Abstract
The syringopeptins are a group of antimicrobial cyclic lipodepsipeptides produced by several plant-associated pseudomonads. A novel syringopeptin, SP508, was shown to be produced as two homologs (A and B) by Pseudomonas syringae pv. lachrymans strain 508 from apple and to structurally resemble syringopeptin SP22. SP508 differed from SP22 and other syringopeptins by having three instead of four α,β-unsaturated amino acids and a longer β-hydroxy acyl chain. Both SP508 and SP22 displayed growth-inhibitory activities against Mycobacterium smegmatis, other gram-positive bacteria, and yeasts but not against gram-negative bacteria. Structure-activity analyses of the SP508 and SP22 homologs indicated chemical structural features that lead to enhanced antimycobacterial activity by these pseudomonad cyclic lipodepsipeptides.
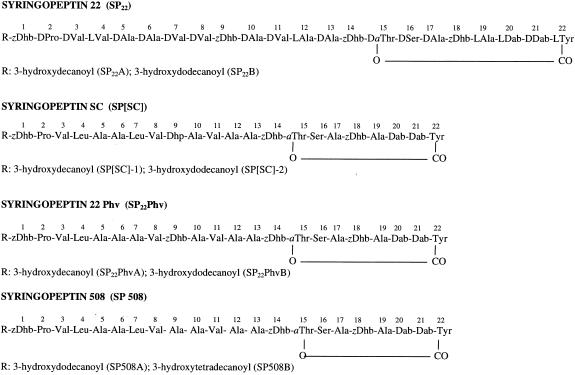
Syringopeptins (SPs) are bacterial secondary metabolites belonging to a class of cyclic lipodepsipeptides produced by certain pathovars of the plant bacterium Pseudomonas syringae (1, 2, 25, 36). Their peptide portions contain either 22 (SP22) or 25 (SP25) amino acids that are predominantly hydrophobic, valine and alanine in particular. About 70% of the chiral residues are of the d configuration, and there are four α,β-unsaturated and two 2,4-diaminobutyric acid residues (2, 17, 21, 30). An N-terminal residue dehydroaminobutyric acid (Dhb) is N acylated by a 3-hydroxylated fatty acid chain containing either 10 or 12 carbon atoms; these two types of chains are designated A and B homologs and are typically the more abundant and less abundant forms, respectively. The C-terminal carboxyl group is esterified by the hydroxyl group of the allo-Thr residue positioned at the distance of 7 residues, thus forming an eight-membered lactone macrocycle. So far, two SP25 and three SP22 forms have been identified. SP25 is produced by P. syringae pv. syringae strains that have been isolated from infected millet (B359), citrus (B427) (2), and wheat (M1) (1) as well as from the wheat pathogen P. syringae pv. atrofaciens (36). An isoform differing in the C-terminal residue, SP25-Phe, was detected in a laurel-infecting strain (30). SP22 is produced by a P. syringae pv. syringae strain isolated from pear (B301) (2), and variants are produced by P. syringae pv. syringae strains from sugar cane (SP[SC]) (21) and bean (SPPhv) (17). Each SP-producing P. syringae strain produces one type of SP together with a smaller, nine-amino-acid-containing cyclic lipodepsipeptide—either syringomycin (13, 32), syringotoxin (3), syringostatin (13), or pseudomycin (5).
The SPs are produced in infected plant tissues (12, 15), and they play roles as virulence factors in plant diseases (31). The phytotoxic physiological effects of the SPs were demonstrated with isolated plant mitochondria (10) and tobacco protoplasts (18). Lipid bilayer studies have revealed that the probable mechanism of action involves insertion into target membranes with formation of ion channels and consequent ion imbalances that lead to cell death (9, 18). In addition to their phytotoxic effects, SPs have prominent antibiotic properties (22). They are strongly inhibitory to gram-positive bacteria, particularly Bacillus spp. (22). Compared to the smaller cyclic lipodepsinonapeptides (such as syringomycin), the SPs generally display low levels of fungicidal activity (20). However, SP25 shows significant growth-inhibitory activity against Botrytis cinera (22).
In the present work, we describe the chemical structure and antimicrobial properties of a new SP—SP508—produced by P.syringae pv. lachrymans strain 508. This organism was isolated in a New York apple orchard and shown to be antagonistic to Venturia inequalis, the causative agent of apple scab (8). We show that SP508 is a novel SP and a structural variant of SP22. In addition, we evaluated its inhibitory activities against a wide spectrum of microbes. Like SP22, SP508 is inhibitory to gram-positive bacteria and fungi. In addition, both SPs, and particularly the A homolog of SP22, are shown to display strong activities against Mycobacterium smegmatis, a nonpathogenic surrogate for Mycobacterium tuberculosis used in the preliminary evaluation of antimycobacterial agents.
MATERIALS AND METHODS
Organisms and culture conditions.
P. syringae pv. lachrymans strain 508 was obtained from T. J. Burr (New York Agricultural Experiment Station) (8) and propagated on potato dextrose agar medium (Difco) at 28°C. P. syringae pv. syringae strain B301D was cultivated in the same way and used as a source for SP22A and SP22B.
For antimicrobial tests, the following organisms were obtained from the microbial culture collection of the Department of Biology, Utah State University: M. smegmatis ATCC 14468, Staphylococcus aureus ATCC 6538, Bacillus megaterium ATCC 14381, Bacillus subtilis ATCC 1965, Streptococcus pyogenes ATCC 19615, Alcaligenes faecalis ATCC 8750, Escherichia coli ATCC 25922, Proteus vulgaris ATCC 13315, Pseudomonas aeruginosa ATCC 15442, Salmonella enterica serovar Typhimurium ATCC 14028, Serratia marcescens ATCC 8100, Citrobacter freundii ATCC 8090, Candida albicans ATCC 10231, Aspergillus fumigatus ATCC 17073, Rhodotorula pilimanae (37), and Rhodotorula rubra ATCC 9449. Listeria monocytogenes ATCC 82302 and a laboratory strain of Listeria innocua were obtained from B. Weimer (Department of Nutrition and Food Sciences, Utah State University). A clinical isolate of Cryptococcus neoformans was obtained from the Fungal Testing Laboratory of the University of Texas Health Sciences Center at San Antonio (34).
M. smegmatis was grown at 37°C in Middlebrook 7H9 broth supplemented with 0.2% glycerol and 10% Middlebrook oleic acid-albumin-dextrose-catalase enrichment (24). S. aureus was grown at 37°C in Mueller-Hinton medium (Difco), and B. megaterium and B. subtilis were grown in Luria-Bertani medium (29) at 37°C. L. monocytogenes and L. innocua were grown on brain heart infusion medium (Difco) at 37°C. A. faecalis, E. coli, P. vulgaris, P. aeruginosa, S. enterica serovar Typhimurium, S. marcescens, and C. freundii were grown on Luria-Bertani medium (29) at 37°C. All fungi were cultivated in potato dextrose medium (Difco) at 28 to 30°C.
SP purification and quantitation.
P. syringae pv. lachrymans strain 508 and P.syringae pv. syringae strain B301D (sources for SP508 and SP22, respectively) were grown in noncommercial potato dextrose Casamino Acids medium in 4- or 8-liter cultures as described previously (37). The SPs were extracted from cultures using acidified acetone and purified using chromatographic methods according to previously described procedures (6). A high-performance liquid chromatography (HPLC) peak eluting with retention typically observed for SPs obtained from P. syringae pv. lachrymans strain 508 was designated SP508. The final purification step was performed using the gradient described in reference 2, and two peaks designated SP508A and SP508B were generated and separately collected. In addition to SP508, P. syringae pv. lachrymans strain 508 produced the small cyclic lipodepsinonapeptide syringomycin (data not shown).
The purified SPs were quantitated by measurement of absorbance at 220 nm (16). A calibration curve was constructed using samples quantified by amino acid analysis after hydrolysis.
MS.
SP508 was subjected to electrospray ionization (ESI) mass spectrometry (MS) using a single quadrupole mass spectrometer ESI interface (Sciex Instruments) coupled to micro-HPLC (Perkin-Elmer). An Aquapore RP300 2.1-mm column was used for separation, and the mobile phase consisted of phase A, 0.2% trifluoroacetic acid in water, and phase B, acetonitrile/isopropanol (80:20, vol/vol), at a flow rate of 200 μl · min−1. Chromatography was monitored simultaneously by UV detection at 220 nm and by MS measuring the total ion current after splitting the flow rate down to 50 μl min−1. Ion source parameters were as follows: capillary voltage, 4.7 kV; declustering potential, 40 V. Positive ion mass spectra were acquired in the range of m/z 500 to 2,500 with a dwell time of 1 ms and a step size of 0.5 Da. Calibration was performed in the same mass range using ammonium adduct ions of polypropylene glycol. Data were processed with BioMultiView software (Sciex).
Collision-induced dissociation tandem MS experiments were performed on a Q-Tof micro (Micromass; Waters) quadrupole time-of-flight instrument using argon as the collision gas. The sample was introduced into the ion source by a syringe pump at a flow rate of 5.0 μl min−1. The ion source was operated using the following parameters: capillary voltage, 2,700.0 V; sample cone, 30.0 V; extraction cone voltage, 2.0 V; desolvation temperature, 180.0°C; source temperature, 80.0°C; cone gas flow, 60 liters h−1; and desolvation gas flow, 400 liters h−1. Spectra were acquired on selected molecular and fragment ions in the m/z range of 100 to 2,200. Collision energy of 35.0 V was applied on doubly charged species at a low-mass resolution of 5.0 and a high-mass resolution. For data acquisition and processing, MassLynx3.5 software was used.
NMR spectroscopy.
Nuclear magnetic resonance (NMR) spectra were obtained using a Bruker AVANCE 600 instrument operating at 600.13 MHz with a z-gradient selection. Samples for NMR study were prepared by dissolving approximately 1 mg of lyophilized SP508A in deuterated trifluoroethanol (TFE; ICN Biochemicals) or TFE/H2O (4:1, vol/vol). 1H NMR experiments were performed using either presaturation of the water signal or a gradient water suppression sequence. Two-dimensional NMR total correlation spectroscopy (TOCSY) and nuclear Overhauser enhancement spectroscopy (NOESY) were performed in the phase-sensitive mode with the time-proportional phase increments method using the WATERGATE sequence. Correlated spectroscopy was performed in the magnitude mode using the presaturation-of-water signal. Heteronuclear multiple-quantum coherence (HMQC) was performed using a presaturation-to-suppress-water signal and the echo-antiecho detection method. The number of scans was optimized to obtain a satisfactory signal/noise ratio. All two-dimensional analyses were acquired with a time domain of 1,024 data points in the F2 dimension and 512 data points in the F1 dimension and a recycle delay of 1 s. In order to improve the resolution, it was necessary to perform a zero-filling processing followed by a fast Fourier transform. 1H-1H TOCSY was acquired with a spin-lock duration of 80 ms; 1H-1H NOESY was acquired with a mixing time of 80 ms; the HMQC was acquired using a coupling constant 1JC-H= 150 Hz.
Antimicrobial activities. (i) Broth microdilution assays.
For C. albicans, C.neoformans, R. pilimanae, R. rubra, and all bacteria except M. smegmatis, the Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS)-approved standard reference methods for broth dilution susceptibility testing were performed to obtain MICs for yeasts (27) and for aerobic bacteria (26). Cells were grown to a final concentration of 108 CFU per ml and suspended at a final concentration of 5 ×105 CFU/ml. Cell suspensions (25 ml) were added to 25-ml aliquots of twofold serial dilutions of SPs (initial undiluted concentrations were either 1 mg or 2.8 mg per ml), and appropriate broth media (26, 27) were dispensed (100-ml total volume) in wells of 96-well polystyrene microtiter plates (Fisher Scientific catalog no. 21-377-203). For bacteria, the plates were incubated at 37°C for 16 to 20 h, and for yeasts, incubation was at 28°C. For A. fumigatus, MICs were determined using the CLSI-approved method for antifungal susceptibility testing of filamentous fungi (M38-A). For the inoculum, a suspension of conidia was prepared by washing 7-day-old cultures with sterile saline containing 0.2% Tween 80. The conidial suspension was diluted to 82% transmittance at 530 nm using saline with 0.2% Tween 80. The suspension was diluted 1:50 with RPMI medium (27) (HyClone Laboratories) to achieve a 2× concentration of conidia. One hundred μl of the conidium suspension was added to each well of 96-well polystyrene microtiter plates to achieve a 1× concentration with 104 conidia per ml. Aliquots (100 μl) of twofold serial dilutions of SP (same initial concentrations as given above) were added to each well (200-μl total volume). The plates were incubated at 35°C, and readings were taken at 24 and 48 h. For M. smegmatis MIC determinations, cells were grown to a density of 1.0 McFarland standard (3 × 108 CFU per ml) (23). Inocula were diluted to 5 × 104 CFU per ml, cells were dispensed, and equal volumes of twofold serial dilutions of SPs were applied to 96-well polystyrene plates (100-μl total volume in each well) as described above. MICs were determined after 48 h of incubation at 37°C (24).
(ii) Disk diffusion assays.
These tests were done according to methods outlined by the CLSI (28). Bacteria and fungi were grown in appropriate media (as specified by CLSI for each microbial species) for 24 to 48 h. The culture densities were adjusted to 0.5 McFarland standards, and the cultures were spread over the medium agar surfaces as thin films. Sterilized paper disks (0.5-cm diameter) were placed on the agar surfaces. SP508 and SP22 samples (10 μl; 1 to 5 mg per ml) were applied to the disks, and the plates were incubated for 16 to 24 h at 28°C (yeasts) or 37°C (bacteria) before examination and measurement of the diameters of the cleared zones of inhibition.
RESULTS
Chemical structure of SP508.
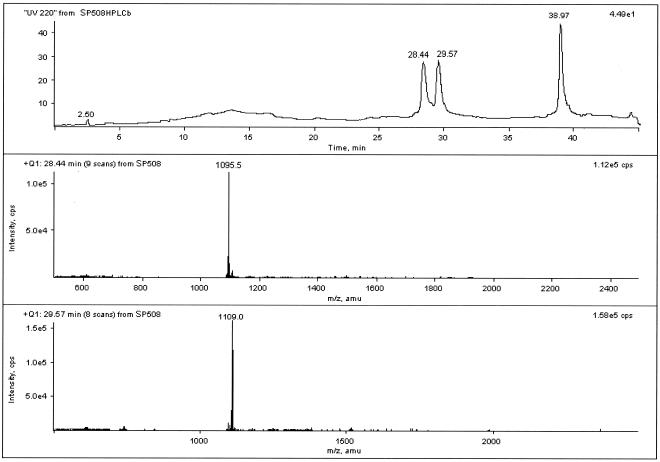
Total ion current micro-HPLC-ESI-MS analysis was performed on the purified material obtained from extracts of P. syringae pv. lachrymans strain 508. Mass spectra recorded at 28.51 and 29.71 min revealed doubly charged molecular ions of m/z 1,095.0 [M + 2H]2+ and 1,109.0 [M + 2H]2+, respectively (Fig. 1). These values indicated the presence of two compounds with molecular masses of 2,187 and 2,215 atomic mass units (average molecular mass values). These masses did not match the molecular masses for any of the known SPs. The difference of 28 atomic mass units suggested homologs with a two-carbon difference in the acyl chain length of the lipid moiety, as observed with other paired SP homologs and originating from a single P. syringae strain (2). These homologs were designated SP508A and SP508B.
FIG. 1.
Total ion current chromatogram of the prepurified culture of P. syringae pv. lachrymans strain 508 filtrates (upper panel) and mass spectra of SP508A (middle panel) and SP508B (lower panel). amu, atomic mass units.
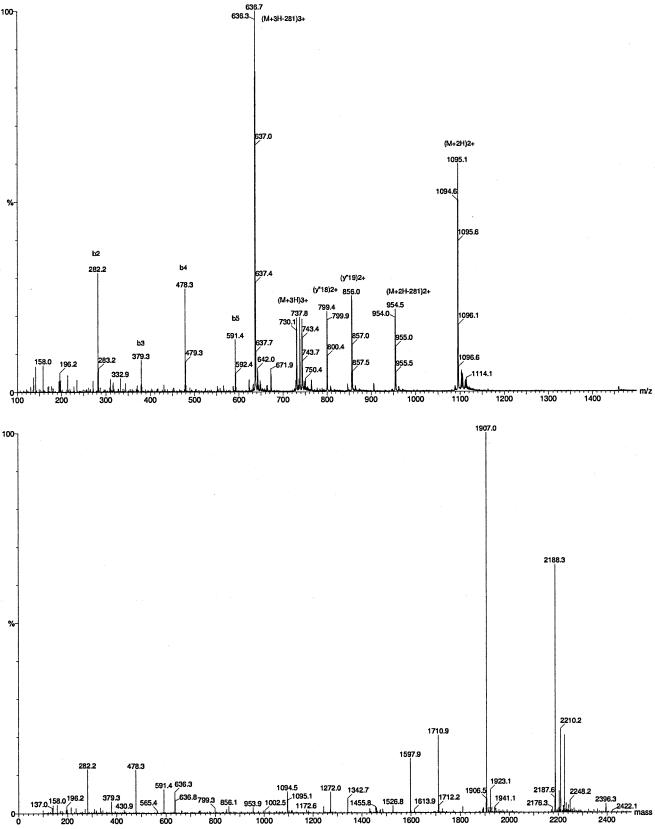
The single-stage ESI-MS result obtained for SP508A is shown in Fig. 2 (upper panel). Besides the doubly and triply charged molecular ion peaks at m/z 1,094.6 and 730.1, respectively, several fragment ions were observed in the spectrum. The MaxEnt3 program was applied to the low-mass, multiply charged continuum spectrum to resolve the multiply charged peaks onto a singly charged axis (Fig. 2, lower panel). The measured molecular mass of the monoisotopic 12C neutral molecule was 2,187.3 Da. The most intense peak in the singly charged mass spectrum was at 1,907.0, corresponding to a 281-Da neutral loss from the molecular ion. Intense bn singly and y”n doubly charged fragment ions revealed the N-terminal partial sequence as R-Dhb-Pro-Val-Leu.
FIG. 2.
MS analyses of SP508A using single-stage ESI MS (upper panel) and with MaxEnt3 deconvolution (lower panel).
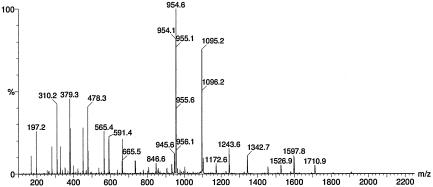
The SP508A tandem MS spectrum of [M + 2H]2+ at m/z 1,094.7 showed a fragmentation pathway very similar to that previously described for SP22PhvA (17). The loss of 281.2 Da corresponding to M-R-Dhb1 resulted in an abundant doubly charged fragment peak ([F + 2H]2+) at m/z 954.1. (Fig. 3). Partial sequence information can be drawn from the bn, bn*, and y”n ion series (Table 1) defining the sequence of the first 14 amino acids as R-Dhb1-Pro2-Val3-Leu/Ile4-Ala5-Ala6-Leu/Ile7-Val8-Ala9-Ala10-Val11-Ala12-Ala13-Dhb14.
FIG. 3.
ESI tandem MS of the [M + 2H]2+ molecular ion at m/z 1,095.2.
TABLE 1.
1H-NMR spectrum resonance assignments of SP508A in H2O/D2O
| bn | bnm/z | bn* | bn* m/z (Δm)b | y"n | y"nm/z (Δm) |
|---|---|---|---|---|---|
| b1 (R-Dhb1) | 282.2 (113) | b1* | NO | (y"21)2+ and y"21 | 954.1 and 1907.0 (282) |
| b2 (Pro2) | 379.2 (97) | b2* (Pro2Val3) | 197.2 | y"20 (Pro2) | 1810.0 (97) |
| b3 (Val3) | 478.3 (99) | b3* (Leu/Ile4) | 310.2 (113) | y"19 (Val3) | 1710.9 (99) |
| b4 (Leu/Ile4) | 591.4 (113) | b4* (Ala5) | 381.3 (71) | y"18 (Leu/Ile4) | 1597.9 (113) |
| b5 (Ala5) | 662.5 (71) | b5* (Ala6) | 452.3 (71) | y"17 (Ala5) | 1526.9 (71) |
| b6 (Ala6) | 733.5 (71) | b6* (Leu/Ile7) | 565.4 (113) | y"16 (Ala6) | 1455.8 (71) |
| b7 (Leu/Ile7) | 846.6 (113) | b7* (Val8) | 664.5 (99) | y"15 (Leu/Ile7) | 1342.7 (113) |
| b8 (Val8) | 945.6 (99) | b8* (Ala9) | 735.5 (71) | y"14 (Val8) | 1243.6 (99) |
| b9 (Ala9) | 1016.7 (71)a | b9* (Ala10) | 806.5 (71) | y"13 (Ala9) | 1172.6 (71) |
| b10 (Ala10) | 1087.7 (71)a | b10* (Val11) | 905.6 (99) | y"12 (Ala10) | 1101.6 (71) |
| b11 (Val11) | 1186.8 (99)a | b11* (Ala12) | 976.6 (71) | y"11 (Val11) | 1002.5 (99) |
| b12 (Ala12) | 1257.8 (71) | b12* (Ala13) | 1047.7 (71) | y"10 (Ala12) | 931.5 (71) |
| b13 | b13* (Dhb14) | 1130.7 (83) | y"9 (Ala13) | 860.4 (71) | |
| y"8 (Dhb14) | 777.4 (83) |
Relative intensity is less than 1%.
NO, not observed.
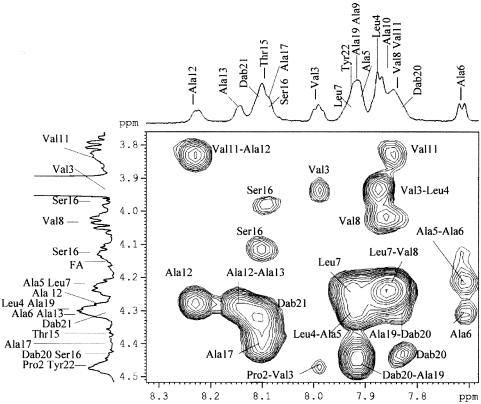
All values were compatible with a structure in which R is a 3-hydroxydodecanoate residue. Consequently, the fatty acid chain of the higher homologue SP508B was concluded to be 3-hydroxytetradecanoate. MS analysis did not allow resolution between Leu and Ile in positions 4 and 7. The complete structure of SP508A was elucidated by two-dimensional NMR spectroscopy procedures. The presence of the following amino acids was shown: three of Dhb, one of Pro, three of Val, two of Leu, eight of Ala, one of Thr, one of Ser, two of Dab, and one of Tyr. The complete assignment of resonances resulted from the chemical shift values (Table 2) reported for common amino acids and from 1H-1H TOCSY, 1H-1H correlated spectroscopy, and 1H-13C HMQC, which allowed identification of all the spin systems. In particular, in the olefinic spectral region, three signals were assigned to the CH protons of three Dhb residues (33). The assignment of the tyrosine aromatic protons was also straightforward. The presence of the hydroxyl group in position 3 of the fatty acid chain was confirmed by TOCSY and HMQC experiments. The length of the fatty acid moiety was determined by the integration of the fatty acid chain resonances. The closure of the lactone ring between the carboxyl of the Tyr and the C-β-HOH of the Thr residue was indicated by the diagnostic downfield shift of the C-β-H-Thr signal (5.495 ppm) in the 1H NMR spectrum (19). The assignments of all the resonances present in the 1H NMR spectrum of SP508A in TFE/H2O (4:1, vol/vol) are shown in Table 2. The NOESY map spectrum showed cross-peaks due to dipolar connectivities. In particular, the C-α-Hi/NH(i + 1) cross-peaks (Fig. 4), where i designates a numerical position of an amino acid, allowed us to obtain the amino acid sequence. In addition, the z-configuration of the Dhb18 residue was assigned on the basis of cross-peaks between the signal due to the C-β proton of the Dbh18 residue and the signal due to the NH proton of the Ala19 residue.
TABLE 2.
Assignments of 1H and 13C resonances of SP508A in TFE/H2Oa
| Acid | Atom | Chemical shift (ppm) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C2 | C3 | C4 | C5 | C6-C9 | C10 | C11 | C12 | ||
| Fatty acid | 1H | 2.574 | 4.143 | 1.587 | 1.356 | 1.345 | 1.298 | 1.325 | 0.892 |
| 13C | 43.8 | 70.4 | 38.0 | 30.5 | 30.5 | 33.1 | 23.8 | 14.5 | |
| Amino acids | NH | C-α | C-β,β′ | C-γ,γ′ | C-δ,δ′ | |
| Dhb1 | 1H | 8.948 | 5.797 | 1.801 | ||
| 13C | 124.7 | 12.2 | ||||
| Pro2 | 1H | 4.484 | 2.345 | 1.979 | 3.632, 3.699 | |
| 13C | 63.2 | 35.5 | 26.1 | 51.4 | ||
| Val3 | 1H | 7.991 | 3.940 | 2.373 | 1.010 | |
| 13C | 61.3 | 30.5 | 19.8 | |||
| Leu4 | 1H | 7.884 | 4.300 | 1.704 | 1.491 | 0.940, 0.924 |
| 13C | 52.8 | 40.6 | 21.9 | |||
| Ala5 | 1H | 7.905 | 4.239 | 1.487 | ||
| 13C | 52.9 | 17.4 | ||||
| Ala6 | 1H | 7.715 | 4.311 | 1.495 | ||
| 13C | 55.6 | 17.4 | ||||
| Leu7 | 1H | 7.930 | 4.235 | 1.884 | 1.702 | 0.980, 0.965 |
| 13C | 54.6 | 40.5 | 23.3 | |||
| Val8 | 1H | 7.849 | 4.029 | 2.222 | 1.035 | |
| 13C | 63.0 | 31.4 | 19.8 | |||
| Ala9 | 1H | 7.917 | 4.361 | 1.464 | ||
| 13C | 52.5 | 17.4 | ||||
| Ala10 | 1H | 7.868 | 4.285 | 1.491 | ||
| 13C | 53.4 | 17.4 | ||||
| Val11 | 1H | 7.852 | 3.835 | 2.175 | 0.984 | |
| 13C | 63.7 | 31.4 | 19.8 | |||
| Ala12 | 1H | 8.227 | 4.280 | 1.525 | ||
| 13C | 51.7 | 17.4 | ||||
| Ala13 | 1H | 8.145 | 4.305 | 1.614 | ||
| 13C | 53.9 | 17.3 | ||||
| Dhb14 | 1H | 9.267 | 6.523 | 1.810 | ||
| 13C | 133.0 | 13.3 | ||||
| Thr15 | 1H | 8.100 | 4.353 | 5.495 | 1.366 | |
| 13C | 61.6 | 72.2 | 19.1 | |||
| Ser16 | 1H | 8.090 | 4.428 | 3.984, 4.125 | ||
| 13C | 58.4 | 63.7 | ||||
| Ala17 | 1H | 8.105 | 4.402 | 1.510 | ||
| 13C | 52.5 | 17.4 | ||||
| Dhb18 | 1H | 9.375 | 6.645 | 1.810 | ||
| 13C | 135.1 | 13.3 | ||||
| Ala19 | 1H | 7.913 | 4.293 | 1.555 | ||
| 13C | 51.5 | 17.4 | ||||
| Dab20 | 1H | 7.838 | 4.443 | 1.965, 2.057 | 3.026 | (NH2) 7.671 |
| 13C | 53.0 | 28.3 | 38.3 | |||
| Dab21 | 1H | 8.116 | 4.325 | 2.318 | 3.137 | (NH2) 7.584 |
| 13C | 52.7 | 28.3 | 38.8 | |||
| Tyr22 | 1H | 7.920 | 4.474 | 3.040, 3.189 | (o) 7.065 | (m) 6.860 |
| 13C | 56.4 | 36.1 | 132.3 | 117.2 |
At 4:1 vol/vol and 300 K.
FIG. 4.
Slice of 1H-1H NOESY map of SP508A in TFE/H2O (4:1, vol/vol) at 300 K. The major connections leading to the amino acid sequence are highlighted. In the F1 and F2 dimensions, the 1H NMR spectrum in TFE/H2O (4:1, vol/vol) at 300 K is reported.
Antimicrobial activities of SP508 and SP22.
SP508A displayed strong inhibitory activities against gram-positive bacteria (B. megaterium and S. aureus) and yeasts (R. pilimanae, R. rubra, and C. albicans) and no activity against several gram-negative bacterial species (Tables 3 and 4). The same organisms were inhibited by SP508B, albeit with higher MICs than with SP508A. Both homologs also showed antimycobacterial activities against M. smegmatis, which has been heretofore unreported for the SPs. In addition, mixtures of the A and B homologs were observed to inhibit gram-positive bacteria B. subtilis and S. pneumonia and the pathogenic fungi C. neoformans and A. fumigatus (data not shown). Because of the structural resemblance of SP508A and SP508B to SP22 homologs, the antimicrobial activities of the latter were also investigated. The antimicrobial activities of SP22A and SP22B (from P. syringae pv. syringae strain B301D) paralleled those of SP508A and SP508B but were more inhibitory against gram-positive bacteria and M. smegmatis (Tables 3 and 4). SP22 activities against yeasts, however, were similar to those of SP508.
TABLE 3.
Antimicrobial MICs of A and B homologs of SP508 and SP22
| Organism | MIC (μg/ml) ofa:
|
|||
|---|---|---|---|---|
| SP508A | SP508B | SP22A | SP22B | |
| M. smegmatis ATCC 14468 | 31.25 | 62.5 | 7.8 | 31.25 |
| S. aureus ATCC 6538 | 7.8 | 21.8b | 3.9 | 31.25 |
| B. megaterium ATCC 14381 | 15.6 | 31.25 | 7.8 | 31.25 |
| E. coli ATCC 25922 | >250 | >250 | >250 | >250 |
| P. vulgaris ATCC 13315 | >250 | >250 | >250 | >250 |
| P. aeruginosa ATCC 15442 | >250 | >250 | >250 | >250 |
| Serovar Typhimurium ATCC 14028 | >250 | >250 | >250 | >250 |
| S. marcescens ATCC 8100 | >250 | >250 | >250 | >250 |
| C. freundii ATCC 8090 | >250 | >250 | >250 | >250 |
| R. rubra ATCC 9449 | 3.9 | 2.7b | 1.95 | 7.8 |
| C. albicans ATCC 10231 | 1.95 | 5.4b | 1.95 | 3.9 |
| R. pilimanae | 1.95 | 10.9b | 1.95 | 7.8 |
Values were obtained from triplicate determinations. For each determination, the initial SP concentration was 1.0 mg per ml, unless otherwise indicated.
The initial SP concentration before twofold serial dilution was 2.8 mg per ml.
TABLE 4.
Antimicrobial disk diffusion inhibitory activities of A and B homologs of SP508 and SP22
| Organism | Zone of inhibition (mm) fora:
|
|||
|---|---|---|---|---|
| SP508A | SP22A | SP508B | SP22B | |
| M. smegmatis ATCC 14468 | 15 | 18 | 12 | 14 |
| S. aureus ATCC 6538 | 11 | 14 | 9 | 9 |
| B. megaterium ATCC 14381 | 9 | 12 | 7 | 10 |
| E. coli ATCC 25922 | <1 | <1 | <1 | <1 |
| P. vulgaris ATCC 13315 | <1 | <1 | <1 | <1 |
| P. aeruginosa ATCC 15442 | <1 | <1 | <1 | <1 |
| Serovar Typhimurium ATCC 14028 | <1 | <1 | <1 | <1 |
| S. marcescens ATCC 8100 | <1 | <1 | <1 | <1 |
| C. freundii ATCC 8090 | <1 | <1 | <1 | <1 |
| R. rubra ATCC 9449 | 15 | 18 | 13 | 12 |
| C. albicans ATCC 10231 | 11 | 14 | 11 | 9 |
| R. pilimanae | 17 | 21 | 12 | 15 |
The error was ± 2 mm, as determined from duplicate determinations.
DISCUSSION
The present work revealed that SP508 is a novel SP and occurs as homologs (SP508A and SP508B) that differ by a two-methylene group in their lipid acyl chains. It represents a structural variation of SP22, with 22 amino acids comprising the peptide moiety. At position 9, SP508 contains alanine instead of an α,β-unsaturated residue present at this position in all other SP22s. SP25s have the equivalent unsaturated residue in position 10. As a result, SP508 is the first case among the several known SPs to possess three α,β-unsaturated amino acids instead of four. Finally, in contrast to other SPs, the SP508 homologs have longer lipid moieties, with 14- and 12-carbon-long fatty acid chains versus 12 and 10 carbons for the homolog equivalents of other SPs. The structure of SP508 is shown in Fig. 5, together with those of other 22-amino-acid SPs. It is significant that the N-terminal three-amino-acid motifs of all these molecules are identical (R-Dhb-Pro-Val) and that the sections from residues 10 to 22, including the lactone macrocycle, are highly conserved.
FIG. 5.
Structures of SPs containing 22-amino-acid residues.
The structural differences among the SP508 and SP22 homologs are expected to account for the differences in their antimicrobial activities. The possession of four versus three unsaturated amino acids is predicted to impart a high conformational rigidity to SP22 compared with SP508 (4). Also, the lipid moiety is hypothesized to play an important role in interactions with target membranes (18, 35), and the longer acyl chain lengths for the SP508 homologs should impart a higher degree of overall hydrophobicity. The SP508 homologs were less active than the SP22 homologs against M. smegmatis and gram-positive bacteria but were not so against yeasts (Tables 3 and 4). The data suggest that the additional α,β-unsaturated residue (in SP22) at position 9 instead of alanine (in SP508) increases the propensity for antimycobacterial activity and inhibition of gram-positive bacteria but not that for antifungal activity. In all cases, the B homologs of both SP508 and SP22 were less active than their corresponding A homologs. Since the B homologs have longer acyl chains and the SP508 homologs are less active than the corresponding SP22 homologs, it also appears that having longer acyl chains (beyond 10 carbons) coincides with less overall antimicrobial activity.
The SP508 and SP22 homologs join other pseudomonad cyclic lipodepsipeptides that display antimycobacterial activities. Massetolide A and viscosin, from Pseudomonas species isolated from a marine alga and a marine tube worm, respectively, were active against Mycobacterium avium and M. tuberculosis (11, 14). Buber et al. (7) reported an antimycobacterial activity of the smaller and less hydrophobic cyclic lipodepsinonapeptide syringomycin from P. syringae. Finally, SP508 inhibits several soil Mycobacteria isolates that degrade polycyclic aromatic hydrocarbons (M. Bensaci and A. J. Anderson, unpublished observations). In the present work, SP22A in particular displayed strong activity against M. smegmatis, and it may be considered as a potential lead compound for the development of therapeutic agents against tuberculosis.
Acknowledgments
This publication is dedicated to Professor Alessandro Ballio in honor of his retirement from the Università “La Sapienza” di Roma and his major contributions to our knowledge of bacterial cyclic lipodepsipeptides.
Research support was provided by the Utah Agricultural Experiment Station (UAES) (Project 607) and by grants from the Italian Ministero dell'Istruzione, Università e Ricerca (MIUR), PRIN2002, and Institute Pasteur—Fondazione Cenci Bolognetti (to I.G.) and Provincia Autonoma di Trento (PAT, Project SyrTox) (to V.F.).
The technical assistance of Katie Sprague and Lynnette Takemoto is acknowledged.
This is UAES publication no. 7764.
REFERENCES
- 1.Adetuyi, F. C., A. Isogai, D. Di Giorgio, A. Ballio, and J. Y. Takemoto. 1995. Saprophytic Pseudomonas syringae strain M1 of wheat produces cyclic lipodepsipeptides. FEMS Microbiol. Lett. 131:63-67. [DOI] [PubMed] [Google Scholar]
- 2.Ballio, A., D. Barra, F. Bossa, A. Collina, I. Grgurina, G. Marino, G. Moneti, M. Paci, P. Pucci, A. Segre, and M. Simmaco. 1991. Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 291:109-112. [DOI] [PubMed] [Google Scholar]
- 3.Ballio, A., F. Bossa, A. Collina, M. Gallo, N. S. Iacobellis, M. Paci, P. Pucci, A. Scaloni, A. Segre, and M. Simmaco. 1990. Structure of syringotoxin, a bioactive metabolite of Pseudomonas syringae pv. syringae. FEBS Lett. 269:377-380. [DOI] [PubMed] [Google Scholar]
- 4.Ballio, A., F. Bossa, D. Di Giorgio, A. Di Nola, C. Manetti, M. Paci, A. Scaloni, and A. L. Segre. 1995. Solution conformation of the Pseudomonas syringae pv. syringae phytotoxic lipodepsipeptide syringopeptin 25-A. Two dimensional NMR, distance geometry and molecular dynamics. Eur. J. Biochem. 234:747-758. [DOI] [PubMed] [Google Scholar]
- 5.Ballio, A., F. Bossa, D. Di Giorgio, P. Ferranti, M. Paci, A. Scaloni, A. Segre, and G. Strobel. 1994. Novel bioactive lipodepsipeptides from Pseudomonas syringae: the pseudomycins. FEBS Lett. 355:96-100. [DOI] [PubMed] [Google Scholar]
- 6.Bidwai, A. P., L. Zhang, R. C. Bachmann, and J. Y. Takemoto. 1987. Mechanism of action of Pseudomonas syringae phytotoxin, syringomycin. Stimulation of red beet plasma membrane ATPase activity. Plant Physiol. 83:39-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buber, E., A. Stindl, N. L. Acan, T. Kocagoz, and R. Zocher. 2002. Antimycobacterial activity of lipodepsipeptides produced by Pseudomonas syringae pv syringae B359. Nat. Prod. Lett. 16:419-423. [DOI] [PubMed] [Google Scholar]
- 8.Burr, T. J., M. C. Matteson, C. A. Smith, M. R. Corral-Garcia, and T. Z. Huang. 1996. Effectiveness of bacteria and yeasts from apple orchards as biological control agents of apple scab. Biol. Control 6:151-157. [Google Scholar]
- 9.Dalla Serra, M., G. Fagiuoli, P. Nordera, I. Bernhart, C. Della Volpe, D. Di Giorgio, A. Ballio, and G. Menestrina. 1999. The interaction of lipodepsipeptide toxins from Pseudomonas syringae pv. syringae with biological and model membranes: a comparison of syringotoxin, syringomycin, and two syringopeptins. Mol. Plant-Microbe Interact. 12:391-400. [DOI] [PubMed] [Google Scholar]
- 10.Di Giorgio, D., P. Lavermicocca, C. Marchiafava, L. Camoni, G. Surico, and A. Ballio. 1996. Effect of syringomycin-E and syringopeptins on isolated plant mitochondria. Physiol. Mol. Plant Pathol. 48:325-334. [Google Scholar]
- 11.El Sayed, K. A., P. B. Batryzel, X. Shen, T. L. Perry, J. K. Zjawiony, and M. T. Hamman. 2000. Marine natural products as antituberculosis agents. Tetrahedron 56.
- 12.Fogliano, V., M. Gallo, F. Vinale, A. Ritieni, G. Randazzo, M. Greco, R. Lops, and A. Graniti. 1999. Immunological detection of syringopeptins produced by Pseudomonas syringae pv lachrymans. Physiol. Mol. Plant Pathol. 55:255-261. [Google Scholar]
- 13.Fukuchi, N., A. Isogai, J. Nakayama, S. Takayama, S. Yamashita, K. Suyama, J. Y. Takemoto, and A. Suzuki. 1992. Structures and stereochemistry of three phytotoxins, syringomycin, syringotoxin and syringostatin, produced by Pseudomonas syringae pv. syringae. J. Chem. Soc. Perkin Trans. 1:1149-1157. [Google Scholar]
- 14.Gerard, J., R. Lloyd, T. Barsby, P. Haden, M. T. Kelly, and R. J. Andersen. 1997. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J. Nat. Prod. 60:223-229. [DOI] [PubMed] [Google Scholar]
- 15.Grgurina, I., N. S. Iacobellis, C. Ippolito, and R. Curci. 1997. Detection of syringomycin in plant tissues infected with Pseudomonas syringae pv. syringae, p. 188-191. In K. Rudolph, T. J. Burr, J. W. Mansfield, D. Stead, A. Vivian, and J. von Kietzell (ed.), Pseudomonas syringae pathovars and related pathogens. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 16.Grgurina, I., and F. Mariotti. 1999. Biosynthetic origin of syringomycin and syringopeptin 22, toxic secondary metabolites of the phytopathogenic bacterium Pseudomonas syringae pv. syringae. FEBS Lett. 462:151-154. [DOI] [PubMed] [Google Scholar]
- 17.Grgurina, I., F. Mariotti, V. Fogliano, M. Gallo, A. Scaloni, N. S. Iacobellis, P. Lo Cantore, L. Mannina, V. van Axel Castelli, M. L. Greco, and A. Graniti. 2002. A new syringopeptin produced by bean strains of Pseudomonas syringae pv. syringae. Biochim. Biophys. Acta 1597:81-89. [DOI] [PubMed] [Google Scholar]
- 18.Hutchison, M. L., and D. C. Gross. 1997. Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol. Plant-Microbe Interact. 10:347-354. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, T.-L., and A. J. Shaka. 1995. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. Ser. A 112:276-279. [Google Scholar]
- 20.Iacobellis, N. S., P. Lavermicocca, I. Grgurina, M. Simmaco, and A. Ballio. 1992. Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol. Mol. Plant Pathol. 40:107-116. [Google Scholar]
- 21.Isogai, A., J. Iguchi, J. Nakayama, A. Kusai, J. Takemoto, and A. Suzuki. 1995. Structural analysis of new syringopeptins by tandem mass spectroscopy. Biosci. Biotech. Biochem. 59:1374-1376. [DOI] [PubMed] [Google Scholar]
- 22.Lavermicocca, P., N. Iacobellis, M. Simmaco, and A. Graniti. 1997. Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol. Mol. Plant Pathol. 50:129-140. [Google Scholar]
- 23.Linde, C. M., S. E. Hoffner, E. Refai, and M. Andersson. 2001. In vitro activity of PR-39, a proline-arginine-rich peptide, against susceptible and multi-drug-resistant Mycobacterium tuberculosis. J. Antimicrob. Chemother. 47:575-580. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., and H. Nikaido. 1999. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. USA 96:4011-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monti, S. M., M. Gallo, R. Ferracane, R. C. Borrelli, A. Ritieni, M. L. Greco, A. Graniti, and V. Fogliano. 2001. Analysis of bacterial lipodepsipeptides by matrix-assisted laser desorption/ionisation time-of-flight and high-performance liquid chromatography with electrospray mass spectrometry. Rapid Commun. Mass Spectr. 15:623-628. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 1993. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A3. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 27.NCCLS. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard—section edition. NCCLS document M27-A2, vol. 22 (15). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.NCCLS. 2003. Susceptibility testing of Mycobacteria, Norcardia, and other aerobic actinomycetes: approved standard. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Scaloni, A., L. Camoni, D. Di Giorgio, M. Scortichini, R. Cozzolini, and A. Ballio. 1997. A new syringopeptin produced by a Pseudomonas syringae pv. syringae strain isolated from diseased twigs of laurel. Physiol. Mol. Plant Pathol. 51:259-264. [Google Scholar]
- 31.Scholz-Schroeder, B. K., M. L. Hutchison, I. Grgurina, and D. C. Gross. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant-Microbe Interact. 14:336-348. [DOI] [PubMed] [Google Scholar]
- 32.Segre, A., R. C. Bachmann, A. Ballio, F. Bossa, I. Grgurina, N. S. Iacobellis, G. Marino, P. Pucci, M. Simmaco, and J. Y. Takemoto. 1989. The structure of syringomycins A1, E and G. FEBS Lett. 255:27-31. [DOI] [PubMed] [Google Scholar]
- 33.Sklenar, M., R. Piotto, V. Leppik, and V. Saudek. 1993. 1H-15N gradient-tailored water suppression for 1H-15N HSQC experiments optimised to retain full sensitivity. J. Magn. Reson. Ser. A 102:241-245. [Google Scholar]
- 34.Sorensen, K. N., K. H. Kim, and J. Y. Takemoto. 1996. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob. Agents Chemother. 40:2710-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takemoto, J. Y., J. G. Brand, Y. A. Kaulin, V. V. Malev, L. V. Schagina, and K. Blasko. 2003. The syringomycins: lipodepsipeptide pore formers from plant bacterium, Pseudomonas syringae, p. 260-271. In G. Menestrina, M. D. Serra, and P. Lazarovici (ed.), Pore forming peptides and protein toxins. Taylor and Francis, London, England.
- 36.Vassilev, V., P. Lavermicocca, C. Di Giorgio, and N. Iacobellis. 1996. Production of syringomycins and syringopeptins by Pseudomonas syringae pv. atrofaciens. Plant Pathol. 45:316-322. [Google Scholar]
- 37.Zhang, L., and J. Y. Takemoto. 1987. Effects of Pseudomonas syringae phytotoxin, syringomycin, on plasma membrane functions of Rhodotorula pilimanae. Phytopathology 77:297-303. [Google Scholar]