Abstract
A transferable plasmid from Klebsiella pneumoniae carried a class 1 integron containing blaVIM-12, a novel blaVIM-type gene, flanked by two copies of aacA7. blaVIM-12 was clustered between blaVIM-1 and blaVIM-2 and differed from blaVIM-1 by 18 nucleotides that were all located at the 3′ end and matched the corresponding nucleotides in blaVIM-2. The blaVIM-12-associated 59-base element was identical to that described in blaVIM-2 alleles.
Gram-negative microorganisms producing acquired metallo-β-lactamases (MBLs), including VIM, IMP, GIM, and SPM types, are increasingly implicated in nosocomial infections (12, 21). VIM-type MBLs have been firstly described in Pseudomonas aeruginosa and thereafter in other nonfermenters and enterobacterial species. So far, 12 VIM variants clustered into three groups represented by VIM-1, VIM-2, and VIM-7 have been described. The VIM-1 group comprises VIM-4, VIM-5, and VIM-11A. Six VIM variants (VIM-3, VIM-6, VIM-8, VIM-9, VIM-10, and VIM-11B) are considered derivatives of VIM-2, which exhibits 90% amino acid sequence identity with VIM-1. VIM-7 is markedly divergent from the remaining VIM types (21 and www.lahey.org/studies/other.asp#table 1). We report here on the identification of blaVIM-12, a novel blaVIM gene classified as intermediate between blaVIM-1 and blaVIM-2.
TABLE 1.
Agar dilution MICs of β-lactams against VIM-12-producing enterobacterial strains
| Antibiotic(s)a | MIC (μg/ml) for strain:
|
||||
|---|---|---|---|---|---|
| K. pneumoniae 2873 (VIM-12 + CMY + TEM-1 + SHV-1) | E. coli(p2873) (VIM-12 + CMY + TEM-1) | E. coli(pB-V12) (VIM-12) | E. coli(pBCSK) | E. coli 26R793 | |
| Amoxicillin | >256 | >256 | >256 | 2 | 2 |
| Amoxicillin + CLA | >256 | >256 | >256 | 2 | 2 |
| Piperacillin | >256 | >256 | >256 | 1 | 1 |
| Piperacillin + TAZ | >256 | >256 | 128 | 0.5 | 0.5 |
| Cefotaxime | 128 | 64 | 16 | ≤0.06 | ≤0.06 |
| Ceftazidime | >128 | >128 | >128 | 0.12 | 0.12 |
| Cefepime | 64 | 32 | 16 | ≤0.06 | ≤0.06 |
| Aztreonam | 16 | 4 | ≤0.06 | ≤0.06 | 0.12 |
| Imipenem | 8 | 2 | 1 | ≤0.06 | ≤0.06 |
| Meropenem | 4 | 0.5 | 0.5 | ≤0.06 | ≤0.06 |
CLA, clavulanic acid (2 μg/ml); TAZ, tazobactam (4 μg/ml).
Klebsiella pneumoniae 2873 was recovered in March 2005 at the Hippokration Hospital (Thessaloniki, Greece) from blood cultures of a 67-year-old surgical patient. The patient had been treated with multiple courses of antibiotics, including imipenem, prior to the isolation of K. pneumoniae. Species identification was performed by the API 20E system (bioMérieux, Marcy l' Étoile, France). Escherichia coli K-12 strain 26R793 (Rifr) was used as the recipient in conjugation experiments. E. coli DH5α was used as a host of recombinant plasmids. The plasmids pMON-38201 (7) and pBCSK(+) (Stratagene, La Jolla, CA) were used for cloning of blaVIM-containing fragments.
Susceptibility to β-lactams was determined by an agar dilution method (11). Phenotypic detection of MBL production was performed using the E-test MBL containing imipenem and EDTA (AB Biodisk, Solna, Sweden). Susceptibility status to non-β-lactam antibiotics was assessed by disk diffusion (10).
Transfer of resistance by conjugation was performed as described previously (19). E. coli transconjugants were selected on MacConkey agar containing rifampin (100 μg/ml) and imipenem (0.5 to 2 μg/ml). Plasmid DNA was extracted with an alkaline lysis procedure (13).
β-Lactamases were extracted by ultrasonic treatment of cell suspensions and clarified by centrifugation. Protein concentration of the extracts was determined with a protein assay kit (Bio-Rad, Richmond, CA). Isoelectric focusing was performed in polyacrylamide gels containing ampholytes (pH range, 3.5 to 9.5) (Pharmacia-LKB, Uppsala, Sweden). β-Lactamases were visualized in situ with nitrocefin (Oxoid Ltd., Basingstoke, United Kingdom).
PCR detection of various bla gene types, including blaVIM, blaTEM, blaSHV, and Citrobacter freundii-derived cmy, was performed using consensus primers and amplification conditions as described previously (1, 22, 23). For integron mapping, PCR assays combining primers specific for 5′ conserved segment (5′CS) and 3′CS sequences (6) with primers specific for blaVIM, aacA, dhfrI, aadA, qacEΔ1, and sul genes were performed. PCR products were purified using a Qiaex gel extraction kit (QIAGEN, Chatsworth, CA) and used as templates for sequencing on both strands with an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA).
Antibiotic susceptibility testing showed that K. pneumoniae 2873 was resistant to penicillin-inhibitor combinations and broad-spectrum cephalosporins and also exhibited reduced susceptibility to carbapenems and aztreonam (Table 1). The isolate was positive by the E-test MBL. K. pneumoniae 2873 was also resistant to gentamicin, netilmicin, tobramycin, amikacin, ciprofloxacin, cotrimoxazole, and tetracycline.
Conjugation experiments showed that resistance to β-lactams was readily transferable to E. coli at a high frequency (8 × 10−2 per donor cell). Plasmid analysis indicated transfer of a single plasmid of approximately 70 kb, designated p2873. Transconjugants exhibited most of the resistance characters of the donor, although MICs of carbapenems and oxyimino-β-lactams were slightly lower (Table 1). Resistance to cotrimoxazole, amikacin, netilmicin, and tobramycin, but not gentamicin, was also transferred.
Isoelectric focusing showed that E. coli(p2873) produced three β-lactamases with pIs of 5.1, 5.4, and 8.9. These bands were also observed in the extracts of K. pneumoniae 2873 along with a β-lactamase focusing at 7.6. Plasmid DNA extracts from both strains were positive in the blaVIM-, blaCMY-, and blaTEM-specific PCRs. Types of bla genes were confirmed by sequencing of the respective amplicons. These findings, taken together with the β-lactam resistance phenotypes, indicated that p2873 encoded a VIM MBL, a C. freundii-originated CMY cephalosporinase, and a TEM-1 β-lactamase. The nontransferable β-lactamase produced by K. pneumoniae was SHV-1, as shown by sequencing of the respective PCR product.
Plasmid DNA extracts from E. coli(p2873) were subjected to PCR using various primer combinations. Assembly of the nucleotide sequences of overlapping PCR products revealed a class 1 integron named In-h12. The variable region of In-h12 was approximately 2.1 kbp and included a novel blaVIM-type gene cassette (designated blaVIM-12) flanked by two copies of an aacA7 gene cassette similar to those encountered frequently among VIM-encoding class 1 integrons. A typical 5′ conserved segment (5′CS) containing an intI1 gene with a strong P1 promoter followed directly by an activated P2 promoter (including a GGG insertion) and an attI1 site was identified. qacEΔ1/sul1 sequences were also detected at the 3′CS of In-h12. The blaVIM-12 gene (798 bp) shared 97.7, 94.5, and 80.2% nucleotide homology with the blaVIM-1, blaVIM-2, and blaVIM-7 genes, respectively. This novel MBL differed from blaVIM-1 (GenBank accession no. Y18050) by 18 nucleotides. Notably, these changes were all located at the 3′ end and matched exactly the nucleotides found at the corresponding positions in the blaVIM-2 gene (GenBank accession no. AF191564). blaVIM-12 could therefore be viewed as a blaVIM-1/blaVIM-2 hybrid being identical to blaVIM-1 from the 5′ end up to nucleotide 663 and to blaVIM-2 from nucleotide 614 up to its 3′ end. Furthermore, the 59-base element of the blaVIM-12 gene cassette (72 bp in length) was identical to the element commonly found in blaVIM-2 cassettes (14-16) and differed significantly from the 59 bp of the blaVIM-1 gene cassettes (5, 8, 18). A PCR product that included the promoter sequence, the left-hand aacA7, and the blaVIM-12 gene cassette was directly cloned to pMON-38201 and subsequently to the polycloning site of pBCSK(+). The resulting recombinant plasmid (pB-V12) was used to transform E. coli DH5α-competent cells. The E. coli(pB-V12) clone exhibited a β-lactam resistance pattern characteristic of laboratory strains carrying cloned blaVIM genes (Table 1).
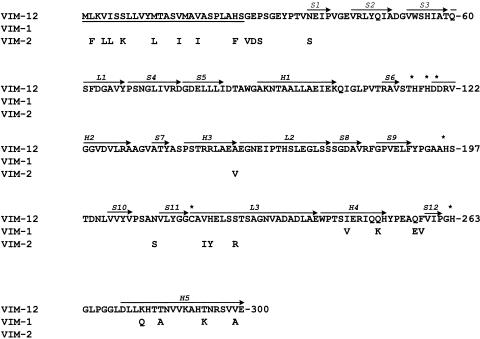
The putative VIM-12 polypeptide (266 amino acids; molecular weight, 28,120) shared 97.0 and 93.6% amino acid identity with VIM-1 and VIM-2, respectively. VIM-12 differed from VIM-1 by 8 amino acid residues at the C terminus and by 18 residues from VIM-2 at the N terminus, 8 of which were in the putative leader peptide (Fig. 1). The latter likely comprised 26 amino acid residues (HS-GE). The mature VIM-12 protein (240 amino acids; molecular weight, 25,417) had a calculated pI of 4.77, close to that determined by isoelectric focusing (5.1). A dendrogram based on a CLUSTAL W multiple alignment of the VIM proteins showed that VIM-12 was clustered between the VIM-1 and VIM-2 groups (data not shown).
FIG. 1.
Amino acid sequence of VIM-12 metallo-β-lactamase and comparison with VIM-1 and VIM-2. Numbering of amino acid residues is according to a standard scheme for MBLs (3). Arrows above sequence regions indicate the secondary structure elements (S, strands; H, helices; L, loops). Asterisks denote zinc-associated amino acid residues. The putative leader peptide is underlined.
In this study, VIM-12, a novel plasmid-mediated VIM-type MBL from a clinical isolate of K. pneumoniae, is described. Emergence of blaVIM-12 in a class 1 integron that is also novel underscores the continuous evolution of these important determinants. It is of interest that the blaVIM-12 gene cassette in its 5′ end is identical to blaVIM-1, whereas in its 3′ end it resembles blaVIM-2. The possibility that blaVIM-12 belongs to a distinct VIM lineage cannot be excluded. However, the pattern of the nucleotide changes of the blaVIM-12 gene, compared with blaVIM-1 and blaVIM-2, leads to the hypothesis that this novel variant might have been formed by a recombination event between blaVIM-1- and blaVIM-2-containing sequences. It is probable that blaVIM-12 has arisen within our hospital environments, where VIM-1- and VIM-2-producing microorganisms are endemic (4, 17, 20).
VIM-12 differs from VIM-1 by 8 amino acid residues at positions 246, 251, 257, 258, 284, 287, 294, and 299 (standard class B beta-lactamases numbering scheme [3]). All eight residues are located at the protein surface and are distant from the active site (2). Therefore, it is unlikely that the substrate specificity of VIM-12 differs significantly from that of VIM-1. The β-lactam resistance pattern of the VIM-12-producing strains does not contradict this hypothesis. Nevertheless, slight differences between the hydrolysis spectra of VIM-12 and VIM-1 cannot be excluded.
The simultaneous production of two potent β-lactamases, VIM-12 and a CMY-type cephalosporinase, by a K. pneumoniae clinical isolate was also of note. Similar to the case of E. coli V541, in a recent clinical isolate from Athens that also produced VIM-1 and CMY-13 (9) the acquired β-lactamases of K. pneumoniae 2873 were encoded by a single transferable plasmid. Given that combination of these enzymes inactivates all clinically available β-lactams, spread of the respective strains or plasmids may have serious consequences in the treatment of nosocomial infections.
Nucleotide sequence accession number.
The nucleotide sequence described here has been submitted to the EMBL and GenBank nucleotide sequence databases under accession number DQ143913.
REFERENCES
- 1.Arlet, G., and A. Philippon. 1991. Construction by polymerase chain reaction and use of intragenic DNA probes for three main types of transferable β-lactamases (TEM, SHV, CARB). FEMS Microbiol. Lett. 66:19-25. [DOI] [PubMed] [Google Scholar]
- 2.Docquier, J. D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J.-M. Frere, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 3.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, J.-M. Frere, and the Metallo-β-Lactamase Working Group. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giakkoupi, P., A. Xanthaki, M. Kanellopoulou, A. Vlahaki, V. Miriagou, S. Kontou, E. Papafraggas, H. Malamou-Lada, L. S. Tzouvelekis, N. J. Legakis, and A. C. Vatopoulos. 2003. VIM-1 metallo-β-lactamase-producing Klebsiella pneumoniae strains in Greek hospitals. J. Clin. Microbiol. 41:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavroidi, A., A. Tsakris, E. Tzelepi, S. Pournaras, V. Loukova, and L. S. Tzouvelekis. 2000. Carbapenem-hydrolysing VIM-2 metallo-β-lactamase in Pseudomonas aeruginosa from Greece. J. Antimicrob. Chemother. 46:1041-1043. [DOI] [PubMed] [Google Scholar]
- 8.Miriagou, V., E. Tzelepi, D. Gianneli, and L. S. Tzouvelekis. 2003. Escherichia coli with a self-transferable, multiresistant plasmid coding for metallo-β-lactamase VIM-1. Antimicrob. Agents Chemother. 47:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miriagou, V., L. S. Tzouvelekis, L. Villa, E. Lebessi, A. C. Vatopoulos, A. Carattoli, and E. Tzelepi. 2004. CMY-13, a novel inducible cephalosporinase encoded by an Escherichia coli plasmid. Antimicrob. Agents Chemother. 48:3172-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial disk susceptibility tests. Approved Standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4 (M100-S7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 13.Olsen, J. E. 1990. An improved method for rapid isolation of plasmid DNA from wild-type gram-negative bacteria for plasmid restriction profile analysis. Lett. Appl. Microbiol. 10:209-212. [DOI] [PubMed] [Google Scholar]
- 14.Pallecchi, L., M. L. Riccio, J. D. Docquier, R. Fontana, and G. M. Rossolini. 2001. Molecular heterogeneity of blaVIM-2-containing integrons from Pseudomonas aeruginosa plasmids encoding the VIM-2 metallo-β-lactamase. FEMS Microbiol. Lett. 195:145-150. [DOI] [PubMed] [Google Scholar]
- 15.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pournaras, S., M. Maniati, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 18.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsakris, A., A. P. Johnson, R. C. George, S. Mehtar, and A. C. Vatopoulos. 1991. Distribution and transferability of plasmids encoding trimethoprim resistance in urinary pathogens from Greece. J. Med. Microbiol. 34:153-157. [DOI] [PubMed] [Google Scholar]
- 20.Tsakris, A., S. Pournaras, N. Woodford, M.-F. I. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan, J.-J., P.-R. Hsueh, W.-C. Ko, K.-T. Luh, S.-H. Tsai, H.-M. Wu, and J.-J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]