Abstract
It has been recently shown that resistance to both imipenem and meropenem in multidrug-resistant clinical strains of Acinetobacter baumannii is associated with the loss of a heat-modifiable 25/29-kDa outer membrane protein, called CarO. This study aimed to investigate the channel-forming properties of CarO. Mass spectrometry analyses of this protein band detected another 25-kDa protein (called Omp25), together with CarO. Both proteins presented similar physicochemical parameters (Mw and pI). We overproduced and purified the two polypeptides as His-tagged recombinant proteins. Circular dichroism analyses demonstrated that the secondary structure of these proteins was mainly a β-strand conformation with spectra typical of porins. We studied the channel-forming properties of proteins by reconstitution into artificial lipid bilayers. In these conditions, CarO induced ion channels with a conductance value of 110 pS in 1 M KCl, whereas the Omp25 protein did not form any channels, despite its suggested porin function. The pores formed by CarO showed a slight cationic selectivity and no voltage closure. No specific imipenem binding site was found in CarO, and this protein would rather form unspecific monomeric channels.
Acinetobacters are nonfermentative gram-negative coccobacilli found in soil and water. The species Acinetobacter baumannii is also part of the normal flora of human skin and the gastrointestinal and upper respiratory tracts (6, 58). This opportunistic pathogen causes a variety of hospital-acquired infections, including pneumonia, urinary tract infections, bacteremia, and meningitis, especially in immunocompromised patients (for a review, see reference 6). Hospital outbreaks due to multidrug-resistant (MDR) A. baumannii strains have become increasingly common in hospitals worldwide and especially in intensive care units (17, 18, 51) and burn units (6, 10, 37, 52, 56, 58). These multidrug-resistant strains are frequently resistant to the usual drug families such as aminoglycosides, fluoroquinolones, and β-lactams (penicillins and cephalosporins), although carbapenems remain the most used antimicrobial drugs. An increasing number of studies, however, reported the emergence of clinical A. baumannii strains that are resistant to imipenem (13, 16, 45). Molecular mechanisms such as acquisition of carbapenemases (1, 8-11, 20, 49), reduced affinity of a penicillin binding protein for the drug (20, 21), or the loss of outer membrane (OM) proteins were described previously (8, 12, 14, 36, 48, 50).
Antibiotic resistance suggests modifications of OM permeability. However, there are very little data available on the membrane proteins of A. baumannii. Of the data available, a channel-forming function was determined for 45.5- and 47-kDa minor OM proteins (55) and for the 37/43-kDa major OM protein (35, 43). This protein was the only porin identified and was demonstrated as belonging to the OmpA-like family of proteins (23). The loss of 33-, 22-, and 25/29-kDa protein bands seen on OM protein extracts of clinical isolates by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) suggested their role in carbapenem resistance (8, 12, 36), although no structural and functional studies were carried out for these proteins.
Here, we investigated the function of the 25/29-kDa protein band of A. baumannii, called CarO, which may allow the influx of carbapenem antibiotics by a predicted transmembrane β-barrel (40). We used mass spectrometry to show that there were two proteins present in this electrophoretic band. Using circular dichroism (CD) spectra, we confirmed that both proteins may adopt β-barrel folding. We also provide evidence that only one of the proteins, CarO itself, was able to form channels in lipid bilayers.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Acinetobacter baumannii strain ATCC 19606 was purchased from the Pasteur's Institute Collection (Paris, France). Bacteria were grown at 37°C in Mueller-Hinton broth (Difco). Strains used for cloning and expression of recombinant proteins were Escherichia coli DH5α (Clontech Laboratories, Inc., Palo Alto, Calif.) and E. coli BL21λ(DE3)(pLysS) (Novagen, R&D Systems Europe Ltd., Abingdon, Oxford, United Kingdom). All E. coli strains were grown and maintained in LB medium at 37°C. When required, media were supplemented with 100 μg/ml ampicillin and/or 50 μg/ml chloramphenicol (Sigma).
SDS-PAGE and N-terminal sequencing.
Discontinuous SDS-PAGE (7% stacking, 14% separating gel) (35a) was used for outer membrane protein separation. Staining was done by silver staining or with 0.1% (wt/vol) Coomassie brilliant blue G 250 solution. N-terminal sequences were obtained using an Applied Biosystems Procise 492A sequencer after protein electroblotting onto polyvinylidene difluoride membranes (Millipore, Bedford, Mass.).
Mass spectrometry.
Stained protein bands were excised from SDS-PAGE gels and subjected to tryptic digestion using the method described previously by Vilain et al. (57). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) was carried out on a Bruker BIFLEX III spectrometer (Bruker Daltonics, Bremen, Germany) equipped with SCOUT High Resolution Optics with an X-Y multisample probe and a gridless reflector. Analyses were carried out using α-cyano-4-hydroxycinnamic acid as a matrix. In positive mode, internal calibration was performed with peptides from the autodigestion of trypsin. Monoisotopic peptide masses were assigned and used for database searches (MASCOT; Matrix Science, London, United Kingdom). The peptide mass error was ±50 ppm.
Tryptic digests were analyzed using nanoscale capillary liquid chromatography (nanoLC)-tandem mass spectrometry (MS/MS) (Q-TOF II; Micromass, Altrincham, United Kingdom). Chromatographic separations were carried out on a reversed-phase capillary column (Pepmap C18 [75-μm internal diameter, 15-cm length]; LC Packings, Amsterdam, The Netherlands) at a flow rate of 200 nl/min. Argon was used as the fragmentation collision gas, with a collision energy profile optimized for various mass ranges of ion precursors. Four ion precursors were fragmented at a time. Mass data collected from the nanoLC-MS/MS analysis were processed and then submitted to de novo sequencing.
Mass spectrometry data analysis.
From the MS/MS spectra, de novo sequences were found using PEAKS Studio (Bioinformatics Solutions Inc., Waterloo, Canada) and analyzed with the BioEdit v5.0.9 software (24) with the Gapped BLAST algorithm (2) using the nonredundant NCBI protein database (http://www.ncbi.nlm.nih.gov) or the Genoscope Acinetobacter sp. ADP1 protein database (http://www.genoscope.cns.fr/agc/mage) (see reference 4).
Amplification, cloning, and sequencing of carO and omp25.
The 678-bp carO gene fragment lacking the code for the presumed amino-terminal signal sequence, with appropriate sites at both ends, was synthesized by PCR amplification. We used A. baumannii ATCC 19606 genomic DNA as a template with the following degenerated primers based on the A. baumannii 15839 sequenced genome (U.S. patent 6562958, May 2003): #249 (5′-TATGGATCCGATGAAGCTGTTGTTCATGACAGC-3′) and #250 (5′-TATGAATTCTTACCAGAAGAAGTTCACACCAAC-3′). This DNA fragment was restricted by BamHI and EcoRI and ligated into vector pETSIG. This was digested with the same enzymes, yielding pETSIG-carO. The pETSIG vector is a pET (Novagen, Madison, Wis.) derivative specially constructed for this study. It allows the expression of the His-tagged protein of interest in the N-terminal fusion with the signal peptide of E. coli OmpA porin to target the proteins to the membrane.
The 702-bp omp25 gene fragment lacking the code for the presumed amino-terminal signal sequence, with appropriate sites at both ends, was synthesized by PCR amplification. We used A. baumannii ATCC 19606 genomic DNA as a template with the following degenerated primers: #251 (5′-TAT GGA TCC TAC CAA GCT GAA GTT GGT GGT CG-3′) and #252 (5′-TAT GAA TTC TTA GAA GCG GTA TGC TGC ACG AAC-3′). This DNA fragment was restricted by BamHI and EcoRI and ligated into vector pETSIG. This was digested with the same enzymes, yielding pETSIG-omp25. The nucleotide sequences of the carO and omp25 genes from A. baumannii ATCC 19606 were obtained using an ABI Prism Dye Terminator cycle sequencing kit (Perkin-Elmer, Foster City, Calif.).
Extraction and purification of native CarO protein.
A. baumannii strain ATCC 19606 was grown and harvested at the late exponential phase (5 × 108 CFU) by centrifugation (8,000 × g, 20 min) (JLA 10.500; Beckman-Coulter, Fullerton, Calif.). Inner and outer membrane samples were separated using the sucrose gradient method described previously by Hancock and Nikaido (28) and then stored at −80°C. The Omp25 protein was first obtained using preparative SDS-PAGE. Outer membrane extracts were solubilized at 1 mg/ml by incubation for 1 h at 4°C in a buffer containing 2% (wt/vol) SDS, 62.5 mM Tris-HCl, pH 6.8, 10% (vol/vol) glycerol, and 0.001% (wt/vol) bromophenol blue and then centrifuged for 3 min at 5,000 × g. The supernatant was loaded onto an acrylamide gel (7% stacking, 14% separating gel [16-cm length]; SE 600; Hoefer, San Francisco, Calif.). Bands corresponding to the Omp25 protein were excised and then electroeluted using a Biotrap system (Schleicher & Schuell, Dassel, Germany) with a solution of 192 mM glycine, 25 mM Trizma base, and 0.1% Triton X-100.
Expression and purification of recombinant CarO and Omp25 proteins.
E. coli BL21λ(DE3)(pLysS) cells were transformed with the pETSIG-carO or pETSIG-omp25 vectors. These recombinant E. coli strains harboring the pETSIG derivatives were used to inoculate LB medium supplemented with ampicillin and chloramphenicol and were incubated at 37°C under shaking until the A600 reached 0.5. IPTG (isopropyl-β-d-thiogalactopyranoside) was then added at a final concentration of 0.5 mM, and growth was continued for an additional 3 h at 37°C with shaking. The cells were harvested by centrifugation at 6,000 × g for 10 min and resuspended in lysis buffer (50 mM Tris-HCl, pH 8, 300 mM NaCl, 10% glycerol, 1 mM mercaptoethanol, antiprotease cocktail [Roche]). The cells were disrupted by sonication, and the resulting suspension was centrifuged at 100,000 × g for 30 min at 4°C. The membrane pellet was resuspended and incubated for 1 h on a rotating wheel at room temperature in extraction buffer (50 mM Tris-HCl, pH 8, 300 mM NaCl, 3% n-octyl-polyoxyethylene (OPOE), antiprotease cocktail [Roche]). The resulting extraction suspension was centrifuged at 100,000 × g for 30 min at 4°C. The supernatant was incubated with Ni-nitrilotriacetic acid agarose suspension (QIAGEN, Venlo, The Netherlands) suitable for purification of His6 fusion proteins. The protein-resin complex was packed into a column for washing and elution. The column was washed extensively with a solution containing 50 mM Tris-HCl, pH 8, 300 mM NaCl, 1% OPOE, and 15 mM imidazole. Protein elution was carried out with a solution containing 50 mM Tris-HCl, pH 8, 300 mM NaCl, 1% OPOE, and 200 mM imidazole. Eluted fractions were analyzed by SDS-PAGE or immunoblotting. Immunoblot analysis was carried out in a 12.5% SDS-polyacrylamide gel, and proteins were electroblotted onto an Immobilon-P membrane (Millipore). INDIA HisProbe-HRP (SuperSignal West HisProbe kit; Pierce Chemicals, Rockford, Ill.), a nickel-activated derivative of horseradish peroxidase, was then used to directly detect the blotted recombinant polyhistidine-tagged fusion proteins. The SuperSignal kit (Pierce) and BioMax film (Kodak, New Haven, Conn.) were used for protein visualization. The concentration of the samples was determined using the Bio-Rad (Hercules, Calif.) protein assay.
CD spectroscopy.
Proteins were dialyzed overnight at 4°C against 1% OPOE-200 mM NaCl-20 mM phosphate buffer (pH 7.5) before measurements were taken. CD spectra were recorded at room temperature for solutions of proteins (0.4 or 0.6 mg/ml) using a CD6 spectropolarimeter (Jobin-Yvon, Longjumeau, France). Ellipticity values were expressed as mean residue molar ellipticity as deg centimeter−2 decimole−1. Each spectrum was acquired between 200 and 250 nm, in 1-nm steps, with a 0.02-cm path length and an integration time of 3 s. Experimental data were baseline corrected by subtracting a blank, i.e., 1% OPOE solution-200 mM NaCl-20 mM phosphate buffer (pH 7.5). Both experimental and blank values were the means of five recordings.
Reconstitution in planar lipid bilayers.
Bilayer membranes were formed from a 0.5-mg/ml hexane solution of DPhPC (Avanti Polar Lipids, Alabaster, Ala.), according to the method described previously by Montal and Mueller (38), over a 100- to 150-μm-diameter orifice in a 10-μm-thick Teflon film (Goodfellow Corp., Devon, Pa.) separating the two chambers of a planar bilayer apparatus. Each chamber contained 2 ml of electrolyte solution of 1 M KCl buffered with 10 mM HEPES (pH 7.4). A potential difference was applied across the bilayer. When a channel was inserted inside the membrane, the movement of ions through the pore created a current which was recorded with a BLM-120 amplifier (Bio-Logic, Claix, France) connected to the chambers via Ag/AgCl electrodes. Data were acquired on a DTR-1201 digital audio tape (Bio-Logic), low-pass filtered with an AF-180 eight-pole filter (Bio-Logic), and sampled by a computer at 3 to 6 kHz. Data were analyzed using Satori 3.01 software from INTRACEL (Royston, United Kingdom). For selectivity experiments, we set up a KCl gradient across the lipid bilayer from 0.1 M in the cis side to 1 M in the trans side. The zero current potential was corrected by subtracting the asymmetric potential resulting from the salt gradient. PK and PCl refer to the permeabilities to potassium and chloride, respectively.
Nucleotide sequence accession numbers.
The nucleotide sequences of the carO and omp25 genes from A. baumannii ATCC 19606 have been deposited in the EMBL nucleotide sequence database under accession numbers AJ938079 and AJ938080, respectively.
RESULTS AND DISCUSSION
The outer membrane permeability of A. baumannii is poorly characterized. There are very few OM proteins that have been identified and definitively characterized as porins. However, their role in antibiotic influx and resistance in other gram-negative bacteria is not in doubt (25, 41).
Analysis of major outer membrane proteins of A. baumannii.
We isolated the OM proteins from the A. baumannii ATCC 19606 envelope using the discontinuous sucrose gradient procedure and analyzed them by SDS-PAGE. We saw three major protein bands. Two of them presented apparent molecular masses of 25 and 37 kDa on the gel (Fig. 1A, lane 1), which, after heating the sample at 95°C for 10 min, shifted to 29 and 45 kDa, respectively (Fig. 1A, lane 2). This shift is a common feature of integral monomeric OM proteins exhibiting a major β-barrel structure (27). The N-terminal sequence of the 37/45-kDa protein was GVTVTPLLLG, which is 100% similar to the N terminus of the heat-modifiable protein (HMP) from A. baumannii (23). The HMP is the most extensively studied porin in A. baumannii and is structurally related to OmpA of E. coli or OprF of Pseudomonas aeruginosa (23, 43, 44, 47). A liposome swelling assay showed that this protein allowed the penetration of β-lactams and saccharides of up to 800 Da but with an efficiency that was much lower than that of the major OmpF porin of E. coli (23, 43, 44). It was suspected that this OmpA-like protein, like its monomeric homologues (42), contributes poorly to the membrane permeability (23).
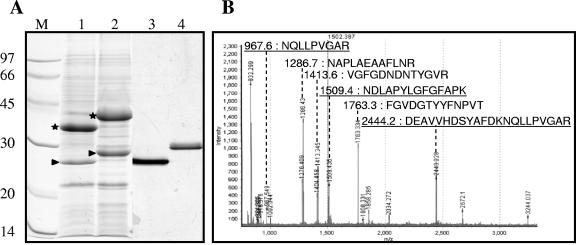
FIG. 1.
Purification and identification of the CarO major OM protein of A. baumannii strain ATCC 19606. (A) SDS-PAGE analysis on 14% polyacrylamide gels (Coomassie blue staining for lanes 1 and 2 and silver staining for lanes 3 and 4). Lane 1, OM fraction; lane 2, OM fraction heated for 10 min at 95°C (positions of major HMP and 25/29-kDa OM proteins are indicated by stars and arrowheads, respectively); lane 3, purified 25/29-kDa protein; lane 4, purified 25/29-kDa protein heated for 10 min at 95°C. The molecular mass and position of each of the different standards (lane M) are shown on the left. (B) Mass spectrometry analysis of the 25/29-kDa protein. MALDI-TOF [MH+] masses and the corresponding sequences are shown for the peptides identified by nanoLC-MS/MS using the Acinetobacter sp. strain ADP1 databank. Underlined peptides correspond to the CarO protein, and the others correspond to Omp25.
The other major protein, the 25/29-kDa protein band, had an N-terminal sequence (DEAVVHDSYA) that was 100% similar to the first 10 residues of the P83446 sequence from the SWISS-PROT knowledge database (36). This sequence corresponded to the protein CarO (GenBank accession number AAV80243). The loss of this protein is thought to be associated with carbapenem resistance in A. baumannii clinical isolates (40). However, the porin function or the role of this protein in the carbapenem influx was suggested but not demonstrated (40). Therefore, we decided to purify this protein for further functional investigations.
Identification and characterization of the native CarO protein.
We purified the protein corresponding to the 25/29-kDa band by preparative electrophoresis followed by electroelution in the nonionic detergent Triton X-100 to study its functional and structural properties (Fig. 1A). After SDS-PAGE, the protein band showed the same temperature migration shift as mentioned above, demonstrating that the protein retained its correct folding (Fig. 1A, lanes 3 and 4). Therefore, we evaluated its channel-forming properties by protein reconstitution in planar lipid bilayers. We carried out several experiments and found conductance increments distributed over a large range of values corresponding to different sizes of ion channels. This heterogeneity led us to reinvestigate the quality of the purified sample. We carried out a tryptic digestion on the excised protein band, and we analyzed the resulting peptides by mass spectrometry. We could not identify the protein by mass fingerprinting obtained from MALDI-MS analysis in Acinetobacter sp. strain ADP1 (an Acinetobacter strain with a completely sequenced and annotated genome) (see reference 4). Therefore, we used tandem mass spectrometry to analyze the digested protein. From the MS/MS data, we determined six sequences for the peptides with m/z's of 967.6, 1,286.7, 1,413.6, 1,509.4, 1,763.3, and 2,444.2 [MH+] (Fig. 1B). These peptides were highly conserved with sequences from two different proteins in Acinetobacter sp. strain ADP1: ACIAD2598, a protein similar to CarO (77 to 86% identity) (sequences underlined in Fig. 1B), and ACIAD3499, a 25-kDa protein (61 to 87% identity), the so-called Omp25. Using the GenBank database, we also found identical sequences for a CarO-like protein (accession number AAQ29344) and for an Omp25-like protein (accession number AAQ29195) in A. baumannii 15839.
Amino acid sequence analyses of CarO and Omp25 in A. baumannii strain 15839 showed very similar physicochemical parameters (MW/pI, 24,422 Da/4.61 and 25,597 Da/4.47, respectively), explaining their copurification. At this point, these data suggest that when resistance to carbapenems is studied, the comparison between OM protein profiles of different A. baumannii strains by SDS-PAGE should be taken very carefully. The separation of these two proteins was very difficult whatever purification method we used. Therefore, we decided to construct and purify their corresponding recombinant proteins to study their individual properties.
Determination and analysis of the nucleotide sequences of carO and omp25.
We determined the N-terminal residues of CarO by Edman sequencing (not shown). This sequence was 100% similar to that already published (36). For the Omp25 protein, the identified peptides (Fig. 1B) were identical with those obtained from a predicted protein of A. baumannii strain 15839 (GenBank accession number AAQ29195). According to this sequence and using the signalP algorithm (5), we predicted the first N-terminal sequence of the mature form of Omp25 to be YQAEVGGSYN. We confirmed it in A. baumannii 19606 by reanalyzing the mass fingerprinting data: the 2,350.1-Da peak referred to the predicted N-terminal sequence YQAEVGGSYNYLDPDNGSSVSK in the 15839 strain (data not shown).
Degenerate primers were designed using the genome database of A. baumannii 15839. The carO and omp25 gene fragments lacking the sequence for their native peptide signal were synthesized by PCR amplification using A. baumannii ATCC 19606 chromosomic DNA. The amplified products were inserted into pETSIG to yield pETSIG-carO and pETSIG-omp25 from which we determined their nucleotide sequences.
Analysis of the primary structure indicated that the mature form of CarO comprises 228 residues, with a theoretical molecular weight of 24,772 Da and an isoelectric point value of 4.50. We found similar CarO sequences in other Acinetobacter species, A. baumannii Ab244 (70% identity), A. baumannii 15839 (74% identity), and Acinetobacter sp. strain ADP1 (71% identity), by using the NCBI protein database and the gapped BLAST algorithm. We found a high overall identity rate (between 58% and 82% overall similarity) between the four strains. However, two regions of about 20 amino acids showed large differences: 111A-R139 and 183T-A205, with 33% and 34% similarity, respectively (numbering begins at the first amino acid of the mature form of CarO). A previous alignment and a secondary structure prediction were performed on the family CarO proteins according to a method described previously by Mussi et al. (40). This suggested that these proteins form β-barrels comprising 10 β-strands. In this model, β-sheets 6 and 7 appeared in the variable area 111A-R139 of the CarO proteins. This is in contrast to usually observed features for β-barrels in which the variable sequences reside in the cell-surface-exposed domains (loops) consistent with their numerous functions, whereas β-strands are more conserved (34). We compared the domains and signatures of the protein families using InterProScan (version 10) (see reference 39) to predict CarO function. We detected the SCOP (structural classification of protein) superfamily (22) OmpA-like signature (SSF56925) consistent with the β-barrel folding for this protein and its suggested porin function (40).
By comparing the sequence of the mature form of Omp25 polypeptide (233 amino acid residues, 25,596 Da, pI 4.47) with sequences in protein databases, we identified eight protein sequences. We found two highly conserved proteins: Omp25 in A. baumannii 15839 (100% identity) and Omp25 in Acinetobacter sp. strain ADP1 (GenBank accession number YP_047967) (67% identity). However, Omp23 of Microbulbifer degradans 2-40 (accession number ZP_00314866) and Omp28 of A. baumannii 15839 (accession number AAQ29140) were less conserved (30% and 38%, respectively). An alignment of those sequences (data not shown) also highlighted, in the Acinetobacter genus, a subcluster of three similar proteins (accession numbers YP_O47932, AAQ29097, and CAH40481) with higher molecular masses (≈30 kDa) due to the insertion of 22 amino acid residues in the medium region of the sequences. Despite their low overall similarity level (28%), these proteins all exhibit a highly conserved pattern, 43PLAEAAFL50, located in the N-terminal domain. This sequence appears to characterize the Omp25 protein family. They also exhibit common features usually described for bacterial OM proteins. These include a significant Ala/Gly-rich content (about 20%), an aromatic amino acid-rich content (11 to 14%), and a C-terminal aromatic residue (Phe). The above-described data and the location of the proteins in the OM suggest β-barrel folding for this family of proteins.
Function prediction for Omp25 using InterProScan identified a SCOP superfamily porin signature (SSF56935) for all eight proteins, consistent with this type of folding. Moreover, M. degradans Omp23 exhibited the OmpT protease signature (SSF69917). It has recently been suggested that one member of this protein family, the Omp33-36 protein (CAH40841), is implicated in the multiresistance of an A. baumannii clinical isolate, as well having a suspected porin function (8, 12).
In conclusion, the Omp25 and CarO primary sequence analyses suggested β-barrel folding and a porin function, suggesting a functional role of these proteins in the bacterial multidrug resistance.
Expression, purification, and conformation of recombinant CarO and Omp25 proteins.
We used recombinant E. coli strains harboring the pETSIG-carO and pETSIG-omp25 vectors to overproduce the CarO and Omp25 His-tagged recombinant proteins fused to the E. coli OmpA signal peptide. The polyhistidine tag made purification easier, and the OmpA signal peptide allowed the fusion protein to maintain proper addressing to the outer membrane. We analyzed the total protein fractions by SDS-PAGE (Fig. 2A, lanes 2, 3, 6, and 7) and immunoblotting (Fig. 2B, lanes 2, 3, 6, and 7) and found that CarO and Omp25 proteins were present in the membrane fraction, whereas we did not detect immunoreactive bands from the noninduced overexpressing strains carrying pETSIG-carO or pETSIG-omp25 (Fig. 2A and B, lane 1). The recombinant proteins were therefore purified to homogeneity using OPOE detergent conditions (Fig. 2A and B, lanes 4, 5, 8, and 9). We used Edman sequencing to determine the sequence of first N-terminal residues of purified recombinant CarO and Omp25 proteins, revealing that the signal peptide had been properly cleaved, generating the mature His-tagged CarO and Omp25 proteins. The purified proteins were then treated with thrombin to remove the His6 tag. We tested several different conditions but were unable to remove the tag, suggesting that CarO and Omp25 were folded in a conformation in which the His6 tag was inaccessible to the thrombin cleavage.
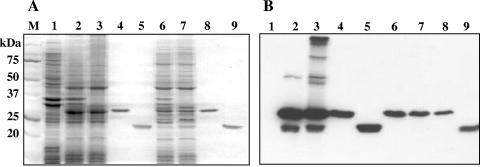
FIG. 2.
Overexpression and purification of the recombinant OM proteins CarO and Omp25 of A. baumannii ATCC 19606. (A) SDS-PAGE (12.5%) with Coomassie blue staining. Lane 1, membrane fraction before overexpression; lanes 2 and 3, membrane fraction containing overproduced CarO heated for 10 min at 100°C or without heating, respectively; lanes 4 and 5, purified CarO with or without heating, respectively; lanes 6 and 7, membrane fraction containing overproduced Omp25 with or without heating, respectively; lanes 8 and 9, purified Omp25 with or without heating, respectively. The molecular mass and position of each of the different standards (lane M) are shown on the left. (B) Immunoblot analysis of the same samples with antibodies directed against the His-tagged CarO and Omp25 OM proteins.
We verified the correct folding of the purified proteins by using their heat-modifiable migration (27). Nonheated purified CarO and Omp25 proteins subjected to electrophoresis (Fig. 2A, lanes 5 and 9) appeared to migrate faster than the related heated denatured monomer (Fig. 2A, lanes 4 and 8), suggesting a folded monomer conformation. Interestingly, we found low levels of immunoreactive species with high molecular masses in the nonheated membrane fraction containing overproduced CarO (Fig. 2B, lane 3). These bands may be dimers, trimers, and higher oligomers. However, the immunoblotting analysis of the purified protein suggested that these oligomeric structures were lost during the purification (Fig. 2B, lane 5). Studies have shown that purified oligomeric OM proteins, such as OprD from P. aeruginosa or major OM protein from Campylobacter jejuni, can also dissociate into monomers upon 0.1% SDS treatment during SDS-PAGE analysis and spontaneously reassemble into trimers upon dilution of SDS below 0.01%, for example, in a native gel (7, 15, 59). However, the CarO protein did not reoligomerize in 0.01% SDS gel (not shown). That these oligomeric species are unstable casts doubts on their biological relevance. We did not detect any of these complexes for the Omp25 protein (Fig. 2B, lanes 6 to 9), and we presume that both CarO and Omp25 proteins were present in the membrane as monomers.
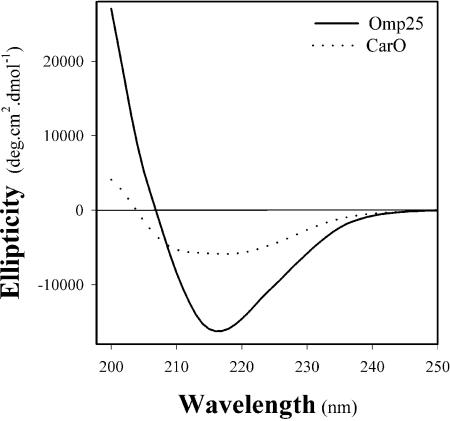
We carried out circular dichroism experiments to examine the secondary structure of the recombinant purified proteins CarO and Omp25. The spectra of both proteins showed a maximum ellipticity between 210 and 220 nm (Fig. 3), characteristic of OM proteins such as porins, which have conformations rich in β-strands and form β-barrel (19). These results confirmed that the CarO and Omp25 purified recombinant proteins folded correctly, making it possible to use them in further functional investigations.
FIG. 3.
Circular dichroism spectra of recombinant CarO (dotted curve) and Omp25 (solid curve). Purified CarO (0.4 mg/ml) and Omp25 (0.6 mg/ml) are in a solution containing 1% OPOE, 200 mM NaCl, and 20 mM NaPi, pH 7.5.
Functional assays using planar lipid bilayers.
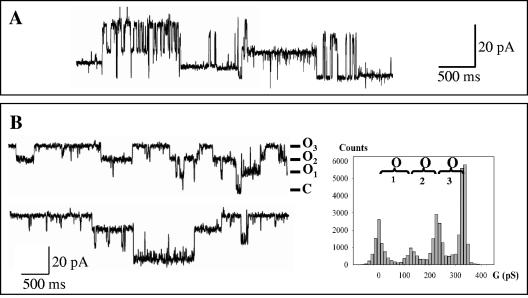
The native 25/29-kDa protein band (Fig. 1A) was first reconstituted in DPhPC membranes. The sample was added into the cis compartment of the measurement cell at a concentration of 10−9 M. After applying a potential, we detected several current increments, which demonstrated channel formation at different conductance values: 100 and 140 pS but also 300 and 400 pS under different reconstitution conditions (Fig. 4A). These multiple conductance increments may be due to various oligomeric forms of the protein or to contamination by other channel-forming proteins. As mass spectrometry analyses showed sample contamination by the Omp25 protein, we carried out separate reconstitutions of recombinant CarO and Omp25 proteins.
FIG. 4.
Single-channel recordings of native and recombinant A. baumannii OM proteins. Proteins (5 × 10−9 M) were reconstituted into DPhPC planar bilayers in 1 M KCl-10 mM HEPES, pH 7.4. (A) Triton X-100-solubilized CarO and Omp25. The recordings show different current levels for an applied potential of 120 mV. (B) Left panel, selected recording showing the single-channel activity of CarO at an applied voltage of 80 mV. O, open state at different current levels; C, closed state. Right panel, histogram associated with the recording showing that the different current levels correspond to similar conductance values: 105 ± 15 pS.
When the purified recombinant CarO was introduced into the measurement cell, the current instantly increased after applying a potential (Fig. 4B), demonstrating typical channel-forming behavior of CarO, with perfectly characterized and reproducible current fluctuations as shown by the associated conductance histogram. The reincorporated CarO protein did not show a threshold potential for channel activation or the voltage-gated behavior in the range of ±170 mV usually seen for unspecific trimeric porins such as E. coli OmpF. Ten independent experiments with the CarO protein resulted in the formation of channels with a major conductance level of 110 ± 20 pS. We did not detect any smaller conductance increments but found infrequent and higher conductance levels (310, 400, and 800 pS), which may correspond to the incorporation into the lipid bilayer of large aggregates. This major conductance value can be compared to the ionophore properties already described for the channel-forming part of proteins such as E. coli OmpA or P. aeruginosa OprF. The N-terminal transmembrane part of these proteins (20 kDa) forms an eight-stranded β-barrel with a very similar conductance value of 75 to 85 pS in 1 M NaCl or 110 pS in 1 M KCl (3, 54). As CarO protein has been described as participating in the influx of carbapenem antibiotics through the A. baumannii membrane, we compared its ionophore properties with those of the P. aeruginosa OprD porin. The OprD porin in reconstituted artificial bilayers showed a much smaller conductance value (20 pS in 1 M KCl) for a predicted 16-stranded β-barrel (30, 32, 33) than for reconstituted CarO (31, 46). Furthermore, the conductivity measurements for this porin are consistent with the presence of a specific binding site to basic amino acids or imipenem (30, 33). Therefore, we carried out assays to determine the putative presence of such a binding site for the CarO channel. Macroscopic conductance inhibition experiments were carried out in 1 M KCl with a CarO concentration 10 times higher than that for the single-channel measurements (10−8 M). After waiting for equilibrium of the porin incorporation, imipenem was added by aliquots to one side and then to the other side of the membrane, giving a final concentration of 1 mM in the bulk. We saw no inhibition of conductance whatever conditions were used: electrolyte composition, imipenem concentration, or duration of the experiment. This demonstrated the lack of an imipenem-specific binding site in this channel. Finally, we investigated the ionic selectivity of the CarO channel. After applying a KCl gradient (0.1 M:1 M, cis:trans), we estimated from the resulting reversal potential, using the Goldman-Hodgkin-Katz equation (29), a PK/PCl ratio of 2.7 ± 0.2 (n = 4). This indicated a slight selectivity for cations, which is consistent for nonspecific porins such as P. aeruginosa OprF (26) and E. coli OmpA (53). All these reconstitution experiments demonstrated a porin function for CarO with ionophore features very similar to those of the channel domain (the N-terminal part) of E. coli OmpA or P. aeruginosa OprF (54).
We also carried out reconstitution assays with the Omp25 protein. However, this protein did not generate discrete channels. Irrespective of the protein concentration (up to 10−7 M) or electrolyte used (NaCl), we found only spikes of very different current amplitudes (data not shown). This result was surprising, considering that the other experimental data suggested a porin function for this Omp25 protein. There are two possible explanations: the presence of the His6 tag may obstruct the channel and prevent the ionic conduction, or the protein has another function, as suggested by the OmpT protease signature found for one of the Omp25 family proteins.
In conclusion, this study contributed to characterize the molecular bases of the A. baumannii OM permeability, which is necessary for a better understanding of the antibiotic resistance mechanisms of this organism. We demonstrated that the 25/29-kDa OM protein band of A. baumannii, until now called CarO, was in fact two proteins that adopted a typical β-barrel conformation. However, only one of these proteins (the CarO protein) displayed pore-forming properties. No binding site for imipenem could be detected in CarO, suggesting an unspecific monomeric channel function rather than a specific function, as previously suggested by Mussi et al. (40).
Acknowledgments
This work was supported by the Haute-Normandie region with a grant attributed to A.S.
We thank the Laboratoire de Spectrométrie de Masse BioOrganique and A. VanDorsselaer, Strasbourg, France, for mass spectrometry facilities, G. Molle, J. M. Pagès, P. Cosette, and K. Glinel for helpful discussions, and L. Coquet for technical support.
REFERENCES
- 1.Afzal-Shah, M., and D. M. Livermore. 1998. Worldwide emergence of carbapenem-resistant Acinetobacter spp. J. Antimicrob. Chemother. 41:576-577. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora, A., D. Rinehart, G. Szabo, and L. K. Tamm. 2000. Refolded outer membrane protein A of Escherichia coli forms ion channels with two conductance states in planar lipid bilayers. J. Biol. Chem. 275:1594-1600. [DOI] [PubMed] [Google Scholar]
- 4.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, S. Cruveiller, C. Robert, S. Duprat, P. Wincker, L. N. Ornston, J. Weissenbach, P. Marliere, G. N. Cohen, and C. Medigue. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Bergogne-Berezin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolla, J. M., E. Loret, M. Zalewski, and J. M. Pages. 1995. Conformational analysis of the Campylobacter jejuni porin. J. Bacteriol. 177:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bou, G., G. Cerveró, M. A. Domínguez, C. Quereda, and J. Martínez- Beltrán. 2000. Characterization of a nosocomial outbreak caused by a multiresistant Acinetobacter baumannii strain with a carbapenem-hydrolyzing enzyme: high-level carbapenem resistance in A. baumannii is not due solely to the presence of β-lactamases. J. Clin. Microbiol. 38:3299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou, G., and J. Martínez-Beltrán. 2000. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 44:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bou, G., A. Oliver, and J. Martínez-Beltrán. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, R. B. 1996. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 38:245-251. [DOI] [PubMed] [Google Scholar]
- 13.Corbella, W., A. Montero, M. Pujol, M. A. Dominguez, J. Ayats, M. J. Argerich, F. Garrigosa, J. Ariza, and F. Gudiol. 2000. Emergence and rapid spread of carbapenem resistance during a large and sustained hospital outbreak of multiresistant Acinetobacter baumannii. J. Clin. Microbiol. 38:4086-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Silva, G. J., G. J. Leitao, and L. Peixe. 1999. Emergence of carbapenem-hydrolyzing enzymes in Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 37:2109-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dé, E., M. Jullien, G. Labesse, J. M. Pages, G. Molle, and J. M. Bolla. 2000. MOMP (major outer membrane protein) of Campylobacter jejuni; a versatile pore-forming protein. FEBS Lett. 469:93-97. [DOI] [PubMed] [Google Scholar]
- 16.Del Mar Tomas, M., M. Cartelle, S. Pertega, A. Beceiro, P. Llinares, D. Canle, F. Molina, R. Villanueva, J. M. Cisneros, and G. Bou. 2005. Hospital outbreak caused by a carbapenem-resistant strain of Acinetobacter baumannii: patient prognosis and risk-factors for colonisation and infection. Clin. Microbiol. Infect. 11:540-546. [DOI] [PubMed] [Google Scholar]
- 17.Denton, M., M. H. Wilcox, P. Parnell, D. Green, V. Keer, P. M. Hawkey, I. Evans, and P. Murphy. 2004. Role of environmental cleaning in controlling an outbreak of Acinetobacter baumannii on a neurosurgical intensive care unit. J. Hosp. Infect. 56:106-110. [DOI] [PubMed] [Google Scholar]
- 18.El Shafie, S. S., M. Alishaq, and M. Leni Garcia. 2004. Investigation of an outbreak of multidrug-resistant Acinetobacter baumannii in trauma intensive care unit. J. Hosp. Infect. 56:101-105. [DOI] [PubMed] [Google Scholar]
- 19.Fasman, G. D. 1996. Differentiation between transmembrane helices and peripheral helices by the deconvolution of circular dichroism spectra of membrane proteins, p. 381-412. In G. D. Fasman (ed.), Circular dichroism and the conformational analysis of biomolecules. Plenum Press, New York, N.Y. [DOI] [PMC free article] [PubMed]
- 20.Fernandez-Cuenca, F., L. Martinez-Martinez, M. C. Conejo, J. A. Ayala, E. J. Perea, and A. Pascual. 2003. Relationship between beta-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 51:565-574. [DOI] [PubMed] [Google Scholar]
- 21.Gehrlein, M., H. Leying, W. Cullmann, S. Wendt, and W. Opferkuch. 1991. Imipenem resistance in Acinetobacter baumanii is due to altered penicillin-binding proteins. Chemotherapy 37:405-412. [DOI] [PubMed] [Google Scholar]
- 22.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 23.Gribun, A., Y. Nitzan, I. Pechatnikov, G. Hershkovits, and D. J. Katcoff. 2003. Molecular and structural characterization of the HMP-AB gene encoding a pore-forming protein from a clinical isolate of Acinetobacter baumannii. Curr. Microbiol. 47:434-443. [DOI] [PubMed] [Google Scholar]
- 24.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 25.Hancock, R. E. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27:S93-S99. [DOI] [PubMed] [Google Scholar]
- 26.Hancock, R. E. 1987. Model membrane studies of porin function, p. 187-225. In M. Inouye (ed.), Bacterial outer membranes as model systems. John Wiley & Sons, New York, N.Y.
- 27.Hancock, R. E., and A. M. Carey. 1979. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock, R. E., and H. Nikaido. 1978. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J. Bacteriol. 136:381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hille, B. 1984. Selective permeability: independence, p. 226-248. In B. Hille (ed.), Ionic channels of excitable membranes. Sinauer Associates, Sunderland, Mass.
- 30.Huang, H., and R. E. Hancock. 1996. The role of specific surface loop regions in determining the function of the imipenem-specific pore protein OprD of Pseudomonas aeruginosa. J. Bacteriol. 178:3085-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, H., D. Jeanteur, F. Pattus, and R. E. Hancock. 1995. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol. Microbiol. 16:931-941. [DOI] [PubMed] [Google Scholar]
- 32.Ishii, J., and T. Nakae. 1993. Lipopolysaccharide promoted opening of the porin channel. FEBS Lett. 320:251-255. [DOI] [PubMed] [Google Scholar]
- 33.Ishii, J., and T. Nakae. 1996. Specific interaction of the protein-D2 porin of Pseudomonas aeruginosa with antibiotics. FEMS Microbiol. Lett. 136:85-90. [DOI] [PubMed] [Google Scholar]
- 34.Jeanteur, D., J. H. Lakey, and F. Pattus. 1991. The bacterial porin superfamily: sequence alignment and structure prediction. Mol. Microbiol. 5:2153-2164. [DOI] [PubMed] [Google Scholar]
- 35.Jyothisri, K., V. Deepak, and M. R. Rajeswari. 1999. Purification and characterization of a major 40 kDa outer membrane protein of Acinetobacter baumannii. FEBS Lett. 443:57-60. [DOI] [PubMed] [Google Scholar]
- 35a.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Limansky, A. S., M. A. Mussi, and A. M. Viale. 2002. Loss of a 29-kilodalton outer membrane protein in Acinetobacter baumannii is associated with imipenem resistance. J. Clin. Microbiol. 40:4776-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montal, M., and P. Mueller. 1972. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA 69:3561-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bradley, P. Bork, P. Bucher, L. Cerutti, R. Copley, E. Courcelle, U. Das, R. Durbin, W. Fleischmann, J. Gough, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, D. Lonsdale, R. Lopez, I. Letunic, M. Madera, J. Maslen, J. McDowall, A. Mitchell, A. N. Nikolskaya, S. Orchard, M. Pagni, C. P. Ponting, E. Quevillon, J. Selengut, C. J. Sigrist, V. Silventoinen, D. J. Studholme, R. Vaughan, and D. H. Wu. 2005. InterPro, progress and status in 2005. Nucleic Acids Res. 33:D201-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mussi, M. A., A. S. Limansky, and A. M. Viale. 2005. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: natural insertional inactivation of a gene encoding a member of a novel family of β-barrel outer membrane proteins. Antimicrob. Agents Chemother. 49:1432-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikaido, H., K. Nikaido, and S. Harayama. 1991. Identification and characterization of porins in Pseudomonas aeruginosa. J. Biol. Chem. 266:770-779. [PubMed] [Google Scholar]
- 43.Nitzan, Y., E. B. Deutsch, and I. Pechatnikov. 2002. Diffusion of beta-lactam antibiotics through oligomeric or monomeric porin channels of some gram-negative bacteria. Curr. Microbiol. 45:446-455. [DOI] [PubMed] [Google Scholar]
- 44.Nitzan, Y., I. Pechatnikova, D. Bar-Ela, and H. Wexler. 1999. Isolation and characterization of heat-modifiable proteins from the outer membrane of Porphyromonas asaccharolytica and Acinetobacter baumannii. Anaerobe 5:43-50. [DOI] [PubMed] [Google Scholar]
- 45.Nordmann, P. 2004. Acinetobacter baumannii, the nosocomial pathogen par excellence. Pathol. Biol. 52:301-303. [DOI] [PubMed] [Google Scholar]
- 46.Ochs, M. M., M. Bains, and R. E. Hancock. 2000. Role of putative loops 2 and 3 in imipenem passage through the specific porin OprD of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:1983-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ofori-Darko, E., Y. Zavros, G. Rieder, S. A. Tarle, M. Van Antwerp, and J. L. Merchant. 2000. An OmpA-like protein from Acinetobacter spp. stimulates gastrin and interleukin-8 promoters. Infect. Immun. 68:3657-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh, E.-J., S. Lee, Y.-J. Park, J. J. Park, K. Park, S.-I. Kim, M. W. Kang, and B. K. Kim. 2003. Prevalence of metallo-β-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-β lactamase. J. Microbiol. Methods 54:411-418. [DOI] [PubMed] [Google Scholar]
- 49.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quale, J., S. Bratu, D. Landman, and R. Heddurshetti. 2003. Molecular epidemiology and mechanisms of carbapenem resistance in Acinetobacter baumannii endemic in New York City. Clin. Infect. Dis. 37:214-220. [DOI] [PubMed] [Google Scholar]
- 51.Rello, J., and E. Diaz. 2003. Pneumonia in the intensive care unit. Crit. Care Med. 31:2544-2551. [DOI] [PubMed] [Google Scholar]
- 52.Roberts, S. A., R. Findlay, and S. D. Lang. 2001. Investigation of an outbreak of multi-drug resistant Acinetobacter baumannii in an intensive care burns unit. J. Hosp. Infect. 48:228-232. [DOI] [PubMed] [Google Scholar]
- 53.Saint, N., E. Dé, S. Julien, N. Orange, and G. Molle. 1993. Ionophore properties of OmpA of Escherichia coli. Biochim. Biophys. Acta 1145:119-123. [DOI] [PubMed] [Google Scholar]
- 54.Saint, N., C. El Hamel, E. De, and G. Molle. 2000. Ion channel formation by N-terminal domain: a common feature of OprFs of Pseudomonas and OmpA of Escherichia coli. FEMS Microbiol. Lett. 190:261-265. [DOI] [PubMed] [Google Scholar]
- 55.Sato, K., and T. Nakae. 1991. Outer membrane permeability of Acinetobacter calcoaceticus and its implication in antibiotic resistance. J. Antimicrob. Chemother. 28:35-45. [DOI] [PubMed] [Google Scholar]
- 56.Sengupta, S., P. Humar, A. M. Ciraj, and P. G. Shivananda. 2001. Acinetobacter baumannii—an emerging nosocomial pathogen in the burns unit Manipal, India. Burns 27:140-144. [DOI] [PubMed] [Google Scholar]
- 57.Vilain, S., P. Cosette, M. Hubert, C. Lange, G. A. Junter, and T. Jouenne. 2004. Comparative proteomic analysis of planktonic and immobilized Pseudomonas aeruginosa cells: a multivariate statistical approach. Anal. Biochem. 329:120-130. [DOI] [PubMed] [Google Scholar]
- 58.Wong, T. H., B. H. Tan, M. L. Ling, and C. Song. 2002. Multi-resistant Acinetobacter baumannii on a burns unit—clinical risk factors and prognosis. Burns 28:349-357. [DOI] [PubMed] [Google Scholar]
- 59.Yoshihara, E., H. Yoneyama, and T. Nakae. 1991. In vitro assembly of the functional porin trimer from dissociated monomers in Pseudomonas aeruginosa. J. Biol. Chem. 266:952-957. [PubMed] [Google Scholar]