7 nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (6). Structural proteins are primarily involved in viral particle formation. Nonstructural proteins are responsible for viral RNA replication, but they may also function in viral assembly (19, 22) and evasion of the immune response (11, 21, 30). During flavivirus infection, the genomic plus-strand RNA is transcribed into a complementary minus-strand RNA, which, in turn, serves as the template for the synthesis of more plus-strand RNA (6). In the presence of viral structural proteins, the plus-sense RNA is packaged to form progeny viruses.
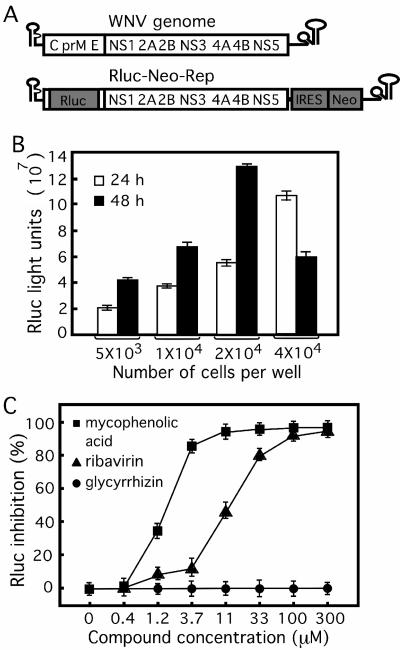
FIG. 1.
Use of a luciferase-expressing replicon cell line in an HTS antiviral assay. (A) WNV genome and a reporting replicon. The replicon contains a Renilla luciferase (Rluc; which substitutes for the deleted viral structural genes) and a neomycin phosphotransferase gene (Neo; driven by an EMCV IRES in the 3′ UTR), resulting in Rluc-Neo-Rep. (B) Titration of Rluc-Neo-Rep cells in a 96-well plate. The indicated number of Rluc-Neo-Rep cells was seeded into each well of a 96-well plate and assayed for Rluc activity at 24 and 48 h postseeding. (C) Antiviral assay validation. Rluc-Neo-Rep BHK-21 cells were incubated with various concentrations, as indicated, of known WNV inhibitors (mycophenolic acid, ribavirin, and glycyrrhizin) for 48 h and assayed for Rluc activity, in which Rluc inhibition (%) = [(Rluc signal without compound − Rluc signal with compound)/Rluc signal without compound] × 100. The data represent the means ± standard deviations (n ≥ 3).
Flavivirus drug discovery requires the development of reliable biological assays. Traditional antiviral assays for flavivirus are based on viral infection of cultured cells, followed by monitoring of compound inhibition of viral replication through observation of cytopathic effects, quantification of viral yields by plaque assay, or measurement of viral RNA by reverse transcription-PCR (15, 28). The low-throughput nature of these assays limits their use for the screening of compound libraries. The availability of reverse genetic systems for many flaviviruses (cDNA clones for infectious full-length genomes and for subgenomic replicons) has made it possible to develop novel assays for high-throughput screening (HTS). In principle, a reporter gene such as luciferase could be engineered into a full-length virus and into a subgenomic replicon. Infection or transfection of susceptible cells with such a reporting virus or replicon results in the expression of luciferase. The level of luciferase activity would reflect the extent of viral replication and could be used to monitor the suppression of viral infection by potential inhibitors (36). Alternatively, viral structural proteins could be supplied in trans to package luciferase-expressing replicon into virus-like particles (VLPs). Infection of cells with such reporting VLPs could lead to luciferase expression, which could be used to monitor the antiviral activities of potential inhibitors.
Although reporter-based subgenomic replicons (10, 14, 17, 37), packaged VLPs (9, 12, 14, 16, 35), and reporting full-length virus (8, 33, 47) have been described for several flaviviruses, their application to antiviral drug discovery has largely been unexplored. We previously showed that a BHK-21 cell line containing a luciferase-expressing replicon of WNV could potentially be used for antiviral screening (24). The replicon contains a Renilla luciferase (Rluc; which is a substitute for the deleted viral structural genes) and a neomycin phosphotransferase gene (Neo; which is driven by an encephalomyocarditis virus [EMCV] internal ribosomal entry site [IRES] in the 3′-UTR [Rluc-Neo-Rep]; see Fig. 1A). Incubation of the Rluc-Neo-Rep-containing cells with a potential WNV inhibitor results in a decrease in Rluc activity (24). In agreement with our results, Rossi et al. recently showed that a similar replicon-bearing cell line was capable of detecting inhibitors of WNV (34). The replicon-based antiviral assay has been widely used for hepatitis C virus (2, 25). Besides the replicon-based approach, we have also shown that a luciferase-expressing full-length WNV could potentially be used for antiviral screening (8). The reporting WNV contains an Rluc gene driven by an EMCV IRES in the 3′-UTR of the genome (Rluc-WNV; see Fig. 3A). However, the assays described above remain to be adapted to an HTS format and to be experimentally validated for large-scale compound screening.
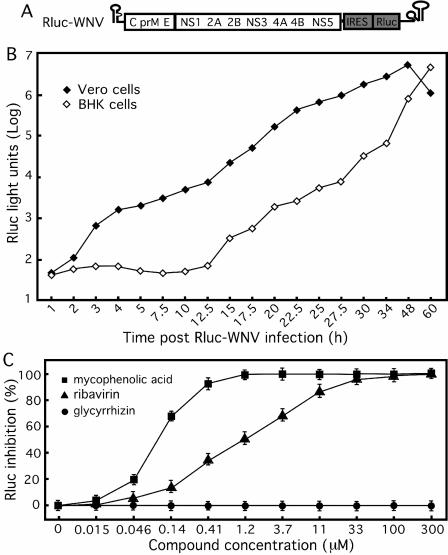
FIG. 3.
Use of luciferase WNV infection as an HTS antiviral assay. (A) A luciferase-reporting full-length WNV. An Rluc gene driven by an EMCV IRES was engineered in the 3′-UTR of the genome, resulting in Rluc-WNV. (B) Time course of Rluc activity in cells infected with Rluc-WNV. Vero and BHK-21 cells were synchronously infected with Rluc-WNV and assayed for their Rluc activities at various time points postinfection. (C) Antiviral assay validation. BHK-21 cells were infected with Rluc-WNV, immediately treated with inhibitors (mycophenolic acid, ribavirin, and glycyrrhizin), and assayed for Rluc inhibition at 24 h after infection and treatment. The data represent the means and standard deviations (n ≥ 3).
In this study, we have compared three cell-based HTS assays for WNV drug discovery: (i) an assay with a replicon-containing cell line that allows screening for inhibitors of viral replication; (ii) a replicon-bearing VLP infection assay that allows screening for inhibitors of viral entry as well as replication; and (iii) A full-length reporting virus infection assay that allows screening for inhibitors of any step(s) of the viral life cycle, including entry, replication, and virion assembly. All three assays were converted into a 96-well format, validated with known WNV inhibitors, and demonstrated to be useful for the screening of a compound library. The development of these assays will greatly facilitate WNV drug discovery.
MATERIALS AND METHODS
Cells and viruses.
Vero and BHK-21cells were cultured as described previously (38). A reporting BHK-21 cell line containing a WNV replicon with an Rluc and a Neo gene (Rluc-Neo-Rep; see Fig. 1A) was prepared previously and maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum (FBS) and 1 mg/ml G418 (24). All cells were maintained in 5% CO2 at 37°C. A full-length reporting WNV (Rluc-WNV; see Fig. 3A) was prepared from an infectious cDNA clone of strain 3356 by inserting an EMCV IRES Rluc fragment into the 3′-UTR of the WNV genome (8).
Construction of expression vector of WNV structural proteins.
A complete coding sequence of C-prM-E of WNV was amplified from an infectious cDNA clone by PCR (38) and engineered into a Semliki Forest virus (SFV) expression vector (Invitrogen) (20) at an unique BamHI site, resulting in SFV-CprME (see Fig. 2A). A Kozak sequence and a stop codon were added to the 5′ and 3′ ends of the C-prM-E fragment, respectively. The resulting construct was then verified by DNA sequencing.
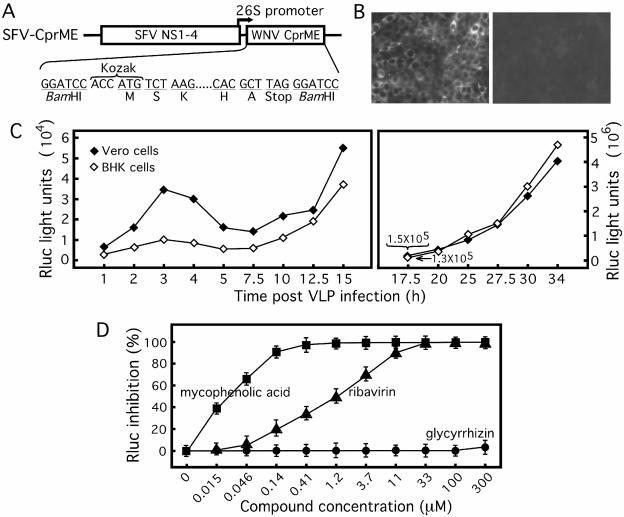
FIG. 2.
Use of VLP infection as an HTS assay. (A) Expression construct of WNV structural proteins. The SFV vector containing viral nonstructural proteins (SFV NS1 to NS4) was used to express WNV structural proteins. The complete coding sequence of WNV C-prM-E was inserted at a unique BamHI site under the control of the SFV 26S subgenomic promoter. A Kozak sequence and a stop codon were added to the 5′ and 3′ ends of the WNV insert, respectively. The amino acids at the N and C termini of the WNV C-prM-E are indicated. (B) WNV protein expression in VLP-infected Vero cells. Vero cells were incubated with culture fluids collected from the SFV-CprME-transfected Rluc-Neo-Rep cells and analyzed for WNV protein expression by IFA (left panel). All Vero cells were IFA positive. As a negative control, no IFA-positive cells were observed when Vero cells were incubated with culture fluid derived from the Rluc-Neo-Rep cells transfected with the SFV vector (without WNV structural genes) (right panel). (C) Time course of Rluc activity in cells infected with VLPs. Vero and BHK-21 cells were synchronously infected with Rluc-Neo-Rep-containing VLPs and assayed for Rluc activities at the indicated time points postinfection. Because of the large range of Rluc activity, the Rluc scale (light units) for the right panel is 2 logs higher than that for the left panel. (D) Assay validation. BHK-21 cells were infected with VLPs and immediately treated with mycophenolic acid, ribavirin, or glycyrrhizin at the indicated concentrations. At 48 h after infection and treatment, the cells were lysed and assayed for Rluc activities. The potencies of the compounds were quantified by the percentage of Rluc inhibition in the presence of compound treatment. The results represent the means and standard deviations derived from three independent experiments.
Packaging of VLPs containing WNV replicon.
For the packaging of VLPs, SFV-CprME RNA was transcribed from linear DNA (digested with the SpeI restriction enzyme) in vitro by using a SP6 mMESSAGE mMACHINE kit (Ambion). Approximately 25 μg of the resulting RNA was electroporated into the Rluc-Neo-Rep BHK-21 cells as described previously (37). The culture fluid of the transfected cells was collected and aliquoted at various time points posttransfection and stored at −80°C prior to VLP quantification. The titer of the VLPs was determined by infecting naïve Vero cells (in a four-chamber Lab-Tek Chamber Slide [Nalge Nunc International]) with serial dilutions of the culture fluid collected, followed by counting immunofluorescence assay (IFA)-positive cell foci at 36 h postinfection (p.i.) (12). A clump of IFA-positive cells was counted as one infectious unit. This is because the VLP infection is single cycled and could not spread to neighboring cells. Thus, the foci of IFA-positive cells are most likely derived from division of a single infected cell. For IFA, WNV immune mouse ascitic fluid (American Type Culture Collection, Manassas, VA) and goat anti-mouse immunoglobulin G conjugated with Texas Red were used as primary and secondary antibodies, respectively (37).
Establishment of Vero cells containing persistently replicating WNV replicon.
Vero cells were infected with Rluc-Neo-Rep-containing VLPs and selected with G418 (1 mg/ml) from day 2 postinfection. Individual foci were detached with a sterile cloning disk (Bel-Art Products, Pequannock, NJ) soaked with trypsin. The cells (absorbed onto the disk) were then transferred into a 24-well plate, amplified to generate independent Rluc-Neo-Rep Vero cell lines, and quantified for their luciferase activities. Both low- and high-level luciferase-expressing Rluc-Neo-Rep Vero cell lines were validated in the antiviral assays with known WNV inhibitors.
Time course analysis of reporting VLP and WNV infections.
For VLP infection, approximately 1 × 105 Vero or BHK cells were seeded per well in a 96-well plate. At 16 h postseeding, the cells were infected with VLP at a multiplicity of infection (MOI) of 2 for 1 h, followed by three washings with phosphate-buffered saline (PBS) to remove unattached VLPs. At various time points postinfection, infected cells were washed once with PBS. The plates (without PBS) were sealed with Parafilm and stored at −80°C. Once samples for all time points had been collected, the 96-well plates were subjected to the luciferase assay in a Turner BioSystems luminometer (Promega). For the luciferase WNV infection time course, a procedure similar to that used for VLP infection was performed, except that a six-well format was used (6 × 105 Vero or BHK-21 cells per well were synchronously infected at an MOI of 2). At every time point postinfection, cells were lysed, collected, and stored at −80°C, as described previously (23). When samples for all time points were collected, luciferase activity was quantified in a single-tube luminometer by using a Renilla luciferase assay kit (Promega).
HTS assays.
All compounds were dissolved in dimethyl sulfoxide (DMSO). If they were not soluble, heating at 30°C facilitated the dissolution of the compounds in DMSO. Three types of antiviral assays were established in a 96-well format. (i) The first was a luciferase-expressing replicon cell line assay (see Fig. 1). Approximately 2 × 104 Rluc-Neo-Rep cells (total volume, 100 μl) were seeded per well in medium without G418. After 16 h of incubation, 1 μl of compound was added to the cells. One microliter of DMSO without compound was added to the cells as a negative control. After treatment with compound for 48 h, the cells were washed once with 300 μl of PBS and lysed with 20 μl of lysis buffer on a shaker for 30 min. The 96-well plate was then assayed for luciferase activity in a Turner BioSystems luminometer (Promega). (ii) The second type of antiviral assay was the VLP infection assay (see Fig. 2). Vero or BHK-21 cells were seeded at 4 × 104 cells per well. At 6 h after seeding of the cells, the cells were infected with VLPs at an MOI of 1. Compounds were added simultaneously to the infected cells at a final concentration of 1% DMSO. At 48 h p.i., cells were washed once with PBS, lysed, and assayed for luciferase activity as described above. (iii) The third type of antiviral assay was the reporting WNV infection assay (see Fig. 3). Vero or BHK-21 cells were seeded at 8 × 104 per well. At 6 h postseeding, the cells were infected with luciferase-expressing WNV (MOI, 1) and treated immediately with the compounds at the indicated concentrations. The 96-well plates were assayed for luciferase activity at 24 h postinfection for each treatment.
Three compounds previously reported to have different efficacies against WNV were selected to validate the HTS assays described above. Mycophenolic acid, ribavirin, and glycyrrhizin (Sigma) were reported to have concentrations that are required to inhibit 50% of the viral activity (EC50s) that ranged from the low to the high micromolar range (7, 15, 28). Based on the luciferase signal and the compound concentration curves, regression analysis (SAS, version 6.12; SAS Institute Inc., Cary, NC) was performed to calculate the EC50 for each inhibitor. All assays were performed in duplicate or triplicate.
Screening of a compound library.
A library of small-molecule compounds was screened to identify potential WNV inhibitors. The library contains 200 small molecules with diverse structures (D. M. Ferguson, unpublished results). As primary screenings, the compounds in the library were tested by the reporting-replicon cell line assay and the full-length reporting WNV infection assay, as described above. Each compound was assayed at 30 μM with 1% DMSO final concentrations in a 96-well format. The 30 μM concentration for screening was selected empirically. Compounds demonstrating greater than 50% inhibition of luciferase activity at 30 μM were assayed for their cytotoxicities by an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, followed by titration experiments to estimate their EC50 values in the three reporting assays as well as an authentic viral titer reduction assay.
Viral titer reduction assay.
Vero cells were seeded in a 12-well plate (9 × 105 per well). At 12 h postseeding, the cells were infected with an epidemic strain of WNV (MOI, 0.1) (38) and immediately treated with compound at 1.2, 3.6, 11, 33, 100, and 300 μM. At 42 h posttreatment, the viral titers in the culture medium were determined by a double-layer plaque assay. Briefly, Vero cells were seeded in a six-well plate (6 × 105 per well) and incubated for 3 days until they were nearly confluent. The cells were infected with 100 μl of 1 to 10 serial dilutions of WNV for 1 h at 37°C. Three milliliters of a first layer (0.6% Oxoid agar, basal medium Eagle medium with 1% FBS, 0.02% DEAE dextran, 0.13% NaHCO3) was added onto the infected cells. Two days later, three milliliters of a second layer (1% Noble agar, basal medium Eagle medium with 1% FBS, 0.02% DEAE dextran, 0.13% NaHCO3, 0.004% neutral red) was added over the first layer. The cells were further incubated for 12 h before the plaques were counted.
MTT cell proliferation assay.
A cell proliferation-based MTT assay (American Type Culture Collection) was used to examine the cytotoxicities of the compounds. Approximately 2 × 104 BHK-21 cells or 8 × 104 Vero cells in 100 μl of medium were seeded per well in a 96-well plate. After 6 h of incubation, 1 μl of compound dissolved in DMSO was added to the cells at 1.2, 3.6, 11, 33, 100, 300, and 600 μM. After 48 h of incubation, 10 μl of MTT reagent was added to each well and the cells were incubated for another 3.5 h, after which 100 μl of detergent reagent was added to each well. The plates were swirled gently and left in the dark at room temperature for 4 h. The absorbance was recorded in a Microtiter plate reader (Molecular Devices Corporation, Sunnyvale, CA) with a 550-nm filter. The compound concentrations required to cause 50% cytotoxicity (CC50s) were estimated from the plots of absorbance versus compound concentration.
RESULTS
Use of a replicon-containing cell line in an HTS assay.
To adapt a luciferase-expressing replicon cell line (Rluc-Neo-Rep) (Fig. 1A) into an HTS assay, we initially determined a linear range of reporting cells for seeding of the 96-well plates. Selection of an appropriate cell number per well is critical because cell density could affect the replication efficiency of the replicon, and analysis of inhibitors requires quantification of a decrease in the steady-state level of replicon RNA. As shown in Fig. 1B, a linear range of the luciferase signal was obtained after 5 × 103 to 2 × 104 Rluc-Neo-Rep-containing cells were seeded per well and incubated for 24 and 48 h. When 4 × 104 cells were seeded, the luciferase signal continued to increase at 24 h but declined at 48 h. Based on the cell density results, we chose to seed 2 × 104 cells per well and incubated the cells for 48 h. Under this experimental condition, the assay window (Rluc signal from replicon-bearing cells divided by the background signal from naïve BHK-21 cells) consistently ranged from 1 × 105- to 2 × 105-fold (data not shown). Next, we evaluated the assay using three known WNV inhibitors (Fig. 1C). Incubation of the reporting cells with mycophenolic acid and ribavirin suppressed the Rluc signals in a dose-responsive manner, whereas glycyrrhizin did not inhibit the Rluc activity within the range of concentrations tested. The EC50s for mycophenolic acid, ribavirin, and glycyrrhizin were estimated to be 1.4, 18, and >300 μM, respectively. The results suggest that the replicon-containing cell line could be used in an HTS assay.
Packaging of VLPs containing WNV replicon.
We packaged the WNV replicon into VLPs by supplying structural proteins in trans (Fig. 2). A complete structural polyprotein (C-prM-E) of WNV was expressed through a 26S subgenomic promoter from the SFV expression vector (SFV-CprME) (Fig. 2A). Transfection of Rluc-Neo-Rep cells with SFV-CprME produced infectious VLPs in the culture fluid. Inoculation of naïve Vero cells with the culture fluid from the SFV-CprME-transfected Rluc-Neo-Rep cells yielded a strong IFA signal, indicating viral protein expression in the inoculated cells (Fig. 2B, left panel). As a negative control, Vero cells inoculated with culture fluid from the Rluc-Neo-Rep cells transfected with the empty SFV vector were entirely IFA negative (Fig. 2B, right panel). These results suggest that WNV structural proteins supplied in trans can package the Rluc-Neo-Rep RNA into infectious VLPs. The infectivity of the packaged VLPs was further demonstrated by increases in the Rluc signals in infected Vero and BHK-21 cells (see below). Optimization of the packaging protocol showed that the VLP titer could consistently reach 1 × 105 to 1 × 106 focus-forming units/ml at 48 h after SFV-CprME transfection (data not shown).
Characterization of VLP-mediated infection.
To characterize the VLP-mediated infection, we performed a time course analysis of the Rluc activity in cells synchronously infected with the Rluc-Neo-Rep VLPs (Fig. 2C). The luciferase curve consisted of three distinct phases: (i) a small peak from 2 to 5 h p.i., (ii) a lag phase from 5 to 12.5 h p.i., and (iii) an exponential phase after 12.5 h p.i. The initial small Rluc peak from 2 to 5 h p.i. (phase I) was consistently observed in the VLP-infected Vero cells. In comparison with Vero cells, the VLP-infected BHK-21 cells exhibited a similar Rluc pattern but had lower signals before 17.5 h p.i., after which point the Rluc signal increased to a level equivalent to or higher than that from the Vero cells. In addition, the initial small Rluc peak derived from the BHK-21 cells was much less apparent than that derived from the Vero cells (Fig. 2C). Overall, the increase in Rluc activity in phase III in both cell types strongly suggests the infectivity of the packaged VLPs.
We examined whether, during VLP packaging, homologous RNA recombination between Rluc-Neo-Rep and structural protein-expressing SFV-CprME RNA had occurred, resulting in infectious full-length WNV. We passaged the culture fluids derived from the VLP-infected cells on naïve Vero cells three times and did not detect either an Rluc signal or cytopathic effects (data not shown). The results agreed with those from previous studies with Kunjin virus (16) and tick-borne encephalitis virus (9) that no detectable homologous recombination occurs during trans packaging of flavivirus replicon RNA.
Use of reporting VLPs in an HTS assay.
The VLP infection assay was converted into a 96-well format and validated with known WNV inhibitors. VLP-infected BHK-21 cells (MOI, 1) were treated with mycophenolic acid, ribavirin, or glycyrrhizin. At 48 h p.i., the antiviral effects were quantified by determination of the reduction in Rluc activity. As expected, the Rluc signal was inversely correlated with the concentrations of mycophenolic acid and ribavirin. The EC50s of mycophenolic acid and ribavirin were approximately 0.04 and 1.1 μM, respectively (Fig. 2D). In contrast, glycyrrhizin did not consistently inhibit Rluc activity, even at 300 μM. The assay window was approximately 1 × 105-fold above the background. The results suggest that VLP infection could be used as an HTS assay.
Use of reporting full-length WNV as an HTS assay.
We adapted a full-length reporting WNV infection assay (Rluc-WNV) (Fig. 3A) into a 96-well format. Initially, a time course analysis was performed to examine the kinetics of Rluc-WNV infection in Vero and BHK-21 cells. As shown in Fig. 3B, after an equal number of cells were planted and synchronously infected with Rluc-WNV, the two cell types displayed dramatically different Rluc patterns. For Vero cells, the Rluc signal increased immediately after infection and peaked at 48 h postinfection. In contrast, the Rluc signals from BHK-21 cells displayed a lag phase during the first 12.5 h of infection, after which point the signal increased substantially and reached a level similar to that from the Vero cells at 60 h postinfection.
Next, we validated the assay using known WNV inhibitors. Based on the kinetics of Rluc (Fig. 3B), we treated the Rluc-WNV-infected BHK-21 cells with compounds and assayed the cells for Rluc activity at 24 h p.i. (Fig. 3C). Both mycophenolic acid and ribavirin suppressed Rluc activity in a dose-dependent manner, with EC50 values of approximately 0.08 μM and 1.2 μM, respectively, whereas glycyrrhizin did not consistently affect the Rluc activity. The assay window is approximately 1 × 104 to 5 × 104 fold above the background (data not shown), indicating that the system is robust and could be used as an HTS assay.
Effects of cell types on assay sensitivity.
To examine the choice of permissive cell type on assay sensitivity, we performed the assays with both BHK-21 and Vero cells using all three systems (Table 1). Among the inhibitors tested, the EC50 values of ribavirin derived from BHK-21 and Vero cells varied most significantly. For all three assays, the EC50 values derived from the Vero cells were severalfold higher than those derived from the BHK-21 cells. For example, the EC50 of ribavirin in Rluc-Neo-Rep Vero cells was as high as 119 μM. To ensure that this was not due to an idiosyncrasy of the particular cell line, we repeated the assays in different Rluc-Neo-Rep Vero cell lines (expressing high or low levels of luciferase). An EC50 value of about 119 μM was consistently obtained (data not shown). It was previously reported that, in an authentic WNV infection assay, ribavirin showed a higher EC50 value in Vero cells than in MA-104 cells (28). A low efficiency of ribavirin monophosphate formation (the form required to inhibit IMP dehydrogenase, which leads to the depletion of intracellular GTP pools) was suggested to contribute to the lower efficacy of ribavirin in Vero cells (in which it was about 13-fold less efficient than it was in other cell types) (40). Although mycophenolic acid is also expected to exert its antiviral activity through the inhibition of IMP dehydrogenase, this compound directly binds to the target enzyme without prior modification (39). Consequently, the effect of the cell type on the EC50 of mycophenolic acid was not as dramatic as that on the EC50 of ribavirin. These results clearly demonstrate that the choice of host cell type can significantly influence assay sensitivity.
TABLE 1.
Comparison of EC50 values of mycophenolic acid, ribavirin, and glycyrrhizin obtained from three HTS assays in BHK-21 and Vero cellsa
| Assay system | EC50 (μM)
|
||
|---|---|---|---|
| Mycophenolic acid | Ribavirin | Glycyrrhizin | |
| Rluc-Neo-Rep BHK-21 cells | 1.4 ± 0.48 | 18 ± 0.68 | >300 |
| Rluc-Neo-Rep Vero cells | 0.08 ± 0.015 | 119 ± 63 | >300 |
| VLP BHK-21 cell infection | 0.04 ± 0.002 | 1.1 ± 0.09 | >300 |
| VLP Vero cell infection | 0.02 ± 0.004 | 34 ± 0.92 | >300 |
| Rluc-WNV BHK-21 cell infection | 0.08 ± 0.01 | 1.2 ± 0.005 | >300 |
| Rluc-WNV Vero cell infection | 0.04 ± 0.02 | 30 ± 2.51 | >300 |
The data represent the means ± standard deviations (n ≥ 3).
HTS of a compound library to identify a new WNV inhibitor.
Using the three assays described above, we performed an HTS of a compound library to identify inhibitors of WNV. Since the choice of cell type could potentially affect assay sensitivity, it would be ideal to screen each compound with three assay systems in both BHK-21 and Vero cells (a total of six assays). However, because the amount of each compound is limited, we could not screen the library in all six assays. Therefore, we decided to screen the library in BHK-21 cells bearing the Rluc-Neo-Rep and in Vero cells infected with the full-length Rluc-WNV. Compounds showing greater than 50% inhibition of luciferase activities at 30 μM were subjected to titration experiments to estimate their EC50 values in different assay systems. Both the reporting cell line and the full-length Rluc-WNV-infected Vero cell assays identified compound CDDMN as an inhibitor, with EC50s of 17 and 23 μM, respectively (Table 2) (the compound structure will be described elsewhere). Further analysis of this compound in the reporting VLP infection assay yielded an EC50 of 3 μM. Furthermore, analysis of compound CDDMN in an authentic viral titer reduction assay showed an EC50 of 28 μM (Table 2). These results demonstrate that comparable EC50s could be obtained from the reporting assay systems and from the authentic WNV infection assay. To exclude the possibility that the antiviral activity observed above was due to cytotoxicity, we analyzed the compound in an MTT assay. The CC50 values were estimated to be 500 μM and >600 μM in naïve BHK-21 and Vero cells, respectively. Accordingly, the therapeutic index (which is equal to CC50/EC50) of the compound was >21 in all four systems. In summary, the results presented above clearly demonstrate that the three reporting assays can be used for HTS of compound libraries.
TABLE 2.
Antiviral activity and cytotoxicity of compound CDDMNe
| Parameter | Rluc-Neo-Rep BHK-21 cells | Rluc-VLP Vero cell infection | Rluc-WNV Vero cell infection | WNV Vero cell infectiona |
|---|---|---|---|---|
| EC50 (μM) | 17 | 3 | 23 | 28 |
| CC50 (μM) | 500b | >600c | >600c | >600c |
| TId | 29 | >200 | >26 | >21 |
Vero cells were infected with an epidemic strain of WNV (MOI, 0.1) and immediately treated with compound CDDMN at concentrations ranging from 1.2 to 300 μM. At 42 h of incubation and treatment, the viral yields in the culture medium were determined by plaque assays on Vero cells, as described in Materials and Methods.
Cytotoxicity was tested in naïve BHK-21 cells.
Cytotoxicity was analyzed in Vero cells, with 600 μM being the highest concentration tested.
TI, therapeutic index, which is the CC50 divided by the EC50.
The results represent the averages of two or more independent experiments.
DISCUSSION
The prevention and treatment of flavivirus infections are public health priorities. The goal of this study was to explore genetic systems for the development of HTS assays for flavivirus drug discovery. Using WNV as a model, we show that three cell-based assays could be used for HTS assays: replicon-harboring cells (Fig. 1), packaged VLPs containing replicon RNA (Fig. 2), and full-length reporting WNV (Fig. 3). Compared to the traditional viral titer reduction assay, our systems offer superior speed and sensitivity and therefore are ideal for the screening of libraries with large numbers of compounds. HTS of a compound library by use of the three assays resulted in a complementary “hit” (Table 2), demonstrating the utility of the systems in antiviral drug discovery.
Comparison of genetic and biochemical approaches for HTS assay development.
Two types of approaches are routinely used for antiviral assay development. One approach is biochemistry based, in which the enzymatic activity of purified viral protein is assayed. For flaviviruses, enzymatic HTS assays can be developed for multifunctional NS3 (protease, helicase, nucleoside triphosphatase, and 5′-RNA triphosphatase) and NS5 (methyltransferase and polymerase). Of the numerous HTS platforms, the scintillation proximity assay and fluorescence resonance energy transfer are well established for protease-, helicase-, and polymerase-based assays (13, 32, 42, 44). The principal advantage of the biochemistry-based assay is that the inhibitors identified possess known targets. The other approach is replicon or infectious virus based and usually involves multiple targets of the viral life cycle. Since these assays are cell based and test the cellular uptake and biochemical modification of each compound, inhibitors identified through such assays have a better success rate in subsequent efficacy studies with animals.
The three assays described in this study provide complementary means for WNV drug discovery. The replicon-harboring cell line allows screening for inhibitors of viral replication, including translation, polyprotein processing, and minus- and plus-strand RNA synthesis. Since the replicon lacks structural genes and does not generate infectious particles, the assay can be performed in a biosafety level 2 laboratory. In contrast, the VLP and full-length reporting WNV infection assays involve infectious particles and therefore should be performed in a biosafety level 3 laboratory. Besides viral replication, the VLP and reporting WNV assays also allow screening for potential inhibitors of viral entry. Furthermore, the full-length reporting WNV could be used to search for inhibitors of virion assembly. However, it should be noted that although the reporting WNV is useful for HTS, the luciferase reporter is not stably retained after multiple passages of the reporting virus (8). Nevertheless, the extra features of the VLP and full-length reporting virus infection assays are particularly important, because envelope proteins of flaviviruses undergo a sequential structural change during the fusion-activating transition and during virion particle maturation (1, 4, 18, 26, 27, 41, 48, 49). These structural transitions are essential for the viral life cycle and, therefore, could potentially be targeted for intervention.
The three HTS assays have been validated with known WNV inhibitors. Comparison of the validation results obtained with mycophenolic acid and ribavirin revealed system-to-system variations in the EC50 values for these known inhibitors (Table 1). Specifically, the reporting assays showed EC50s of >300 μM and 1.1 to 119 μM for glycyrrhizin and ribavirin, respectively. These values were similar to those derived from an authentic viral infection assay, 486 μM and 3 to 729 μM for glycyrrhizin and ribavirin, respectively (7, 15, 28). The broad range of EC50 values obtained for ribavirin is most likely due to different efficiencies of ribavirin monophosphate formation among various cell types (40). For mycophenolic acid, the EC50 ranged from 0.02 to 1.4 μM among the three assays; these values are lower than those (0.3 to 8.4 μM) derived from an authentic WNV infection assay (28). Similarly, for compound CDDMN, the EC50s obtained from the reporting systems (3 to 23 μM) were also slightly lower than that derived from the authentic WNV infection assay (28 μM) (Table 2). The discrepancies among EC50 values derived from different assay systems are not surprising because of intrinsic differences among the systems. For example, the RNA replication level in the replicon-containing cells may differ from that in the VLP- or full-length virus-infected cells. Also, insertion of the luciferase reporter into the replicon or full-length virus could negatively affect viral replication efficiency (37). These differences could contribute to the system-to-system variation of EC50 values obtained for a given compound. In spite of these discrepancies, the overall results clearly demonstrated that the assays described here could be used for HTS drug discovery.
The assays described here could also be used to study the modes of action of potential inhibitors. Because each assay involves multiple but partially overlapping steps of the viral life cycle, analysis of an inhibitor (with unknown mechanism) by all three assays could discriminate among an inhibitory role in viral entry, replication, and virion assembly. Furthermore, the luciferase kinetics derived from the Vero cells infected with VLPs could potentially be used to distinguish between viral translation (2 to 5 h p.i.) and RNA replication (after 12.5 h p.i.; Fig. 2C). Differentiation of compound-mediated inhibition between viral translation and RNA synthesis could further be validated by a transient reporting replicon system (see below) (43). Enzymatic assays of WNV NS3 and NS5 (3, 31, 45) could also be used for more specific analysis. Alternatively, the mode of action of the compound could be identified by selection of resistant virus, followed by mapping the mutated gene and back-engineering of specific mutations into an infectious clone (38, 46) for phenotypic verification.
Viral particle-mediated enhancement of WNV RNA replication.
The establishment of a reporting WNV, packaged VLPs, and a replicon has allowed us to use luciferase signals to compare the replication kinetics upon the delivery of viral RNA into host cells. We previously showed that, upon electroporation of BHK-21 cells with a luciferase-expressing replicon, three phases of luciferase activity were observed (Fig. 4) (23, 43). Phase I includes a small Rluc peak from 1 to 10 h posttransfection, which represents the initial translation of input RNA. Phase II is a lag stage from 10 to 20 h posttransfection, when luciferase signals decrease to a background level, which represents input RNA under initial amplification. Phase III is characterized by a substantial Rluc increase after 25 h posttransfection, which represents exponential viral RNA replication. This transient system allows differentiation between viral translation (phase I) and RNA synthesis (phase III) and therefore has been successfully used to dissect the mechanisms of action of inhibitors (8).
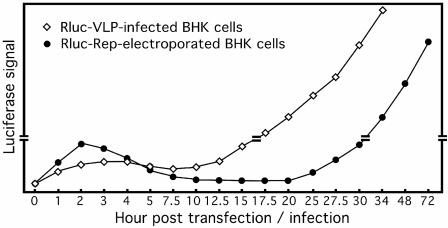
FIG. 4.
Comparison of replication kinetics of the WNV replicon delivered through electroporation and that delivered through VLP-mediated infection. BHK-21 cells were either infected with luciferase VLPs (Rluc-VLP) or electroporated with the luciferase replicon (Rluc-Rep) and assayed for luciferase activities at the indicated time points after electroporation or infection. The luciferase curve for Rluc-Rep-transfected cells was described previously (43). The luciferase curve for Rluc-VLP-infected cells was based on Fig. 2C. The double lines on the vertical axis indicate that the scales of the top and bottom portions of the diagram are different. The drawing is not to scale.
Why was the lag phase (10 to 20 h posttransfection) so long for the electroporated replicon? In this study, we observed a similar three-phase luciferase curve upon VLP infection of Vero cells (Fig. 2C). However, as directly compared in Fig. 4, overlay of the luciferase curve derived from the VLP-infected BHK-21 cells with that derived from the replicon-electroporated BHK-21 cells reveals a significantly shortened lag phase (5 to 12.5 h p.i.) during the VLP infection. Since the electroporated replicon and the VLP replicon contain the luciferase gene engineered at an identical location in the genome (Fig. 1A), the shortened lag phase for the latter system suggests that VLP-mediated cellular delivery of replicon RNA enhances viral replication. This notion was further supported by the shortened lag phase when BHK-21 cells were infected with the luciferase WNV (Fig. 3B). One simple explanation for the enhancement of viral replication observed is that virion particles function as a chaperone for the delivery of the enclosed RNA precisely to the translation apparatus on the endoplasmic reticulum membrane. In contrast, RNA delivered through electroporation is subjected to cellular RNase degradation, and only a few intact molecules likely reach the replication site on the endoplasmic reticulum. Alternatively, the delay in replication of the transfected replicon could result from a traumatic effect of electroporation on host cells. Consequently, a longer time is required for the division of the surviving transfected cells after electroporation. At this time, we do not know what cellular factor(s) contributes to the immediate increase in the luciferase signal (without a lag phase) in Vero cells upon reporting WNV infection. The difference might in part be due to the different defects in the interferon pathway between the Vero and BHK-21 cells.
In summary, the HTS systems described in this study will greatly facilitate WNV drug discovery by serving as primary and secondary screening assays. Although the assays were optimized in the 96-well format in this study, the robustness of the assays should allow these systems to be adapted to a 384-well format. The approach should be applicable to the development of cell-based HTS assays for other flaviviruses. These assays should also be useful for the study of many aspects of WNV, including viral replication, packaging, and pathogenesis.
Acknowledgments
We thank the Molecular Genetics Core and the Cell Culture Facility at the Wadsworth Center for DNA sequencing and for maintenance of BHK-21 and Vero cells, respectively.
The work was supported by grants AI061193, AI065562, and contract N01 AI25490 from the National Institutes of Health and a developmental grant from the Northeast Biodefense Center.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Borowski, P., M. Lang, A. Haag, H. Schmitz, J. Choe, H. M. Chen, and R. S. Hosmane. 2002. Characterization of imidazo[4,5-d]pyridazine nucleosides as modulators of unwinding reaction mediated by West Nile virus nucleoside triphosphatase/helicase: evidence for activity on the level of substrate and/or enzyme. Antimicrob. Agents Chemother. 46:1231-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D. S., and T. P. Monath. 2001. Flaviviruses. Lippincott William & Wilkins, Philadelphia, Pa.
- 6.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 7.Crance, J. M., N. Scaramozzino, A. Jouan, and D. Garin. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 58:73-79. [DOI] [PubMed] [Google Scholar]
- 8.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P.-Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrke, R., M. Ecker, S. W. Aberle, S. L. Allison, F. X. Heinz, and C. W. Mandl. 2003. Incorporation of tick-borne encephalitis virus replicons into virus-like particles by a packaging cell line. J. Virol. 77:8924-8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gehrke, R., F. Heinz, N. Davis, and C. Mandl. 2005. Heterologous gene expression by infectious and replicon vectors derived from tick-borne encephalitis virus and direct comparison of this flavivirus system with an alphavirus replicon. J. Gen. Virol. 86:1045-1053. [DOI] [PubMed] [Google Scholar]
- 11.Guo, J., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harvey, T., W. Liu, X. Wang, R. Linedale, M. Jacobs, A. Davidson, T. Le, I. Anraku, A. Suhrbier, P. Shi, and A. Khromykh. 2004. Tetracycline-inducible packaging cell line for production of flavivirus replicon particles. J. Virol. 78:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ingravallo, P., F. Lahser, E. Xia, B. Sodowich, V. C. Lai, Z. Hong, and W. Zhong. 2001. Characterization of monoclonal antibodies that specifically recognize the palm subdomain of hepatitis C virus nonstructural protein 5B polymerase. Virus Res. 75:179-187. [DOI] [PubMed] [Google Scholar]
- 14.Jones, C., C. Patkar, and R. Kuhn. 2005. Construction and applications of yellow fever virus replicons. Virology 331:247-259. [DOI] [PubMed] [Google Scholar]
- 15.Jordan, I., T. Briese, N. Fischer, J. Y. Lau, and W. I. Lipkin. 2000. Ribavirin inhibits West Nile virus replication and cytopathic effect in neural cells. J. Infect. Dis. 182:1214-1217. [DOI] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 21.Liu, W., X. Wang, V. Mokhonov, P. Shi, R. Randall, and A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo, L., M. Tilgner, K. Bernard, and P.-Y. Shi. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus using a reporting replicon that differentiates between viral translation and RNA replication. J. Virol. 77:10004-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo, L., M. Tilgner, and P.-Y. Shi. 2003. A potential high-throughput assay for screening inhibitors of West Nile virus replication. J. Virol. 77:12901-12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 28.Morrey, J., D. Smee, R. Sidwell, and C. Tseng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 30.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nomaguchi, M., T. Teramoto, L. Yu, L. Markoff, and R. Padmanabhan. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J. Biol. Chem. 279:12141-12151. [DOI] [PubMed] [Google Scholar]
- 32.Parniak, M. A., K. L. Min, S. R. Budihas, S. F. Le Grice, and J. A. Beutler. 2003. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 322:33-39. [DOI] [PubMed] [Google Scholar]
- 33.Pierson, T., M. Diamond, A. Ahmed, L. Valentine, C. Davis, M. Samuel, S. Hanna, B. Puffer, and R. Doms. 2005. An infectious West Nile virus that expresses a GFP reporter gene. Virology 334:28-40. [DOI] [PubMed] [Google Scholar]
- 34.Rossi, S., Q. Zhao, V. O'Donnell, and P. Mason. 2005. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology 331:457-470. [DOI] [PubMed] [Google Scholar]
- 35.Scholle, F., Y. A. Girard, Q. Zhao, S. Higgs, and P. W. Mason. 2004. trans-Packaged West Nile virus-like particles: infectious properties in vitro and in infected mosquito vectors. J. Virol. 78:11605-11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi, P.-Y. 2003. Genetic systems of West Nile virus and their potential applications. Curr. Opin. Investig. Drugs 4:959-965. [PubMed] [Google Scholar]
- 37.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 38.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sintchak, M. D., M. A. Fleming, O. Futer, S. A. Raybuck, S. P. Chambers, P. R. Caron, M. A. Murcko, and K. P. Wilson. 1996. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell 85:921-930. [DOI] [PubMed] [Google Scholar]
- 40.Smee, D. F., M. Bray, and J. W. Huggins. 2001. Antiviral activity and mode of action studies of ribavirin and mycophenolic acid against orthopoxviruses in vitro. Antivir. Chem. Chemother. 12:327-335. [DOI] [PubMed] [Google Scholar]
- 41.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taliani, M., E. Bianchi, F. Narjes, M. Fossatelli, A. Urbani, C. Steinkuhler, R. De Francesco, and A. Pessi. 1996. A continuous assay of hepatitis C virus protease based on resonance energy transfer depsipeptide substrates. Anal. Biochem. 240:60-67. [DOI] [PubMed] [Google Scholar]
- 43.Tilgner, M., and P. Y. Shi. 2004. Structure and function of the 3′ terminal six nucleotides of the West Nile virus genome in viral replication. J. Virol. 78:8159-8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson, K. F., B. D. Rush, S. K. Sharma, D. B. Evans, M. J. Ruwart, J. M. Friis, M. J. Bohanon, and P. K. Tomich. 1993. Development of activity assays for high-volume evaluation of human immunodeficiency virus (HIV) protease inhibitors in rat serum: results with ditekiren. Pharm. Res. 10:562-566. [DOI] [PubMed] [Google Scholar]
- 45.Wong, S. J., R. H. Boyle, V. L. Demarest, A. N. Woodmansee, L. D. Kramer, H. Li, M. Drebot, R. A. Koski, E. Fikrig, D. A. Martin, and P.-Y. Shi. 2003. An immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J. Clin. Microbiol. 41:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 47.Yun, S. I., S. Y. Kim, C. M. Rice, and Y. M. Lee. 2003. Development and application of a reverse genetics system for Japanese encephalitis virus. J. Virol. 77:6450-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]