Abstract
The macrolide resistance determinants and genetic elements carrying the mef(A) and mef(E) subclasses of the mef gene were studied with Streptococcus agalactiae isolated in 2003 and 2004 from 7,084 vaginorectal cultures performed to detect carrier pregnant women. The prevalence of carriage was 18% (1,276 isolates), and that of erythromycin resistance 11.0% (129 of the 1,171 isolates studied). erm(B), erm(A) subclass erm(TR), and the mef gene, either subclass mef(A) or mef(E), were found in 72 (55.8%), 41 (31.8%), and 12 (9.3%) erythromycin-resistant isolates, while 4 isolates had more than 1 erythromycin resistance gene. Of the 13 M-phenotype mef-containing erythromycin-resistant S. agalactiae isolates, 11 had the mef(E) subclass gene alone, one had both the mef(E) and the erm(TR) subclass genes, and one had the mef(A) subclass gene. mef(E) subclass genes were associated with the carrying element mega in 10 of the 12 mef(E)-containing strains, while the single mef(A) subclass gene found was associated with the genetic element Tn1207.3. The nonconjugative nature of the mega element and the clonal diversity of mef(E)-containing strains determined by pulsed-field gel electrophoresis suggest that transformation is the main mechanism through which this resistance gene is acquired.
Streptococcus agalactiae (Lancefield group B Streptococcus) is a commensal bacterium of the human digestive and genital tracts and remains an important cause of perinatal morbidity and mortality. Prevention strategies based on the detection of vaginal and rectal colonization with S. agalactiae followed by intrapartum administration of antibiotics to pregnant women have been demonstrated to reduce early-onset neonatal infection in the newborn (29, 32). S. agalactiae also causes bacteremia and skin and soft tissue infections in adults, as well as other less-common but severe infections, such as endocarditis and meningitis (15, 30).
Due to the uniform susceptibility of S. agalactiae, penicillin and ampicillin are currently the drugs of choice for the treatment and intrapartum prevention of neonatal infections, while clindamycin and erythromycin are recommended alternatives in cases of penicillin allergy (29, 32). There is concern about the increase of macrolide and clindamycin resistance in S. agalactiae associated in some countries with increases in macrolide usage (10). In most recent studies performed in Spain and other countries, erythromycin resistance rates ranged between 10% and 20% (6, 10, 15).
Among the various mechanisms of macrolide resistance found in streptococci (20), the most frequently found in human isolates are methylation of streptococcal 23S rRNA at the erythromycin-binding nucleotide target due to erm genes (erythromycin ribosome methylase) and a proton-dependent efflux of the drug mediated by the mef genes (macrolide efflux) (13, 17). The presence of the erm genes results in the macrolide-lincosamide-streptogramin B resistance phenotype, which may be constitutively or inducibly expressed. In S. agalactiae, erm(B) and erm(A) subclass erm(TR) genes are nearly always responsible for the macrolide-lincosamide-streptogramin B phenotype of macrolide resistance (6, 10, 15). Erythromycin resistance in streptococci mediated by the mef genes confers resistance only to 14- and 15-membered-ring macrolides, resulting in the M phenotype of resistance (20, 33). The mef(A) subclass gene was initially identified in Streptococcus pyogenes and the mef(E) subclass gene in Streptococcus pneumoniae, and these two subclass genes are 90% identical. Both the mef(A) and mef(E) gene subclasses have been described for erythromycin-resistant S. agalactiae clinical isolates (3).
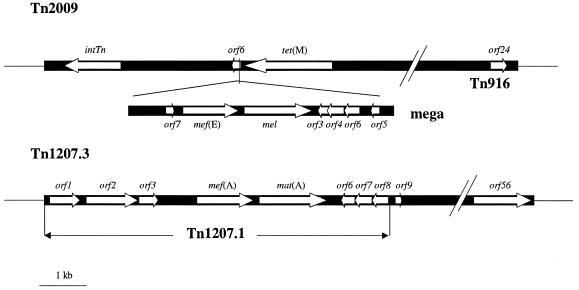
In other streptococcal species, the mef(A) subclass gene is carried in the defective transposon Tn1207.1 and mef(E) in mega (macrolide efflux genetic assembly) (18, 26, 28). In S. pneumoniae, mega can be inserted at different sites in the chromosome or into a transposon Tn916-like genetic element, forming a new composite named Tn2009, which also contains the tetracycline resistance tet(M) gene (12, 26). Tn1207.1 has been found in S. pneumoniae inserted within the competence celB gene (11, 28), and in S. pyogenes, Tn1207.1 has been described as integrated into the conjugative transposon Tn1207.3 and into a conjugative tet(O)-mef(A) element (4, 7, 9, 26).
The primary aim of the present study was to determine the macrolide resistance genes responsible for erythromycin resistance in S. agalactiae isolated from pregnant women and to study the genetic elements carrying the mef genes in the 13 M-phenotype erythromycin-resistant S. agalactiae strains found. As a secondary aim, the presence of tet(M) and other specific genes of the newly described transposon Tn2009 was also investigated to determine whether the genetic elements carrying the mef(E) gene were integrated into this transposon.
MATERIALS AND METHODS
Bacterial isolates.
Throughout 2003 and 2004, all pregnant women in San Sebastián, Basque Country (Spain), were screened at weeks 35 to 37 for vaginorectal carriage of S. agalactiae using the selective and differential “Granada medium” (27). Duplicate specimens were not included in the study. The characteristic red-orange colonies produced by S. agalactiae in Granada medium were tested for erythromycin susceptibility by the disk diffusion method according to CLSI (formerly NCCLS) methods and criteria (24). MICs of erythromycin and other antimicrobial agents were determined by the broth microdilution method using Sensititre microtiter trays (Sensititre; Trek Diagnostics Systems, West Sussex, England) and cation-adjusted Mueller-Hinton broth supplemented with 3% lysed horse blood and were interpreted according to the CLSI criteria (23).
Serotyping was performed by coagglutination according to the manufacturer's instructions (ESSUM group B Streptococcus serotyping test; Bacterum AB, Umeå, Sweden).
PCR.
The presence of the erythromycin resistance genes erm(B), erm(A) subclass erm(TR), and mef was studied with erythromycin-resistant isolates by PCR (Table 1). S. pneumoniae ATCC 700676 and ATCC 700677 were used as PCR-positive controls for the mef and erm(B) genes, respectively. S. agalactiae clinical isolate B222703 was used as a positive control in the PCR used to detect the erm(A) subclass erm(TR) gene. The presence of the erm(TR) subclass gene in the control isolate B222703 was confirmed after sequencing both strands of the amplification product with the same primers used for the amplification.
TABLE 1.
Primers used for detection of erythromycin resistance determinants and genetic carrying elements in Streptococcus agalactiae strains
| Primer group or target gene | Forward and reverse primers (5′-3′) | Amplification size (bp) | Nucleotide positions for genetic element analyzedb
|
|||
|---|---|---|---|---|---|---|
| Tn1207.3 (AY657002) | Mega (AF274302) | Tn916 (U09422) | Tn2009 (AF376746) | |||
| erm(B)a | ATTGGAACAGGTAAAGGGC | 442 | ||||
| GAACATCTGTGGTATGGCG | ||||||
| erm(TR) | AACTTGTGGAAATGAGTCAACGG | 375 | ||||
| CAGAATCTACATTAGGCTTAGGG | ||||||
| Primers for analysis of Tn1207.3 and mega | ||||||
| mef(A), mef(E) | ATGGAAAAATACAACAATTGG | 1,218 | 3271-3291 | 1125-1145 | 1372-1392 | |
| TTATTTTAAATCTAATTTTCTAA(C/T)C | 4464-4488 | 2318-2342 | 2565-2589 | |||
| matA, mela | TGCCTATATTCCCCAGTT | 708 | 4808-4825 | 2662-2679 | 2909-2926 | |
| TAATTTCCGCACCGACTA | 5497-5514 | 3351-3368 | 3598-3615 | |||
| orf8, orf5 | ATGACCGCCGATAAGTTTACTAC | 291 | 6870-6892 | 5074-5094 | 5319-5339 | |
| GACCCAGCAAATCTTCCAAAAAC | 7138-7160 | 5342-5364 | 5586-5608 | |||
| orf9 | ATGACCGCCGATAAGTTTACTAC | 699 | 6870-6892 | |||
| GTATGGGCCATTCCAAAACTAAC | 7546-7568 | |||||
| orf56 | ATATGCGAGGGTATCTGATAGTC | 495 | 49036-49058 | |||
| GGCTCAATGACATAATTCTCACC | 49508-49530 | |||||
| Primers for analysis of mega and Tn2009 | ||||||
| orf7 | TCAATTTATCAATAAATGAAATGAG | 205 | 516-540 | 781-805 | ||
| ATACTTCCAATTCGATGCCAG | 700-720 | 964-984 | ||||
| intTn | GGTCTTCGTATTTCAGAGTTTGG | 473 | 17307-17329 | |||
| GTTGCATGTGCGTAATAGTTCAG | 17766-17788 | |||||
| orf24 | TCAAAAGGTCGTTCCCCACTC | 344 | 260-280 | |||
| ACGAGTGCCGATTTTGTAGCC | 582-602 | |||||
| orf5-orf6-tet(M) | ORF5-F: TAATACAAGTACATGACTCACC | 585 | 5316-5337 | 5561-5582 | ||
| TetM-R: TCCTAAATATTGTGCGAACATC | 6123-6144 | |||||
From reference 1.
GenBank accession numbers are given in parentheses.
For mef-containing strains, another PCR was performed to study the presence of the msr(A) homologue, a gene encoding a protein that mediates resistance to macrolides and streptogramin B in staphylococci (18). These homologues are named matA and mel according to the names provided in GenBank and in other streptococcal species are located downstream of mef(A) and mef(E), respectively (Fig. 1). After DNA extraction using QIAamp spin columns (QIAGEN, Chatsworth, CA), amplification was performed using the specific primers described in Table 1. PCRs were performed at a final volume of 50 μl using approximately 30 to 50 ng of genomic DNA as a template, 1 U of AmpliTaq Gold DNA polymerase (Roche, Branchburg, NJ), 200 μM deoxynucleoside triphosphates, 1× PCR buffer, 3 mM MgCl2, and 100 ng of each primer in a GeneAmp PCR system 2700 thermocycler (PE Applied Biosystems, Foster City, California). PCR conditions for amplification of the these four genes comprised an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and elongation at 72°C for 1 min. A final elongation step at 72°C for 10 min was carried out after amplification cycles.
FIG. 1.
Schematic representation of mega inserted into Tn2009 and of Tn1207.1 integrated into Tn1207.3, adapted from references 12 and 26, respectively. Relevant ORFs, to which PCRs were performed in each genetic element (see Table 1), are represented by white arrows.
On mef-containing isolates, distinct regions of the genetic elements carrying the mef(A) and mef(E) genes were studied by means of PCR. The presence of transposon Tn1207.3 was studied using primers that hybridized with its orf8, orf9, and orf56 genes. orf8 is also present in Tn1207.1, but orf9 and orf56 are exclusive to transposon Tn1207.3 (Fig. 1).
The presence of mega was detected using specific primers that hybridized with its orf5 and orf7 genes on the basis of the S. pneumoniae sequences of mega and Tn2009. To study whether mega was inserted into a transposon Tn916-like element, a PCR with two specific sets of primers that hybridized with the intTn (the characteristic integrase gene of transposons of the Tn916-Tn1545 family) and orf24 genes from the Tn916 family was performed. With strains showing these genes, another PCR was performed with primers that hybridized the 3′-end sequence of mega (orf5 gene) and the 5′-end sequence of tet(M) on the basis of the transposon Tn2009 sequence (Fig. 1). This PCR amplifies a 585-bp fragment of DNA including fragments of the orf5, orf6, and tet(M) genes, a fragment that in this study was arbitrarily named “orf5-orf6-tet(M).” The conditions for all of these last PCRs included 35 cycles of denaturation at 94°C for 30 s, annealing at 60°C or 65°C for 30 s, and elongation at 72°C for 1 min.
The presence of the tet(M) and tet(O) genes of tetracycline resistance was determined using previously described primers and conditions (14, 25).
A previously described S. pneumoniae E-1824-J erythromycin-resistant clinical isolate (22) was used as a positive control for the PCRs detecting the orf5 and orf7 genes from mega, the intTn and orf24 genes of the Tn916 family, and the orf5-orf6-tet(M) fragment of Tn2009.
When the PCR products yielded the expected size, the specificities of the amplicons obtained were assessed by sequencing, using the same primers as those used for amplification. The sequences were then compared with the S. pyogenes sequence of Tn1207.3 (GenBank accession number AY657002), with the S. pneumoniae sequences of Tn1207.1 (GenBank accession number AF227520), mega (GenBank accession number AF274302), and Tn2009 (GenBank accession number AF376746), and with the Enterococcus faecalis sequence of Tn916 (GenBank accession number U09422) using the BLAST software available at the web site of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed on mef(E)-containing isolates as described previously in protocols (5) with minor modifications. Briefly, genomic DNA was extracted in agarose plugs after incubation with lysozyme, mutanolysin, and proteinase K. The DNA-containing agar plugs were treated with the restriction endonuclease SmaI. The resultant DNA fragments were separated by PFGE in 1% agarose gels in a CHEF-DRIII system (Bio-Rad, Hercules, Ca) with pulse times increasing from 2 to 30 s over 23 h at a voltage gradient of 6 V/cm. The gels were stained with ethidium bromide, and analysis of SmaI restriction profiles was performed with Diversity Database fingerprinting software version 2 (Bio-Rad). A dendrogram was constructed by the unweighted pair group method with arithmetic averages, the Dice coefficient, and a position tolerance of 1%.
RESULTS
Erythromycin resistance and mechanism of resistance.
From January 2003 to December 2004, vaginorectal screening for S. agalactiae was performed on 7,084 pregnant women (3,629 in 2003 and 3,455 in 2004), and S. agalactiae was detected in 1,276 women, 657 in 2003 and 619 in 2004 (overall prevalence of carriage, 18.0%). As determined by disk diffusion, 129 of the 1,171 S. agalactiae isolates available for study (11.0%) were resistant to erythromycin.
Of the 129 erythromycin-resistant S. agalactiae isolates, 72 (55.8%) had the erm(B) gene, 41 (31.8%) had the erm(A) subclass erm(TR) gene, 12 (9.3%) had the mef gene, 3 (2.3%) had both the erm(B) and erm(A) subclass erm(TR) genes, and 1 (0.8%) isolate had both the erm(A) subclass erm(TR) and mef(E) subclass genes. Of the 12 isolates with only the mef gene, 11 had the mef(E) subclass gene and one had the mef(A) subclass gene. The broth microdilution method showed that all of these 13 mef-containing S. agalactiae isolates were susceptible to penicillin, trimethoprim-sulfamethoxazole, and rifampin and were resistant to tetracycline (Table 2).
TABLE 2.
Serotypes and antibiotic MICs of 13 S. agalactiae strains with the M phenotype of macrolide resistance isolated from vaginorectally colonized pregnant women
| Isolate | Strain no. | Isolation date (mo/day/yr) | Serotype | mef gene | MIC (μg/ml) of antibiotica
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pen | Amp | Ery | Azi | Cli | Tet | Rif | Chl | Sxt | Cip | Lev | |||||
| 1 | B223605 | 2/07/2003 | 1a | mef(E) | <0.03 | <0.25 | 4 | 4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 2 | B220857 | 6/05/2003 | 1b | mef(E) | <0.03 | <0.25 | 4 | 4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 3 | B221331 | 7/17/2003 | 1b | mef(E) | <0.03 | <0.25 | 8 | 4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 4 | B222561 | 11/20/2003 | 1b | mef(E) | <0.03 | <0.25 | 4 | 4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 5 | B222696 | 12/04/2003 | 1a | mef(A) | <0.03 | <0.25 | 4 | 4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 2 | 2 |
| 6 | B220756 | 6/24/2004 | III | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | >8 | <0.5/9.5 | 1 | 1 |
| 7 | B220783 | 6/28/2004 | 1b | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 2 | 1 |
| 8 | B220815 | 6/30/2004 | III | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 9 | B220840 | 7/04/2004 | IV | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | <4 | <0.5/9.5 | 1 | 1 |
| 10 | B220867 | 7/06/2004 | III | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | <2 | <0.5/9.5 | 1 | 1 |
| 11 | B221495 | 9/03/2004 | II | mef(E) | 0.06 | <0.25 | 8 | 4 | <0.25 | >4 | <1 | <2 | <0.5/9.5 | 1 | 1 |
| 12 | B222129 | 11/04/2004 | V | mef(E) | <0.03 | <0.25 | 4 | >4 | <0.25 | >4 | <1 | <2 | <0.5/9.5 | 1 | 1 |
| 13 | B222261 | 11/18/2004 | 1b | mef(E) | <0.03 | <0.25 | 8 | >4 | <0.25 | >4 | <1 | <2 | <0.5/9.5 | <0.5 | <0.5 |
Pen, penicillin; Amp, ampicillin; Ery, erythromycin; Azi, azithromycin; Cli, clindamycin; Tet, tetracycline; Rif, rifampin; Chl, chloramphenicol; Sxt, trimethoprim-sulfamethoxazole; Cip, ciprofloxacin; Lev, levofloxacin.
Detection of mega genetic element in mef(E)-containing strains.
In the 12 erythromycin-resistant S. agalactiae strains with the mef(E) gene, the mel and orf5 genes from mega were also detected (Table 3). Two strains failed to amplify orf7 from mega. The sequences of the mef(E), mel, orf5, and orf7 genes of these S. agalactiae strains demonstrated a similarity of >99% to the corresponding sequences of mega and Tn2009 described for S. pneumoniae.
TABLE 3.
PCR amplification of specific DNA fragments and tet(M) and tet(O) genes in M-phenotype erythromycin-resistant S. agalactiae strains
| Isolate | Strain no. | Presence of:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORFs of Tn1207.3
|
ORFs of mega
|
ORFs of Tn916 and Tn2009
|
tet(M) | tet(O) | |||||||||||
| mef(A) | matA | orf8 | orf9 | orf56 | mef(E) | mel | orf5 | orf7 | intTn | orf24 | orf5-orf6-tet(M) | ||||
| 1 | B223605 | − | − | − | − | − | + | + | + | + | − | − | − | + | − |
| 2 | B220857 | − | − | − | − | − | + | + | + | + | − | − | − | − | + |
| 3 | B221331 | − | − | − | − | − | + | + | + | + | − | − | − | + | − |
| 4 | B222561 | − | − | − | − | − | + | + | + | + | + | + | − | + | − |
| 5 | B222696 | + | + | + | + | + | − | − | − | − | − | − | − | + | − |
| 6 | B220756 | − | − | − | − | − | + | + | + | − | + | + | − | + | − |
| 7 | B220783 | − | − | − | − | − | + | + | + | + | + | − | − | + | − |
| 8 | B220815 | − | − | − | − | − | + | + | + | + | − | − | − | + | − |
| 9 | B220840 | − | − | − | − | − | + | + | + | + | − | − | − | + | − |
| 10 | B220867 | − | − | − | − | − | + | + | + | + | − | − | − | + | − |
| 11 | B221495 | − | − | − | − | − | + | + | + | + | + | + | − | + | − |
| 12 | B222129 | − | − | − | − | − | + | + | + | − | − | − | − | + | − |
| 13 | B222261 | − | − | − | − | − | + | + | + | + | + | + | − | + | − |
All mef(E)-containing isolates were also tetracycline resistant. Of these, 12 had the tet(M) gene and one had the tet(O) gene. In four of the tet(M)-containing strains, the intTn and orf24 genes of the transposon Tn916 family were detected, but no amplification was obtained with the primers designed to amplify the “orf5-orf6-tet(M)” fragment of transposon Tn2009 that was detected in the S. pneumoniae strain used as a control.
Detection of Tn1207.3 in the mef(A)-containing strain.
In the single S. agalactiae isolate that contained the mef(A) subclass gene—strain B222296—the following genes of Tn1207.3 were detected: mef(A), matA, orf8 (encoding an UmuC/MucB-like protein), orf9 (encoding an unknown product), and orf56 (encoding a site-specific recombinase). All of these genes demonstrated a similarity of >99% to the sequences of the same corresponding fragments of Tn1207.3 described for S. pyogenes at GenBank. All these sequences were also found in a mef(A)-containing S. pyogenes clinical isolate used as control for these PCRs (data not shown).
Serotyping and PFGE.
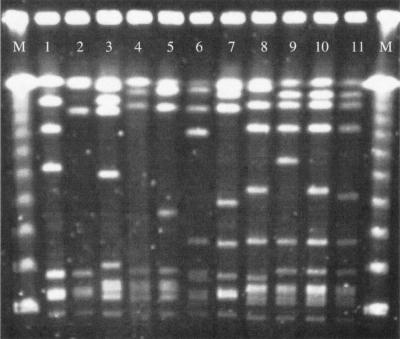
Serotyping showed that serotypes 1b and III were the most frequent among mef(E)-containing isolates (Table 2). By PFGE, the 11 strains that had the mef(E) gene alone showed a different pattern (Fig. 2). Homology among their PFGE patterns was ≤80%.
FIG. 2.
PFGE after SmaI restriction of mef(E) containing S. agalactiae isolates. M: molecular weight DNA marker (48.5-Kb Lambda ladder, Amersham Biosciences, Piscataway, NJ USA). Lines 1 to 11: PFGE patterns of isolates 2, 7, 6, 10, 8, 13, 11, 4, 3, 1, 12 as described in Table 2, respectively.
DISCUSSION
A collection of 1,276 commensal S. agalactiae isolates isolated from healthy carrier pregnant women were studied for the prevalence of erythromycin resistance, the presence of the most frequent determinants of macrolide resistance, and themef(A) and mef(E) genes carrying genetic elements. Both the prevalence of vaginorectal healthy carriers (18%) and the rate of macrolide-resistant isolates (11%) were in agreement with the results reported in other studies (10, 15, 19). The most frequent gene involved in erythromycin resistance was erm(B), followed by erm(A) subclass erm(TR), while, as found in other studies, the prevalence of mef-containing S. agalactiae was low (12, 16, 17). The high prevalence of S. agalactiae isolates containing the erm(B) and erm(A) subclass erm(TR) genes in carrier pregnant women is of concern, since for these patients preventive treatment with either macrolides or clindamycin could lead to therapeutic failures with a consequent risk of neonatal infection in the newborn.
Using serotyping and PFGE after restriction with SmaI, no homology was found among mef(E)-containing strains, suggesting that, as occurs with other gram-positive bacterial species, horizontal transfer may be the main route through which this mechanism of erythromycin resistance is acquired (8, 31). For several bacterial species found in the intestinal tract, including Enterococcus spp., viridans group streptococci, and other gram-positive and even gram-negative bacteria, the presence of the mef genes has been described (2, 21). These species could act as reservoirs and donors of the mef genes of resistance to S. agalalatiae.
Amplification by PCR of different open reading frame (ORF) genes of the mef(A)- and mef(E)-carrying genetic elements was performed to determine whether these elements, which have been found in other streptococci, were present in S. agalactiae strains. In the 12 strains with the mef(E) subclass gene, mef(E) was always associated with the mel resistance efflux gene described for mega. In 10 strains, the orf7 gene, located upstream of mef(E) at the 5′ end of the mega sequence, and the orf5 gene, located at the 3′ end of the mega sequence, were also detected. Adding the four DNA fragments of mega sequenced in mef(E)-containing S. agalactiae strains, orf7, mef(E), mel, and orf5, nearly half of the mega element was sequenced, showing a similarity of >99% with the mega sequence of S. pneumoniae at GenBank. The detection of different parts of the mega element, together with the mef(E) and mel genes, strongly suggested that, as occurs in S. pneumoniae and other streptococcal species containing the mef(E) subclass gene (1, 12, 18, 26), in the majority of the mef(E)-containing S. agalactiae isolates studied the mef(E) subclass gene was carried in mega.
The absence of the intTn and orf24 genes in 8 of the 12 mef(E)-containing strains ruled out the presence of a Tn916-like element. In the four remaining mef(E)-containing S. agalactiae strains in which the intTn and orf24 genes were detected, the failure to demonstrate the location of mega upstream of tet(M) indicated that mega was not carried, at least not in the same orientation as that described for the Tn2009-like transposon.
Erythromycin-resistant S. agalactiae isolates with the M phenotype usually have the mef(E) subclass gene, while mef(A) subclass-containing isolates are rarely found (3). We found only one strain with mef(A), in which the matA and the orf8 genes described for the Tn1207.1 genetic carrying element were also detected. The detection of the orf9 and orf56 genes of the 5′- and 3′-end regions of transposon Tn1207.3 in this mef(A)-containing strain indicated that Tn1207.1 was integrated into the conjugative transposon Tn1207.3. In addition, the detection of the tet(M) gene and not of tet(O) in this erythromycin- and tetracycline-resistant strain ruled out the carriage of Tn1207.1 into the recently described tet(O)-mef(A) element (7).
As far as we know, this is the first report of the presence of mega and Tn1207.1 integrated into Tn1207.3 in S. agalactiae. These two genetic elements have been described previously for other pathogenic and non-pathogenic streptococcal species, such as S. pneumoniae, S. pyogenes, and viridans group streptococci (4, 12, 18, 26, 28).
In conclusion, in S. agalactiae isolated from pregnant carrier women, the rates of prevalence, erythromycin resistance, and determinants of macrolide resistance found were similar to those described in other studies. In erythromycin-resistant S. agalactiae with the M phenotype, the presence of the mef(E) and mef(A) genes was associated with the mega- and Tn1207.3-carrying elements, respectively. The clonal diversity of mef(E)-containing strains and the described lack of conjugation of the mega element (18, 26) suggest that transformation might be the main mechanism through which this genetic element of resistance is acquired.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpin, C., M. H. Canron, J. Maugein, and C. Quentin. 1999. Incidence of mefA and mefE genes in viridans group streptococci. Antimicrob. Agents Chemother. 43:2335-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arpin, C., H. Daube, F. Tessier, and C. Quentin. 1999. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 43:944-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks, D. J., S. F. Porcella, K. D. Barbian, J. M. Martin, and J. M. Musser. 2003. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J. Infect. Dis. 188:1898-1908. [DOI] [PubMed] [Google Scholar]
- 5.Benson, J. A., and P. Ferrieri. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betriu, C., E. Culebras, M. Gomez, I. Rodriguez-Avial, B. A. Sanchez, M. C. Agreda, and J. J. Picazo. 2003. Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob. Agents Chemother. 47:1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenciani, A., K. K. Ojo, A. Monachetti, S. Menzo, M. C. Roberts, P. E. Varaldo, and E. Giovanetti. 2004. Distribution and molecular analysis of mef(A)-containing elements in tetracycline-susceptible and -resistant Streptococcus pyogenes clinical isolates with efflux-mediated erythromycin resistance. J. Antimicrob. Chemother. 54:991-998. [DOI] [PubMed] [Google Scholar]
- 8.Cerda-Zolezzi, P., L. M. Laplana, C. R. Calvo, P. G. Cepero, M. C. Erazo, and R. Gomez-Lus. 2004. Molecular basis of resistance to macrolides and other antibiotics in commensal viridans group streptococci and Gemella spp. and transfer of resistance genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:3462-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 10.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Grosso, M., A. Scotto d'Abusco, F. Iannelli, G. Pozzi, and A. Pantosti. 2004. Tn2009, a Tn916-like element containing mef(E) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 48:2037-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diekema, D. J., J. I. Andrews, H. Huynh, P. R. Rhomberg, S. R. Doktor, J. Beyer, V. D. Shortridge, R. K. Flamm, R. N. Jones, and M. A. Pfaller. 2003. Molecular epidemiology of macrolide resistance in neonatal bloodstream isolates of group B streptococci. J. Clin. Microbiol. 41:2659-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 16.Figueira-Coelho, J., M. Ramirez, M. J. Salgado, and J. Melo-Cristino. 2004. Streptococcus agalactiae in a large Portuguese teaching hospital: antimicrobial susceptibility, serotype distribution, and clonal analysis of macrolide-resistant isolates. Microb. Drug. Resist. 10:31-36. [DOI] [PubMed] [Google Scholar]
- 17.Fitoussi, F., C. Loukil, I. Gros, O. Clermont, M. Mariani, S. Bonacorsi, I. Thomas, D. Deforche, and E. Bignen. 2001. Mechanism of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob. Agents Chemother. 45:1889-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gay, K., and D. S. Stephens. 2001. Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184:56-65. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez, J. J., A. Andreu, and the Spanish Group for the Study of Perinatal Infection from the Spanish Society for Clinical Microbiology and Infectious Diseases. 2005. Multicenter study of the mechanisms of resistance and clonal relationships of Streptococcus agalactiae isolates resistant to macrolides, lincosamides, and ketolides in Spain. Antimicrob. Agents Chemother. 49:2525-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 21.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. Nguyen, and M. C. Roberts. 1999. A variety of gram-positive bacteria carry mobile mef genes. J. Antimicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 22.Marimon, J. M., L. Iglesias, D. Vicente, and E. Perez-Trallero. 2003. Molecular characterization of erythromycin-resistant clinical isolates of the four major antimicrobial-resistant Spanish clones of Streptococcus pneumoniae (Spain23F-1, Spain6B-2, Spain9V-3, and Spain14-5). Microb. Drug Resist. 9:133-137. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.National Committee for Clinical Laboratory Standards. 2004. Performance standards for antimicrobial susceptibility testing: 14th informational supplement. Document M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Poyart, C., L. Jardy, G. Quesne, P. Berche, and P. Trieu-Cuot. 2003. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob. Agents Chemother. 47:794-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pozzi, G., F. Iannelli, M. R. Oggioni, M. Santagati, and S. Stefani. 2004. Genetic elements carrying macrolide efflux genes in streptococci. Curr. Drug Targets Infect Disord. 4:203-206. [DOI] [PubMed] [Google Scholar]
- 27.Rosa-Fraile, M., J. Rodriguez-Granger, M. Cueto-Lopez, A. Sampedro, E. B. Gaye, J. M. Haro, and A. Andreu. 1999. Use of Granada medium to detect group B streptococcal colonization in pregnant women. J. Clin. Microbiol. 37:2674-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrag, S., R. Gorwitz, K. Fultz-Butts, and A. Schuchat. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. Morb. Mort. Wkly. Rep. Recomm. Rep. 51:1-22. [PubMed] [Google Scholar]
- 30.Schuchat, A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin. Microbiol. Rev. 11:497-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seppala, H., M. Haanpera, M. Al-Juhaish, H. Jarvinen, J. Jalava, and P. Huovinen. 2003. Antimicrobial susceptibility patterns and macrolide resistance genes of viridans group streptococci from normal flora. J. Antimicrob. Chemother. 52:636-644. [DOI] [PubMed] [Google Scholar]
- 32.Spanish Society of Obstetrics and Gynecology, Spanish Society of Neonatology, Spanish Society of Infectious Diseases and Clinical Microbiology, Spanish Society of Chemotherapy, and Spanish Society of Family and Community Medicine. 2003. Prevention of perinatal group B streptococcal disease. Spanish revised guidelines. Enferm. Infecc. Microbiol. Clin. 21:417-423. [PubMed] [Google Scholar]
- 33.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for the erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]