Abstract
We investigated a multiresistant strain of Acinetobacter baumannii isolated in our hospital. Analysis of the N-terminal peptide sequence of the outer membrane proteins (OMPs) purified from the strain allowed us to clone and sequence the nucleotides of the gene encoding the 33- to 36-kDa OMP associated with carbapenem resistance in A. baumannii
Acinetobacter spp. are recognized as opportunistic pathogens of increasing relevance in nosocomial infections (4, 5).
Antimicrobial treatment of serious infections caused by Acinetobacter spp., particularly those caused by Acinetobacter baumannii, is complicated by the widespread multidrug resistance pattern of this microorganism (1, 6, 20, 23). The most common mechanism of resistance to β-lactam antibiotics is by synthesis of a naturally occurring AmpC-type cephalosporinase (7), although low permeability of the outer membrane has also been involved (22). Regarding carbapenems, penicillin-binding proteins (PBPs) with reduced affinities for outer membrane proteins (OMPs) (11, 13) and a loss or a reduction of OMPs of 22 and 29 kDa and one of 31 to 36 kDa have been implicated in carbapenem resistance (6, 9-10, 16, 20). Among these, only the nucleotide sequence of the 29-kDa OMP gene has been described (3, 20).
Between October 2001 and August 2002, 30 patients admitted to the Juan Canalejo Hospital became infected or colonized by a multiresistant epidemic strain of A. baumannii (23). The main aims of the present study were to clone the gene encoding the OMP, previously described by different authors as being from 31 to 36 or 33 to 36 kDa, of A. baumannii and to demonstrate its involvement in resistance to carbapenems in the epidemic strain JC10/01 (from the index case of the outbreak).
Susceptibility testing of strain JC10/01 was performed by microdilution (21), and MICs were confirmed by Etest (AB Biodisk, Solna, Sweden). The antibiotic susceptibility profiles of all strains included in the present study are shown in Table 1. Strain JC10/01 was resistant to all β-lactam antibiotics tested, including carbapenems (Table 1).
TABLE 1.
MICs of the Acinetobacter isolates included in this study
| Antibiotic(s) | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| JC10/01b | EG-1c | EG-1revd | JC10/01 (pAT-RA-34s)e | JC10/01 (pAT-RA-34r)e | JC10/01 (pAT-RA-c)e | |
| Amoxicillin | >256 | >256 | >256 | >256 | >256 | >256 |
| Amoxicillin-clavulanatea | >256 | 48 | >256 | 32 | 48 | >256 |
| Piperacillin | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefalothin | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefuroxime | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefoxitin | >256 | >256 | >256 | >256 | >256 | >256 |
| Cefotaxime | >256 | >256 | >256 | 256 | 256 | >256 |
| Ceftazidime | 128 | 48 | 128 | 32 | 48 | 128 |
| Cefepime | 48 | 12 | 48 | 12 | 12 | 32 |
| Aztreonam | 256 | 48 | 256 | 48 | 48 | 128 |
| Imipenem | 32 | 4 | >32 | 2 | 4 | 16 |
| Meropenem | >32 | 12 | >32 | 8 | 12 | >32 |
| Tobramycin | 4 | 4 | 4 | 4 | 4 | 4 |
| Amikacin | 128 | 128 | 128 | 128 | 128 | 128 |
| Ciprofloxacin | >32 | >32 | >32 | >32 | >32 | >32 |
| Tetracycline | >256 | >256 | >256 | >256 | >256 | >256 |
Clavulanic acid was used at a concentration of 4 μg/ml.
JC10/01, A. baumannii epidemic strain from the index case of the outbreak.
EG-1, A. baumannii revertant from the JC10/01 strain.
EG-1rev, A. baumannii revertant from the EG-1 strain.
JC10/01 strain was transformed with the indicated plasmids. MICs were determined in the presence of 50 μg/ml of rifampin. Identical MICs were obtained with three different transformants in each case.
β-Lactamases were analyzed by isoelectric focusing, as described by Matthew et al. (19); and with strain JC10/01, only one band with a pI of >8.5 was detected, strongly indicating the presence of the previously described chromosomal cephalosporinase (7), which was confirmed by using ampC-specific primers P1 and P2 (Table 2) in a PCR assay (data not shown). To rule out a carbapenemase enzyme, a disk assay was performed (18), which yielded a negative result with both imipenem and meropenem. Semipurified protein extracts of the JC10/01 strain were also used in a spectrophotometric assay of these antibiotics, in which no carbapenem hydrolysis was detected. Furthermore, a PCR with consensus oligonucleotides specific for VIM-type, IMP-type, and OXA-type carbapenemase genes was performed; and negative results were obtained. The overall results suggested that strain JC10/01 did not contain any carbapenemases.
TABLE 2.
Oligonucleotides included in this study
To investigate the molecular basis of carbapenem resistance in the JC10/01 isolate, a spontaneous revertant (strain EG-1) derived from strain JC10/01 was obtained after 11 passages on Luria-Bertani (LB) agar plates (9). The imipenem and meropenem MICs for the EG-1 strain were reduced from 32 to 4 μg/ml and from >32 to 12 μg/ml, respectively (Table 1). Therefore, we suggest that carbapenem resistance may be caused by differences in the OMPs in the two strains.
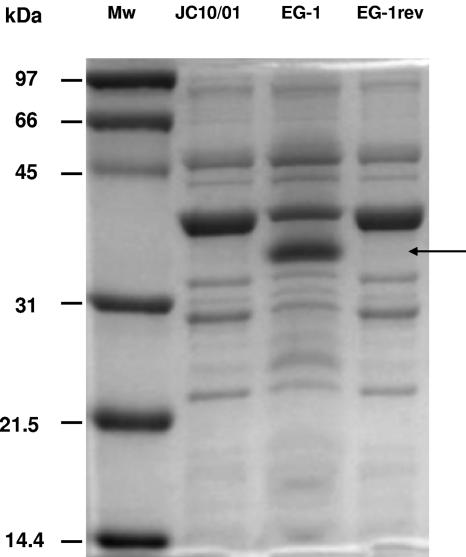
OMPs of both the A. baumannii JC10/01 and EG-1 strains were analyzed by previously described methods (6, 11-12). Analysis of the OMPs of the Acinetobacter isolates revealed a band profile similar to that reported in a previous study (11) (Fig. 1) and showed the presence of an additional 33- to 36-kDa protein in the EG-1 isolate, suggesting that the loss of this OMP is involved in carbapenem resistance (Fig. 1). Moreover, a second imipenem-resistant revertant was obtained from EG-1 by successive selection in media containing different concentrations of imipenem. For this, EG-1 was first grown overnight in LB broth containing 4 μg/ml of imipenem, and then 0.1-ml aliquots of this culture were spread on LB agar plates containing the same amount of antibiotic. Clones of resistant acinetobacters were reisolated on a second agar plate with the same concentration of imipenem; and the procedure was repeated by using 8, 16, and 32 μg/ml of imipenem. After repeated isolation a clone resistant to carbapenems (strain EG-1rev) (Table 1) was chosen for further studies. The OMP pattern of this strain indicated the disappearance of the 33- to 36-kDa protein, thus strongly suggesting that the loss of this polypeptide is linked to resistance to carbapenems (Fig. 1).
FIG. 1.
Electrophoretic analysis of OMPs (ca. 30 μg each) with a 10% sodium dodecyl sulfate-polyacrylamide gel with 6 M urea stained with Coomassie brilliant blue R-250. Lanes: Mw, molecular mass markers of the indicated sizes; JC10/01, purified OMPs from strain JC10/01; EG-1, purified OMPs from strain EG-1; EG-1rev, purified OMPs from strain EG-1rev. The arrow indicates the position of the 33- to 36-kDa OMP.
After 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the resolved OMPs were transferred to polyvinylidene difluoride paper. Edman degradation analysis of the N-terminal sequence was carried out with a Procise 494 analyzer from Applied Biosystems (Foster City, CA); the 33- to 36-kDa polypeptide yielded the following amino acid sequence of the N-terminal region: Tyr-Gln-Phe-Glu-Val-Gln-Gly-Gln-Ser-Glu.
A search by using the release of the Acinetobacter sp. strain ADP1 genome (http://www.genoscope.cns.fr) indicated that the 10-amino-acid peptide sequence of the 33- to 36-kDa OMP shared 90% identity with a region spanning residues 25 to 34 ofa hypothetical protein (EMBL protein database accession no. YP_047932), which is 300 amino acids long and which has a theoretical molecular mass of 31,987 Da. Moreover, use of the protein-protein BLAST algorithm (2) to compare this protein sequence produced significant alignments with a set of porins and OMPs from a Ralstonia sp., OmpF from Serratia marcescens, a Wolbachia sp., and Burkholderia cepacia, strongly suggesting the first identification of the gene encoding the 33- to 36-kDa OMP.
The next step was to attempt to clone this gene in A. baumannii. For this, two oligonucleotides encoding the intergenic sequence surrounding the 33- to 36-kDa OMP gene, P3 and P4 (which include part of the promoter and the 3′ untranslated region, respectively) (Table 2), were used in a PCR assay in which the chromosome of the A. baumannii JC10/01 strain and a carbapenem-susceptible clinical strain of A. baumannii (JC7/04) (identified by amplified ribosomal DNA restriction analysis) that lacks any genetic relationship (repetitive extragenic palindromic-PCR tested) with the JC10/01 isolate were used as the template. A band of ca. 1 kb was obtained in both cases, and this sequence was cloned into the Topo plasmid (Topo TA cloning kit; Invitrogen, Carlsbad, CA). Nucleotide sequencing of the cloned fragment showed an open reading frame (ORF) of 900 nucleotides encoding a protein of 299 amino acids with a theoretical molecular mass of 32,115 Da with the JC10/01 strain, whereas a polypeptide of 293 amino acids long with a theoretical molecular mass of 31,435 Da was obtained with the JC7/04 isolate.
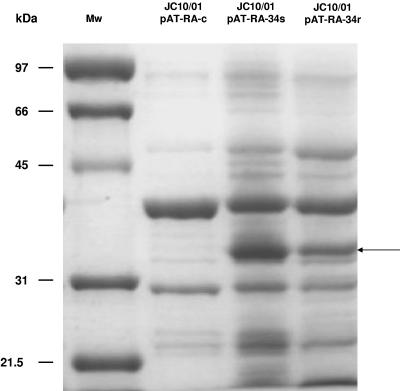
We aimed to determine whether carbapenem resistance is linked to the loss of the 33- to 36-kDa OMP noted above. For this the 33- to 36-kDa OMP genes from isolates JC10/01 and JC7/04 were cloned into the pAT-RA plasmid (made of part of pUC18 and pWH1266), which harbors a replication origin for A. baumannii (15) and which codes for rifampin resistance. The 33- to 36-kDa OMP ORFs linked to the promoter from the CTX-M-14 gene (positions 1502 to 1740 of the sequence with EMBL database accession no. AF252622) were amplified with oligonucleotides P5 and P6 (Table 2), thus generating an unique amplicon, and afterwards were cloned into SmaI and EcoRI restriction sites of pAT-RA, yielding the pAT-RA-34r and pAT-RA-34s recombinant plasmids, respectively. These plasmids were then electroporated into the JC10/01 isolate, and transformants were selected on LB agar plates with 50 μg/ml of rifampin. The pAT-RA plasmid harboring solely the CTX-M-14 promoter (pAT-RA-c) was used as a negative control. The MICs to different antibiotics of JC10/01 with pAT-RA-34r, pAT-RA-34s, and pAT-RA-c as a negative control are shown in Table 1. OMP analysis confirmed expression of the 33- to 36-kDa OMP in pAT-RA-34r/s transformants but not in the negative control (Fig. 2).
FIG. 2.
Electrophoretic analysis of OMPs (ca. 30 μg each) of JC10/01 strain transformed with the indicated plasmids with a 10% sodium dodecyl sulfate-polyacrylamide gel with 6 M urea stained with Coomassie brilliant blue R-250. Lanes: Mw, molecular mass markers of indicated sizes; JC10/01 pAT-RA-c, purified OMPs from strain JC10/01 as negative control; JC10/01 pAT-RA-34s, purified OMPs from strain JC10/01 expressing the 33- to 36-kDa OMP from JC7/04 isolate; JC10/01 pAT-RA-34r, purified OMPs from strain JC10/01 expressing the 33- to 36-kDa OMP from JC10/01 isolate. The arrow indicates the position of the 33- to 36-kDa OMP.
In the present study we have reported on the cloning, and functional analysis of the gene encoding the 33- to 36-kDa OMP of A. baumannii. This OMP has previously been associated with carbapenem resistance by different authors (6, 9-10). Here we report that resistance to carbapenems is also associated with the loss of this 33- to 36-kDa OMP in the epidemic strain under study.
It should be emphasized that the amino acid sequence and composition of the 33- to 36-kDa OMP of A. baumannii was typical of those of gram-negative bacterial porins (14) because of the following features: (i) a high glycine content; (ii) the absence of cysteine residues; (iii) a total negative charge (theoretical pI of 4.84); (iv) an overall average hydropathicity of −0.171 and an instability index of 4.75 (stable protein) (both parameters were determined following the instructions of ProtParam [http://www.expasy.ch]); (v) the absence of hydrophobic residue stretches; and (vi) protein functional analysis of the 33- to 36-kDa OMP, performed with the InterProScan program (www.ebi.ac.uk), which revealed similarity with transmembrane β barrels as well as with bacterial membrane and cell surface proteins.
Indeed, JC10/01 harboring the 33- to 36-kDa OMP genes from two different A. baumannii isolates showed a clear reduction in the MICs of imipenem and meropenem compared with those of JC10/01 (Table 1). Therefore, both experimental approaches presented here, (i) genetic OMP-plasmid transformation and (ii) biochemical studies by OMP profile analysis, show that this OMP is involved in resistance to carbapenems (and other β-lactam antibiotics) (Table 1) in A. baumannii.
Indeed, it has previously been described in members of the family Enterobacteriaceae that the loss of OMPs is associated with imipenem resistance, although this was in addition to other mechanisms (8, 17). Nonetheless, we have provided here strong evidence showing that the restored production of the OMP is necessary to compromise the carbapenem resistance of the JC10/01 isolate.
In summary, although we did not evaluate any possible role of PBPs in our epidemic strain, carbepenem resistance in the epidemic strain under study was caused by the loss of this new 33- to 36-kDa OMP.
Nucleotide sequence accession number.
The nucleotide sequences of the 33- to 36-kDa OMP genes from JC10/01 and anA. baumannii clinical isolate (JC7/04) were submitted to theEMBL database and have been assigned accession nos. AM039631 and AJ831523, respectively.
Acknowledgments
This study was financially supported by Direccion Xeral de I+D, Xunta de Galicia (grant PGIDI4BTF916028PR), Fondo de Investigaciones Sanitarias (grants PI021415 and PI040514), and RESITRA (grant G03/75). Alejandro Beceiro is a receipt of a scholarship from SEIMC.
We thank P. Nordmann for the kind gift of pAT-RA plasmid and Merck & Co., Inc., for the kind of gift of imipenem.
REFERENCES
- 1.Afzal, M. S., and D. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J. Antimicrob. Chemother. 41:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbe, V., D. Vallenet, N. Fonknechten, and A. Kreimeyer. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann, P. 1968. Isolation of Acinetobacter from soil and water. J. Bacteriol. 96:39-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou, G., G. Cervero, M. A. Domínguez, C. Quereda, and J. Martínez- Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bou, G., and J. Martínez-Beltrán. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, P. A., C. Urban, N. Mariano, S. J. Projan, J. J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 10.Costa, S. F., J. Woodcock, M. Gill, R. Wise, A. A. Barone, H. Caiaffa, and A. S. Levin. 2000. Outer-membrane proteins pattern and detection of beta-lactamases in clinical isolates of imipenem-resistance Acinetobacter baumannii from Brazil. Int. J. Antimicrob. Agents 13:175-182. [DOI] [PubMed] [Google Scholar]
- 11.Fernández-Cuenca, F., L. Martínez-Martínez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between β-lactamase production, outer membrane protein, and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 12.Fernández-Cuenca, F., A. Pascual, L. Martínez-Martínez, M. C. Conejo, and E. J. Perea. 2003. Evaluation of SDS-polyacrylamide gel systems for the study of outer membrane protein profiles of clinical strains of Acinetobacter baumannii. J. Basic Microbiol. 43:194-201. [DOI] [PubMed] [Google Scholar]
- 13.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumannii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 14.Gribun, A., Y. Nitzan, I. Pechatnikov, G. Hershkovitz, and D. J. Katcoff. 2003. Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47:434-443. [DOI] [PubMed] [Google Scholar]
- 15.Hunger, M., R. Schmucker, V. Kishan, and W. Hillen. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45-51. [DOI] [PubMed] [Google Scholar]
- 16.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kDa outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Alvárez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Roles of β-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda, G., S. Tomioka, and M. Hasegawa. 1976. Detection of β-lactamase production by gram-negative bacteria. J. Antibiot. (Tokyo) 29:662-664. [DOI] [PubMed] [Google Scholar]
- 19.Matthew, M., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 20.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Sato, K., and T. Nakae. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J. Antimicrob. Chemother. 28:33-45. [DOI] [PubMed] [Google Scholar]
- 23.Tomas, M., M. Cartelle, S. Pertega, A. Beceiro, P. Llinares, D. Canle, F. Molina, R. Villanueva, J. M. Cisneros, and G. Bou. 2005. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin. Microbiol. Infect. 11:540-546. [DOI] [PubMed] [Google Scholar]