Abstract
Human papillomaviruses (HPVs) are the causative agents of benign and malignant lesions of the epithelium. Despite their high prevalence, there is currently no antiviral drug for the treatment of HPV-induced lesions. The ATPase and helicase activities of the highly conserved E1 protein of HPV are essential for viral DNA replication and pathogenesis and hence are considered valid antiviral targets. We recently described novel biphenylsulfonacetic acid inhibitors of the ATPase activity of E1 from HPV type 6 (HPV6). Based on kinetics and mutagenesis studies, we now report that these compounds act by an allosteric mechanism. They are hyperbolic competitive inhibitors of the ATPase activity of HPV6 E1 and also inhibit its helicase activity. Compounds in this series can also inhibit the ATPase activity of the closely related enzyme from HPV11; however, the most potent inhibitors of HPV6 E1 are significantly less active against the type 11 protein. We identified a single critical residue in HPV6 E1, Tyr-486, substituted by a cysteine in HPV11, which is primarily responsible for this difference in inhibitor potency. Interestingly, HPV18 E1, which also has a tyrosine at this position, could be inhibited by biphenylsulfonacetic acid derivatives, thereby raising the possibility that this class of inhibitors could be optimized as antiviral agents against multiple HPV types. These studies implicate Tyr-486 as a key residue for inhibitor binding and define an allosteric pocket on HPV E1 that can be exploited for future drug discovery efforts.
Papillomaviruses infect the squamous and mucosal epithelia of many different mammals, including humans, often resulting in the development of benign and sometimes malignant lesions (reviewed in references 16, 31, and 42). There are over 100 types of human papillomavirus (HPV), each exhibiting a particular tropism for specific tissue types (8). For example, HPV1 causes plantar warts, HPV6 and -11 cause anogenital warts (condyloma acuminata), and infection with HPV16 and -18, among others, can lead to cervical cancer (2, 42). Among the HPV types that infect the anogenital region, those that are associated with cancer are collectively referred to as “high-risk” types, whereas those that cause only benign warts are known as “low-risk” types (42). Despite the medical burden associated with treating and screening for HPV infections, an HPV-specific antiviral drug is still lacking, and there are only a few reports of HPV-specific inhibitors which could serve as potential leads for drug discovery. To our knowledge, the E1 ATPase inhibitors described in this report and our previously published series of E2 inhibitors (37, 39) are the only potent and selective small molecules targeting HPV DNA replication proteins ever to be reported.
All papillomaviruses have a small circular double-stranded DNA genome which encodes for only eight well-characterized proteins (for a recent review, see reference 21). The most highly conserved protein, and the only one with enzymatic activity, is the E1 helicase (reviewed in references 33 and 40). E1 is an attractive target for the development of anti-HPV drugs because it is essential for viral replication and pathogenesis (24, 34). Indeed, it has been shown in the cottontail rabbit papillomavirus (CRPV) infection model that frameshift mutations in the E1 open reading frame abrogate the ability of the CRPV genome to induce papillomas when inoculated into the skin of domestic rabbits (41).
E1 is the replicative helicase of papillomaviruses. It binds cooperatively to the origin of replication in conjunction with the E2 protein (12, 22, 27, 30). Formation of the E1-E2-origin complex involves not only the binding of both proteins to specific DNA elements in the origin but also a protein-protein interaction between the N-terminal transactivation domain of E2 and the helicase/ATPase domain of E1 (1, 3, 4, 36). We recently reported a class of small-molecule inhibitors of HPV DNA replication that bind to the transactivation domain of E2 and prevent its interaction with E1 (37, 39). Assembly of the E1-E2-ori complex facilitates the recruitment of additional E1 molecules to the origin, which assemble into hexamers in a reaction that is stimulated by ATP binding (11, 26, 35). These hexamers are the catalytically active form of E1 capable of melting the origin and unwinding the viral DNA ahead of the replication fork (28). As is the case for most helicases, the DNA-unwinding activity of E1 is powered by the hydrolysis of ATP. We previously characterized the enzymatic activities of highly purified recombinant HPV6 and -11 E1 proteins, produced with a baculovirus expression system (38). These studies revealed that HPV6 and -11 E1 proteins have similar Km values for ATP (12 and 6 μM, respectively) and that the ATPase and unwinding activities are contained within the C-terminal half of E1 (amino acids 353 to 649), the same region that binds to E2 (38). Interestingly, we found that E2 hinders the ATPase activity of E1 by raising its Km for ATP approximately sevenfold. Conversely, we observed that ATP impairs the cooperative binding of E1 and E2 to the origin, most likely by weakening the E1-E2 protein-protein interaction (38). These results and others led to the proposal that ATP not only powers the helicase activity of E1 but also acts as a molecular trigger during the initiation of viral DNA replication to help sever the interaction of E1 with E2 and promote its hexamerization at the origin (26, 38).
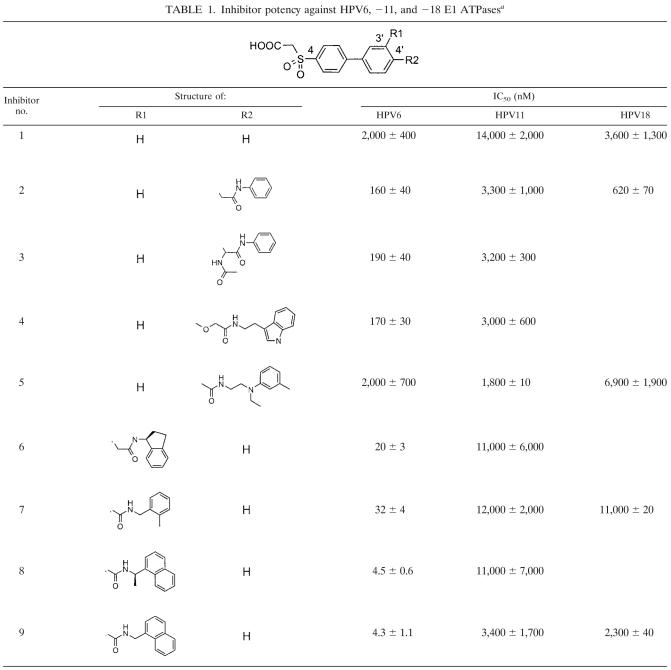
We recently reported on the discovery, by high-throughput screening of our compound collection, of a biphenylsulfonacetic acid inhibitor of HPV6 E1 (compound 1; Table 1) (10). This compound inhibits the ATPase activity of HPV6 E1 with a 50% inhibitory concentration (IC50) of 3 μM at an ATP concentration of 1 μM. More potent derivatives of this original hit were identified which inhibited activity with IC50 values as low as 4 nM. Here we show that these more potent inhibitors of HPV6 E1 are less active against the related HPV11 E1 protein and that a single amino acid difference between the two enzymes accounts for most of this differential inhibition. Furthermore, we describe the kinetic mechanism of action for these compounds and the activity of more potent compounds against the helicase activity of E1. Together, these results define a pocket in E1 that can be exploited for small-molecule modulation of E1 activity.
TABLE 1.
Inhibitor potency against HPV6, −11, and −18 E1 ATPasesa
HPV6 and −11 E1 ATPase assays were carried out with purified full-length E1 proteins and 1 μM ATP. Values are averages of at least duplicate and in most cases four or more experiments. HPV18 E1 ATPase assays were carried out with in vitro-translated helicase domain (amino acids 357 to 659) and 1 μM ATP (see Materials and Methods). Values are averages of duplicate experiments.
MATERIALS AND METHODS
Proteins and inhibitors.
HPV6a and HPV11 E1 proteins were expressed in insect cells using a baculovirus system and purified by nickel affinity chromatography as described previously (38). E1 helicase domains were synthesized in vitro by coupled transcription/translation using the TNT reticulocyte lysate system (Promega). Plasmids encoding the helicase domains of HPV16, HPV18, and CRPV E1 were constructed by PCR amplification from viral genomic DNA obtained from ATCC, followed by ligation into plasmid pTM1. pTM1 contains the encephalomyocarditis virus internal ribosome entry site region downstream of the bacteriophage T7 promoter (23). A sequence encoding a C-terminal FLAG epitope (Met Asp Tyr Lys Asp Asp Asp Asp Lys; Kodak) was added during the PCR. Plasmids encoding HPV6-11 chimeras were constructed by PCR-mediated ligation as described previously (6) and cloned into pTM1. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene) according to instructions supplied by the manufacturer. Additional details on the construction of these plasmids are available on request. Most inhibitors used in this study have been reported previously (10). Compounds 3, 4, and 5 were synthesized using similar procedures.
E1 ATPase assays in solution.
Most solution assays were performed using the previously reported scintillation proximity method (19). Briefly, 45-μl reactions were carried out in assay buffer (20 mM HEPES, pH 7.5, 1 mM dithiothreitol, 0.05 mM EDTA, 0.005% IGEPAL-CA630 [vol/vol]; Sigma [equivalent to Nonidet P-40]) supplemented with 2% dimethyl sulfoxide. Reaction mixtures contained 1 μM ATP, unless stated otherwise, and a concentration of Mg(OAc)2 500 μM greater than that of ATP. [γ-33P]ATP was included either at 30 nCi/well in reactions performed at 1 μM of total ATP or at 100 μCi per μmol of unlabeled ATP in initial velocity studies. Detection was performed by adding to reaction mixtures 40 μl of a suspension of scintillation proximity beads (10 mg/ml; Amersham Biosciences) in ammonium molybdate (0.67%) and 0.8 M HCl, followed by a solution of 100 mM citric acid and 7 M cesium chloride (80 μl). Scintillation signals were quantified using a TopCount NXT microplate scintillation counter (Packard Instruments). To verify inhibitor specificity, reactions were carried out with either purified herpes simplex virus helicase primase (2 nM) (9) or SV40 T antigen (30 nM) (32) under the same conditions used for E1. Approximately 20% conversion of the ATP substrate was obtained at these concentrations of enzymes and under these reaction conditions.
Kinetic parameters were determined by initial velocity experiments. Reactions were performed in total volumes of 160 μl rather than 45 μl. Twenty-microliter samples were removed at multiple time points and quenched with 40 μl of ammonium molybdate-scintillation proximity bead suspension followed by 80 μl of citric acid-cesium chloride. The concentration of product formed at each time point was estimated by determining, for each experiment, a conversion factor relating nM of phosphate product (determined from the ratio of ATP to ADP measured by thin-layer chromatography) to cpm detected by scintillation proximity. Because of the multiple steps involved in this procedure and the relatively high variability in quantification by thin-layer chromatography, estimates of the absolute concentrations of phosphate were found to vary by a factor of 2, although the relative concentrations obtained for different time points or reaction wells were determined much more accurately. This procedure has been described in detail previously (19). Kinetic data were analyzed using the equations for linear or hyperbolic competitive inhibition (29) with the program GraFit 3.0 (Erithacus Software Ltd.).
E1 helicase assays.
Helicase assays were carried out using the same buffer used for ATPase assays and the method described previously (38). Briefly, reaction mixtures contained HPV6 E1 (10 nM), a partial duplex substrate consisting of a 33P-labeled 24-base oligonucleotide covalently linked to biotin and annealed to M13 single-stranded DNA, ATP at 300 μM unless stated otherwise, and Mg(OAc)2 at a concentration 500 μM greater than the ATP concentration. Reaction mixtures were incubated at 37°C for 30 min, and products were detected by addition of streptavidin-coated scintillation proximity beads.
In the experiment used to measure inhibition by ATP-γ-S, helicase assays were carried out essentially as described above, except that ATP and Mg(OAc)2 concentrations were 300 μM and 1 mM, respectively. Helicase and ATPase activities were detected by removing two 20-μl aliquots from 60-μl reaction mixtures. One aliquot was used to quantify helicase activity by measuring the amount of unwound oligonucleotide product following separation from the substrate by electrophoresis on 20% acrylamide gels, as described previously (38). The other aliquot was used to quantify ATP hydrolysis by a colorimetric method using ammonium molybdate and malachite green (18).
ATPase assays using immobilized E1.
We previously showed that the helicase domain of HPV11 E1 (amino acids 353 to 649) made by in vitro transcription/translation is oligomeric (35) and has ATPase activity comparable to that of the full-length protein (38). HPV6 and -11 E1(353-649) were immobilized on plates through an affinity-purified rabbit antiserum directed against the C-terminal 14 amino acids of HPV11 E1. The purified antibody (1 μg/well) was immobilized in white Maxi-Sorp 96-well plates (Nunc) by incubation for 1 h at room temperature (or overnight at 4°C) in 100 μl antibody coating buffer (100 mM potassium phosphate, pH 6.5, 0.02% sodium azide, 0.001% glutaraldehyde). After removal of the coating solution, the plates were blocked for 1 h at room temperature (or 4°C overnight) with 200 μl/well of antibody blocking buffer (50 mM phosphate-buffered saline, 1% sucrose, 0.05% casein, 0.05% sodium azide). Blocking buffer was removed by aspiration, and coated plates were stored at 4°C until needed, up to 3 months. For reactions with FLAG-tagged E1 proteins, the monoclonal antibody M2 (Kodak) was immobilized by the same procedure. On the day of the experiment, plates were washed once with 200 μl of E1 coating buffer (20 mM Tris, pH 7.6, 500 mM NaCl, 0.02% NP-40 [vol/vol], 5 mg/ml bovine serum albumin, 1 mM dithiothreitol). In vitro-translated helicase domains (2 μl/well of the translation reaction for the HPV6 and -11 proteins or 8 μl/well for the others) were immobilized by incubation for 1 h at room temperature in E1 coating buffer (100-μl total volume). Negative control wells contained 2 μl or 8 μl per well of an in vitro translation reaction mixture programmed with pTM1 plasmid. Wells were then washed three to five times in wash buffer (same as ATPase buffer described above but supplemented with 100 mM NaCl and 0.1% [vol/vol] Triton X-100). Thorough washing was needed to remove proteins from the in vitro translation mixture, other than E1, that have ATPase activity. Efficient removal of these contaminating ATPases was verified by comparing the signals in negative control wells to those in wells in which no in vitro translation mixture was added. To determine the activity of the immobilized E1, 45-μl ATPase reactions were carried out and products were detected exactly as in the solution-reaction protocol. The activities of the HPV6 and HPV11 proteins, and of the chimeras and mutants derivatives thereof, varied by a factor of 3, but we did not determine whether this variation was caused by differences in catalytic efficiency of the enzymes or differences in the level of protein obtained by in vitro translation. Activity for the HPV16, HPV18, and CRPV enzymes was lower. In control experiments, we determined that immobilization of purified HPV6 and HPV11 E1 did not affect their activities or their levels of inhibition by the ATPase inhibitors (data not shown).
RESULTS
Mechanism of action of E1 ATPase inhibitors.
We previously reported on the identification of a class of specific, small-molecule inhibitors of the ATPase activity of HPV6 E1 (10). The properties of representative members of this class are indicated in Table 1. Unless stated otherwise, the ATPase assays used in this study were performed at 1 μM ATP, a concentration well below Km (12 μM), and using a recently described scintillation proximity method (19). Under these conditions, the original lead inhibitor, compound 1, had an IC50 of 2 μM for HPV6 E1 and a slightly higher IC50 of 14 μM for inhibition of the closely related HPV11 E1. At 80 μM, it had no effect on the ATPase activity of either HSV helicase primase or the more closely related SV40 T antigen, highlighting its specificity towards E1. Compound 1 served as a lead for a medicinal chemistry investigation (10), and IC50 values for related inhibitors are given in Table 1. As reported previously (10), significantly improved affinity for HPV6 E1 was obtained through the addition of substituted amides at the 3′ and 4′ positions of the biphenyl moiety. However, these substitutions provided at best modest improvements in affinity for the closely related HPV11 E1.
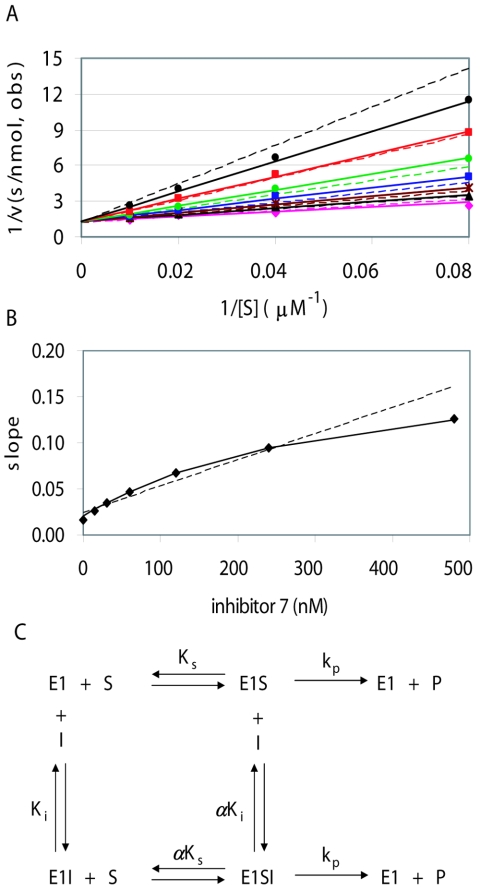
Preliminary experiments indicated that inhibitor 1 and other compounds in this series were less active at higher ATP concentrations, suggesting that they might be competitive inhibitors. Data from initial velocity kinetic experiments using a range of ATP and inhibitor concentrations were collected and analyzed by nonlinear regression as well as by visual examination of Lineweaver-Burke double-reciprocal plots. Systematic deviations of the residuals were observed after fitting to a linear competitive mechanism (Fig. 1A, dotted lines). One hallmark of the standard linear competitive mechanism is that a replot of the slopes for each inhibitor concentration from the Lineweaver-Burke plot should yield a straight line versus inhibitor concentration. This was not observed for compound 7 and other inhibitors in this series. Rather, a replot of their slopes yielded a hyperbolic curve (Fig. 1B), which is the defining characteristic of hyperbolic inhibition. As expected, a much better fit was observed for the hyperbolic competitive mechanism (Fig. 1A, solid lines), which allows formation of both the binary enzyme-inhibitor and ternary enzyme-inhibitor-substrate complexes but assumes that both complexes turn over substrate at the same rate (Fig. 1C) (29). Fitting to a more complex mechanism, in which the substrate turnover of these two complexes is different (hyperbolic noncompetitive inhibition (29), did not improve the fit. Values for the parameter α, the degree to which saturation with inhibitor raises the apparent value of Km for substrate or conversely the degree to which saturation by substrate raises the apparent Ki of the inhibitor, ranged from 10 to greater than 100 for different inhibitors in this series (Table 2). One consequence of this mechanism is that at high concentrations of substrate (i.e., near or above αKm), the enzyme is active even at saturating inhibitor concentrations. In practice, inhibitors giving high values of α may appear to act by the simpler linear competitive mechanism. An important conclusion from these results is that compounds in this series do not bind directly at the ATP-binding site, but rather affect substrate binding through an allosteric mechanism. Other examples of this kinetic mechanism have been reported for both synthetic and natural allosteric inhibitors (see Discussion).
FIG. 1.
Kinetic mechanism of compound 7. (A) Lineweaver-Burke double reciprocal plots for initial velocity data using 10 nM HPV6 E1 and 12.5 to 100 μM ATP. Data points correspond to observed initial velocities for inhibitor concentrations of 480 nM (black triangles), 240 nM (red squares), 120 nM (green circles), 60 nM (blue squares), 30 nM (brown asterisks), and 15 nM (black circles), as well as those observed (obs) without inhibitor (magenta diamonds). Corresponding lines were determined by nonlinear regression using the full data set to the equations for linear competitive or hyperbolic competitive inhibition (dotted or solid lines, respectively). The kinetic parameters obtained for compound 7 from the hyperbolic competitive fit are given in Table 2 (first of the two experiments). (B) Slope replot. For each inhibitor concentration in panel A, the slope of the 1/v versus 1/[S] plot was determined by linear regression, and these slopes are plotted versus inhibitor concentration (diamonds). Lines are plotted for the predicted slopes after fitting to the linear or hyperbolic competitive mechanisms (dotted or solid lines in both panels A and B). The solid line corresponds closely to the experimentally determined slopes. (C) Scheme for the hyperbolic competitive mechanism (29). The parameter α indicates the degree to which inhibitor binding affects substrate binding, or vice versa. Values of α greater than 1 indicate that binding of one ligand weakens binding of the other. As α approaches infinity, the lower line becomes insignificant and the mechanism becomes indistinguishable from linear competitive inhibition. For a relatively slow enzyme such as E1, it is expected but not rigorously proven that the value of Km is approximately equal to the substrate dissociation constant Ks and the value of kcat is approximately equal to the rate constant for conversion of bound substrate to product, kp.
TABLE 2.
Kinetic parameters for inhibition of HPV6 E1 ATPase activitya
| Inhibitor no. | kcat (min−1) | Km (μM) | Ki (nM) | α |
|---|---|---|---|---|
| 4 (1) | 3.5 ± 0.1 | 12 ± 1 | 190 ± 20 | 70 ± 30 |
| 4 (2) | 6.0 ± 0.1 | 16 ± 1 | 130 ± 8 | 220 ± 120 |
| 6 (1) | 3.0 ± 0.1 | 11 ± 1 | 26 ± 5 | 11 ± 2 |
| 6 (2) | 6.3 ± 0.2 | 15 ± 2 | 17 ± 2 | 15 ± 2 |
| 7 (1) | 4.6 ± 0.1 | 16 ± 1 | 35 ± 5 | 10 ± 1 |
| 7 (2) | 3.9 ± 0.1 | 17 ± 1 | 59 ± 8 | 13 ± 2 |
Kinetic parameters were determined from ATPase assays using 10 nM purified recombinant HPV6 E1, ATP concentrations of 12.5, 25, 50, and 100 μM, and twofold serial dilutions of inhibitors ranging from 100 to 3,200 nM (inhibitor 4), or 15 to 480 nM (inhibitors 6 and 7). Initial rates were determined from multiple time points corresponding to <20% conversion and fit to the hyperbolic competitive inhibition model using GraFit 3.0 (Erithacus Software). For each compound, fitted values obtained from two independent runs are presented separately, together with standard errors from the fit. The experiment-to-experiment variability in kcat values is due primarily to the indirect method used to quantify the absolute concentration of product formed (see Materials and Methods).
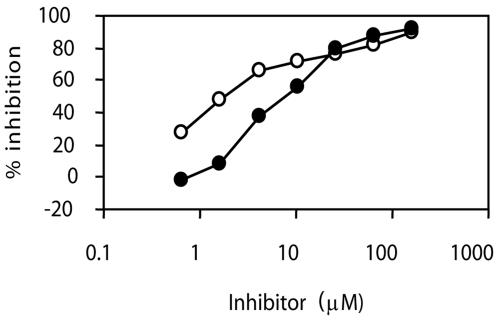
Consistent with ATP hydrolysis being required for helicase activity, the biphenylsulfonacetic acid inhibitors did inhibit the ability of HPV6 E1 to unwind a partial-duplex DNA substrate comprised of a 24-base-long oligonucleotide annealed to M13 DNA (Table 3 and Fig. 2). The degree of inhibition was dependent on the concentration of ATP used in the assay, as anticipated from the mechanism of action of these inhibitors. The potency of the inhibitors in helicase assays could not be assessed at ATP concentrations as low as those used in ATPase assays because, as we reported previously, ATP concentrations much greater than 1 μM (typically 300 μM) are needed to observe robust unwinding activity in vitro (38). The higher ATP concentrations used in unwinding assays is certainly a major factor underlying the increased IC50 values of the inhibitors in these assays relative to those measured in ATPase assays. However, it is not the only factor since the potency of these inhibitors, as well as that of the ATP analog ATP-γ-S, was 3- to 15-fold lower than that predicted solely on the basis of the higher ATP concentration. For example, we obtained an IC50 for compound 8 of 1.4 μM in a helicase assay performed at 100 μM ATP (Table 3) but a lower value of 100 nM for inhibition of ATPase activity at the same ATP concentration. Hence, other factors related to differences in assay kinetics and/or conditions (e.g., temperature or the presence of nucleic acids) probably also influence inhibitor potency. Further experiments will be required to pinpoint these additional contributing factors. Nevertheless, it remains that the main conclusion from the above results is that the biphenylsulfonacetic acid class of inhibitors is capable of abrogating the helicase activity of E1. We also tested whether these inhibitors could affect other activities of E1 that are modulated by ATP, specifically its ability to oligomerize into hexamers and its cooperative binding with E2 to the origin (see introduction). In neither case, however, did we observe any significant inhibition (data not shown), perhaps because high ATP concentrations are needed in both of these assays.
TABLE 3.
Inhibitor potency against HPV6 E1 helicase activitya
| Inhibitor no. | IC50 (μM) for ATP concn of:
|
||
|---|---|---|---|
| 300 μM | 100 μM | 30 μM | |
| 2 | 26 | 6.6 | 3.6 |
| 8 | 3.2 | 1.4 | 0.45 |
Helicase assays were performed at 37°C as described in Materials and Methods, using 42 nM purified HPV6 E1, 0.05 nM radiolabeled partial duplex substrate, the indicated concentration of ATP, and Mg(OAc)2 at 800 μM, 600 μM, or 530 μM (500 μM excess over the ATP concentration). IC50 values were derived using averages of duplicate data points. The curves for 100 μM ATP are shown in Fig. 2.
FIG. 2.
Inhibition of HPV6 E1 helicase activity. Effect of compounds 2 (closed circles) and 8 (open circles) on HPV6 E1 helicase activity. An ATP concentration of 100 μM and serial 2.5-fold dilutions of the inhibitors (160 to 0.7 μM) were used. Data points are averages of duplicate values and are connected by lines to aid in visualization. The IC50 values obtained from these curves are given in Table 3.
Identification of an amino acid responsible for the lower potency of biphenylsulfonacetic acid inhibitors against HPV11 E1.
The difference in sensitivity to inhibition we observed between the HPV6 and -11 proteins was initially surprising because the helicase domains (residues 353 to 649) (38) of these two proteins differ by only 21 amino acids. Furthermore, we verified that these inhibitors act against HPV11 E1 with the same hyperbolic competitive mechanism as observed for the HVP6 enzyme (data not shown). Altogether, this information suggested that differences in a very limited number of residues account for most of the observed difference in inhibitor affinity. We sought to identify these residues by a series of mutagenesis experiments.
To facilitate these studies, we adapted our standard scintillation proximity method to measure the ATPase activity of the isolated helicase domain of E1 (amino acids 353 to 649), made by in vitro translation in a reticulocyte lysate. We found that the ATPase activity of the helicase domains of HPV6 and -11 E1 proteins could be measured following immobilization of the proteins in 96-well plates with an antibody specific for the C terminus of the protein and removal, by several washes, of the contaminating ATPase activities present in the reticulocyte lysate.
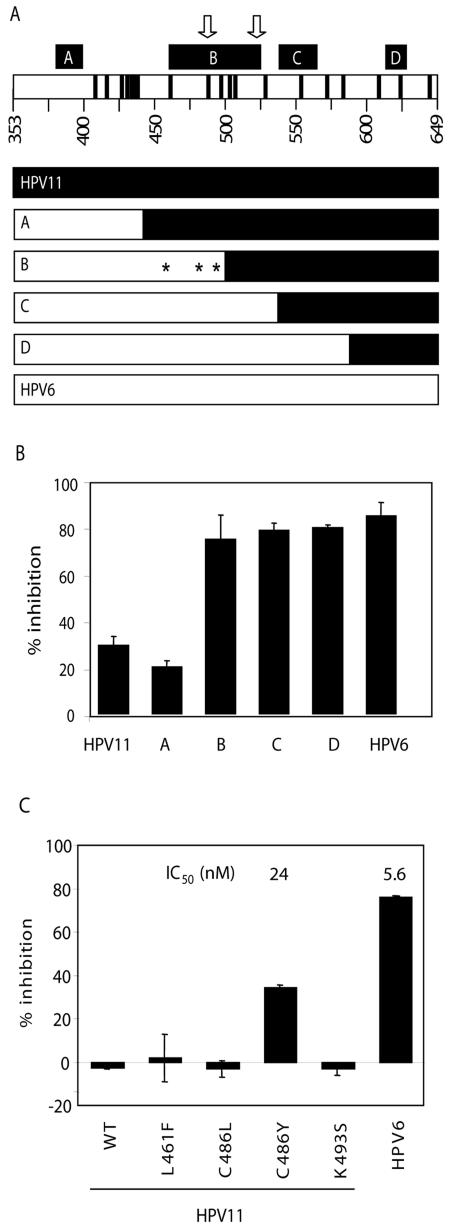
This methodology was used to quickly narrow down a region in the helicase domain of HPV11 E1 responsible for its lower sensitivity to the biphenylsulfonacetic acid inhibitors. Specifically, genes encoding the four HPV6-HPV11 E1 chimeric proteins shown in Fig. 3A were constructed, and the corresponding in vitro-translated proteins were tested for inhibition by compound 3 (Fig. 3B). Whereas relatively weak inhibition was observed for the HPV11 protein and for chimera A, this compound inhibited the activity of chimeras B, C, and D to a similar extent as that of the HPV6 helicase domain. Similar results were obtained with other inhibitors of the same class (data not shown). These results indicated that the region of E1 between amino acids 438 and 493 plays a substantial role in inhibitor sensitivity. Within this region, only three residues, at positions 461, 486, and 493, are not conserved between HPV6 and HPV11 E1. To test if one or more of these residues were responsible for the observed potency differences, they were individually mutated in the context of the isolated helicase domain of HPV11 E1 to the corresponding residue in HPV6 E1. The substitution of Cys-486 for Tyr substantially increased the sensitivity of HPV11 E1 to inhibition by the more potent inhibitor, 9 (Fig. 3C), as well as by other biphenylsulfonacetic acids with various substituents at C-3′ or C-4′ (data not shown). The IC50 value determined for HPV11 E1 C486Y was 24 nM, only fourfold higher than that measured for the HPV6 protein (5.6 nM). Note also that the IC50 of compound 9 against the helicase domain of HPV6 E1 made in vitro is very similar to the value of 4.3 nM obtained with purified full-length HPV6 E1 (Table 1), further supporting the notion that the N-terminal domain of E1 plays no significant role in ATP hydrolysis. From these results, we conclude that a tyrosine at position 486 of the E1 helicase domain is important for inhibitor binding.
FIG. 3.
Amino acid 486 of HPV6 and HPV11 E1 is a key determinant of inhibitor potency. (A, top) Schematic representation of the E1 helicase domain, with the four regions (labeled A to D) (5) of similarity to SV40 T antigen indicated by black boxes. Arrows indicate the positions of the highly conserved Walker A and B motifs, involved in ATP binding and hydrolysis. Positions of residues differing in the HPV6 and -11 sequences are indicated by black bars. (Bottom) Schematic representations of chimeras A to D, with portions derived from HPV11 in black and those derived from HPV6 in white. Chimeras A to D have residues corresponding to HPV6 up to amino acids 437, 493, 531, and 582, respectively. Amino acids 461, 486, and 493, the three amino acid differences between chimeras A and B, are indicated by asterisks. (B) Inhibition observed in ATPase assays using 2 μM compound 3 for HPV11 and -6 E1 as well as the four chimeras. Values are averages of duplicate data points with the standard deviations indicated. Reactions without inhibitors gave signals from 2,000 to 6,000 cpm depending on the protein. Signals from blank reactions (360 cpm) were subtracted to determine percent inhibition. (C) Inhibition observed in ATPase assays using 10 nM of inhibitor 9 for HPV11 and -6 E1 as well as HPV11 mutants L461F, C486L, C486Y, and K493S. (HPV16 E1 encodes a leucine at position 486 but the same residue as HPV11 at the other two positions.) Values are averages of duplicate or quadruplicate data points with the standard deviations indicated. In this experiment, reactions without inhibitor had 3,000 to 9,000 cpm, whereas blanks, subtracted before calculating percent inhibition, had 300 cpm. IC50 values obtained for two of the proteins in a separate experiment are also given.
Activity of biphenylsulfonacetic acid inhibitors against E1 from other papillomavirus types.
The method described above using in vitro-translated E1 helicase domains for ATPase assays also provided a facile procedure for testing the activity of this series of inhibitors against the E1 proteins of various papillomavirus types. Constructs for the E1 helicase domains of high-risk types 16(352-648) and 18(357-659) as well as that from the CRPV E1(306-649) were used for in vitro translation (see Materials and Methods). Due to significant variation in the C-terminal sequences, the antibody raised against HPV11 E1 did not bind to these proteins (data not shown). Thus, we made constructs which appended the FLAG epitope to the C terminus to facilitate immobilization with an anti-FLAG antibody. For comparison, a similar C-terminal FLAG-tagged HPV11 E1 helicase domain was constructed and found to give activity similar to that obtained using the untagged protein immunoprecipitated with the anti-E1 antibody (data not shown).
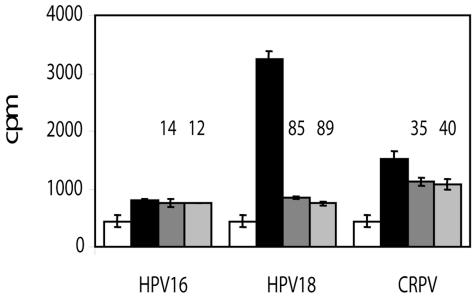
ATPase activity observed with the high-risk E1 or CRPV protein was lower than that observed for HPV11 and -6, and significant activity could only be observed using larger amounts of protein (data not shown). The enzymatic properties of E1 from these other HPV types have not been well characterized in the literature, and further experiments will be required to determine whether this lower activity was due to lower protein expression levels, a lower degree of oligomerization, or differences in kinetic parameters. Regardless, compound 9 at 80 μM clearly inhibited the HPV18 enzyme, but had only a modest effect on CRPV E1 and no discernible activity against the HPV16 enzyme (Fig. 4). Several inhibitors had similar or even greater activity against HPV18 E1 than we had observed against the HPV11 protein (Table 1). Interestingly, for both HPV18 and CRPV, the residue corresponding to Tyr-486 is also a tyrosine, whereas it is a leucine in HPV16. These observations are consistent with the need for a tyrosine at position 486 for optimal compound binding (Fig. 3C).
FIG. 4.
Determination of ATPase activity and compound inhibition for HPV16, HPV18, and CRPV E1 proteins. Scintillation proximity ATPase assays were performed using in vitro-translated E1 helicase domains immobilized via the anti-FLAG antibody. Raw cpm are shown for negative controls (in vitro translation reactions programmed with empty pTM1 vector alone; white bars), and E1 reactions in the absence of inhibitor (black) or in the presence of 80 μM compound 5 (dark gray) or 9 (light gray). Error bars correspond to standard deviations for quadruplicate values. Values for percent inhibition were calculated by comparing blank-subtracted reactions in the presence and absence of inhibitor and are given above the corresponding gray bars.
DISCUSSION
Many viral pathogens, including HPV, hepatitis C virus, and herpes simplex virus, encode helicases which are essential for viral replication and pathogenesis and which therefore have been considered attractive drug targets (13). However, despite screening campaigns by multiple companies, relatively few helicase inhibitors have been reported (13). We have shown here that a series of biphenylsulfonacetic acid compounds, identified using an assay optimized to discover inhibitors of ATP hydrolysis, can inhibit the helicase activity of HPV6 E1. Although the activity of these inhibitors was affected by the concentration of ATP, kinetic studies revealed that they are hyperbolic competitive inhibitors. This mechanism implies that the biphenylsulfonacetic acid inhibitors must bind in another location than ATP and thus affect ATP binding through an allosteric mechanism. Inhibitors acting by such a mechanism would be effective if they can increase the apparent Km of ATP in vivo sufficiently to bring about a significant reduction in viral helicase activity in infected cells. They should be inherently selective since they do not bind to the highly conserved ATP pocket. Several examples of inhibitor with this mechanism of action have been reported in the literature. These include lavendustin-A, a tight-binding peptidic inhibitor of the epidermal growth factor receptor tyrosine kinase (17), and peptide A-183, an inhibitor of factor VIIa identified from a phage-display peptide library (7, 25).
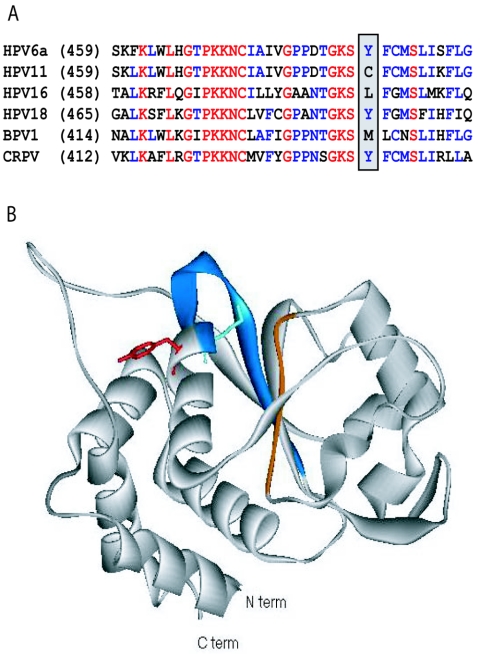
The crystal structure of the HPV18 E1 helicase domain has recently been reported (1), and it is interesting to use this structure to speculate on the location of the biphenylsulfonacetic acid binding site. The residue identified in our studies, Tyr-486 in HPV6 E1 (corresponding to Tyr-492 in HPV18; Fig. 5A), is close to the ATP binding site, on the same α-helix as the highly conserved Lys-484 (Lys-490 in HPV18), which interacts with ATP and is essential for catalysis (15). This crystallized domain of E1 is monomeric, but based on the structure of hexameric SV40 T antigen (14, 20), which is homologous to E1, Tyr-486 should be near to but not at the monomer-monomer interface. Although we have not provided evidence that the biphenylsulfonacetic acids interact directly with Tyr-486, this is a likely possibility. The tyrosine side chain extends from the α-helix in the opposite direction as does the lysine (Fig. 5B). Inhibitors interacting with this tyrosine could cause a conformational change that would affect binding of ATP, consistent with the allosteric mechanism observed for this series. Using the Site Finder application of the MOE software package (Chemical Computing Group Inc., Montreal, Canada), we found no potential small-molecule binding sites on the surface of HPV18 E1 except for the ATP binding site, so it is conceivable that these compounds bind through an induced-fit mechanism.
FIG. 5.
Structure of HPV18 showing the possible binding region of biphenylsulfonacetic acid inhibitors. (A) Sequence alignment of part of the helicase domains of the E1 proteins from two low-risk (HPV6 and -11) and two high-risk (HPV16, and -18) HPV types as well as those of bovine papillomavirus 1 (BPV1) and CRPV. Fully conserved residues are red, highly conserved residues are blue, and others are black. The positions of Tyr-486 in HPV6a E1 and of the corresponding amino acids in other E1 proteins are highlighted. For the entire helicase domain, the two low-risk sequences are 93% identical, whereas the two high-risk sequences are 65% identical. For other possible pair-wise combinations, the sequence identity ranges from 50% to 60%. (B) Ribbon diagram for the HPV18 E1 helicase domain (1). The Walker A and B motifs involved in ATP binding are blue and orange, respectively. The side chains of the catalytic Lys-490 and Tyr-492 are shown extending in opposite directions from the α-helix (equivalent to Lys-484 and Tyr-486, respectively, in HPV6 E1). This illustration was created using Viewer Lite version 5.0 (Accelrys).
The importance of Tyr-486 for compound binding provides some rationalization for the differences in inhibitor affinity observed for the different HPV types. Both HPV18 and CRPV E1 have a tyrosine at this position, whereas HPV16 has a leucine. The HPV18, HPV16, and CRPV helicase domains share 50 to 60% sequence identity with HPV6 E1, compared to 93% for HPV11. It appears that along with this tyrosine, enough critical residues are conserved in the HPV18 protein to maintain some affinity for many compounds in this series, in contrast to the HPV16 and CRPV proteins. A better understanding of the type specificity of these inhibitors and other aspects of their binding will be possible when a high-resolution structure of an inhibitor-E1 complex becomes available.
The work presented here has highlighted the presence of a small-molecule binding pocket on at least some HPV E1 proteins. Guided by structural information, it may now be possible to develop compounds active against not only HPV6 and -11 E1 but also HPV18 and possibly other high-risk proteins. Although the novel ATPase inhibitors presented in this work currently lack the necessary physicochemical properties to be good drug candidates (10), they clearly show the potential for compounds targeting this site to inhibit E1 ATPase and helicase activities and thus potentially to serve as antivirals for HPV.
Acknowledgments
We thank our colleagues from Boehringer Ingleheim (Canada) Ltd., Christian Brochu, Chantal Grand-Maitre, Gulrez Fazal, and Dominik Wernic for the synthesis of compounds used in this study, as well as Steve Titolo for determining that this class of compounds does not inhibit the oligomerization and DNA-binding activity of HPV E1.
REFERENCES
- 1.Abbate, E. A., J. M. Berger, and M. R. Botchan. 2004. The X-ray structure of the papillomavirus helicase in complex with its molecular matchmaker E2. Genes Dev. 18:1981-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman, J. G., and L. A. Koutsky. 2005. The epidemiology of human papillomavirus infections. J. Clin. Virol. 32(Suppl. 1):S16-S24. [DOI] [PubMed] [Google Scholar]
- 3.Benson, J. D., and P. M. Howley. 1995. Amino-terminal domains of the bovine papillomavirus type 1 E1 and E2 proteins participate in complex formation. J. Virol. 69:4364-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, M., and A. Stenlund. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clertant, P., and I. Seif. 1984. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature 311:276-279. [DOI] [PubMed] [Google Scholar]
- 6.Darveau, A., A. Pelletier, and J. Perreault. 1995. PCR-mediated synthesis of chimeric molecules. Methods Neurosci. 26:77-85. [Google Scholar]
- 7.Dennis, M. S., M. Roberge, C. Quan, and R. A. Lazarus. 2001. Selection and characterization of a new class of peptide exosite inhibitors of coagulation factor VIIa. Biochemistry 40:9513-9521. [DOI] [PubMed] [Google Scholar]
- 8.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. Zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 9.Dracheva, S., E. V. Koonin, and J. J. Crute. 1995. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 270:14148-14153. [DOI] [PubMed] [Google Scholar]
- 10.Faucher, A. M., P. W. White, C. Brochu, C. Grand-Maitre, J. Rancourt, and G. Fazal. 2004. Discovery of small-molecule inhibitors of the ATPase activity of human papillomavirus E1 helicase. J. Med. Chem. 47:18-21. [DOI] [PubMed] [Google Scholar]
- 11.Fouts, E. T., X. Yu, E. H. Egelman, and M. R. Botchan. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447-4458. [DOI] [PubMed] [Google Scholar]
- 12.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frick, D. N. 2003. Helicases as antiviral drug targets. Drug News Perspect. 16:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gai, D., R. Zhao, D. Li, C. V. Finkielstein, and X. S. Chen. 2004. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119:47-60. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 16.Howley, P. M. 1996. Papillomaviridae: the viruses and their replication, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 17.Hsu, C. Y., P. E. Persons, A. P. Spada, R. A. Bednar, A. Levitzki, and A. Zilberstein. 1991. Kinetic analysis of the inhibition of the epidermal growth factor receptor tyrosine kinase by Lavendustin-A and its analogue. J. Biol. Chem. 266:21105-21112. [PubMed] [Google Scholar]
- 18.Itaya, K., and M. Ui. 1966. A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta 14:361-366. [DOI] [PubMed] [Google Scholar]
- 19.Jeffery, J. A., J. R. Sharom, M. Fazekas, P. Rudd, E. Welchner, L. Thauvette, and P. W. White. 2002. An ATPase assay using scintillation proximity beads for high-throughput screening or kinetic analysis. Anal. Biochem. 304:55-62. [DOI] [PubMed] [Google Scholar]
- 20.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 21.Longworth, M. S., and L. A. Laimins. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68:362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohr, I. J., R. Clark, S. Sun, E. J. Androphy, P. MacPherson, and M. R. Botchan. 1990. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 23.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 24.Phelps, W. C., J. A. Barnes, and D. C. Lobe. 1998. Molecular targets for human papillomaviruses: prospects for antiviral therapy. Antivir. Chem. Chemother. 9:359-377. [DOI] [PubMed] [Google Scholar]
- 25.Roberge, M., L. Santell, M. S. Dennis, C. Eigenbrot, M. A. Dwyer, and R. A. Lazarus. 2001. A novel exosite on coagulation factor VIIa and its molecular interactions with a new class of peptide inhibitors. Biochemistry 40:9522-9531. [DOI] [PubMed] [Google Scholar]
- 26.Sanders, C. M., and A. Stenlund. 1998. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 17:7044-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sedman, J., and A. Stenlund. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal, I. H. 1975. Rapid equilibrium partial and mixed-type inhibition, p. 161-226. In I. H. Segal (ed.), Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley & Sons, New York, N.Y.
- 30.Seo, Y. S., F. Muller, M. Lusky, E. Gibbs, H. Y. Kim, B. Phillips, and J. Hurwitz. 1993. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 protein to the BPV replication origin. Proc. Natl. Acad. Sci. USA 90:2865-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shah, K. V., and P. M. Howley. 1996. Papillomaviruses, p. 2045-2076. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 32.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88-100. [DOI] [PubMed] [Google Scholar]
- 33.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell Biol. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 34.Sterlinko, G. H., and L. Banks. 2004. HPV proteins as targets for therapeutic intervention. Antivir. Ther. 9:665-678. [PubMed] [Google Scholar]
- 35.Titolo, S., A. Pelletier, A.-M. Pulichino, K. Brault, E. Wardrop, P. W. White, M. G. Cordingley, and J. Archambault. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 74:7349-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Titolo, S., A. Pelletier, F. Sauvé, K. Brault, E. Wardrop, P. W. White, A. Amin, M. G. Cordingley, and J. Archambault. 1999. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 73:5282-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., R. Coulombe, D. R. Cameron, L. Thauvette, M. J. Massariol, L. M. Amon, D. Fink, S. Titolo, E. Welchner, C. Yoakim, J. Archambault, and P. W. White. 2004. Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J. Biol. Chem. 279:6976-6985. [DOI] [PubMed] [Google Scholar]
- 38.White, P. W., A. Pelletier, K. Brault, S. Titolo, E. Welchner, L. Thauvette, M. Fazekas, M. G. Cordingley, and J. Archambault. 2001. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J. Biol. Chem. 276:22426-22438. [DOI] [PubMed] [Google Scholar]
- 39.White, P. W., S. Titolo, K. Brault, L. Thauvette, A. Pelletier, E. Welchner, L. Bourgon, L. Doyon, W. W. Ogilvie, C. Yoakim, M. G. Cordingley, and J. Archambault. 2003. Inhibition of human papillomavirus DNA replication by small molecule antagonists of the E1-E2 protein interaction. J. Biol. Chem. 278:26765-26772. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, V. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275-290. [DOI] [PubMed] [Google Scholar]
- 41.Wu, X., W. Xiao, and J. L. Brandsma. 1994. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J. Virol. 68:6097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zur Hausen, H., and E. M. de Villiers. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]