Abstract
Genome-wide screening of sequence databases for human endogenous retroviruses (HERVs) has led to the identification of 18 coding env genes, among which two—the syncytin genes—encode fusogenic ENV proteins possibly involved in placenta physiology. Here we show that a third ENV, originating from the most “recent” HERV-K(HML2) family, is functional. Immunofluorescence analysis of env-transduced cells demonstrates expression of the protein at the cell surface, and we show that the protein confers infectivity to simian immunodeficiency virus pseudotypes. Western blot analysis of the pseudotyped virions further discloses the expected specific cleavage of the ENV precursor protein. This functional ENV could play a role in the amplification—via infection of the germ line—of the HERV-K genomic copies, all the more as coding HERV-K gag and pol genes can similarly be found in the human genome, which could therefore generate infectious virions of a fully endogenous origin.
Human endogenous retroviruses (HERVs) comprise approximately 8% of the human genome. Most of the identified elements are defective due to mutations and/or deletions within their genes, but some elements have conserved full-length open reading frames. A systematic search has led to the identification of 18 coding env genes (5, 11, 23). The most important contributor to this list is the HERV-K(HML2) family (2, 14), with six coding env genes. The status of this family is rather unique: although some elements first entered the primate genome more that 40 million years ago, new proviral copies have been generated in the recent past that are human specific (17, 20). Actually, the entry dates in the human genome of all six copies with a coding env have been estimated to be less than 5 million years (3, 11), with two of them being even polymorphic in the human population (allele frequencies of 0.04 and 0.19) (22), thus strongly suggesting recent amplification by intracellular transposition and/or infection. Among the other HERV families, it has previously been demonstrated that two env genes, from the HERV-W and HERV-FRD families, are indeed functional since they can induce cell-cell fusion when expressed in cells possessing the corresponding receptors (7, 9, 18) and since they render lentiviral particles infectious in pseudotype experiments (1, 8). In this study, we show that one member of the HERV-K(HML2) family encodes a functional ENV protein.
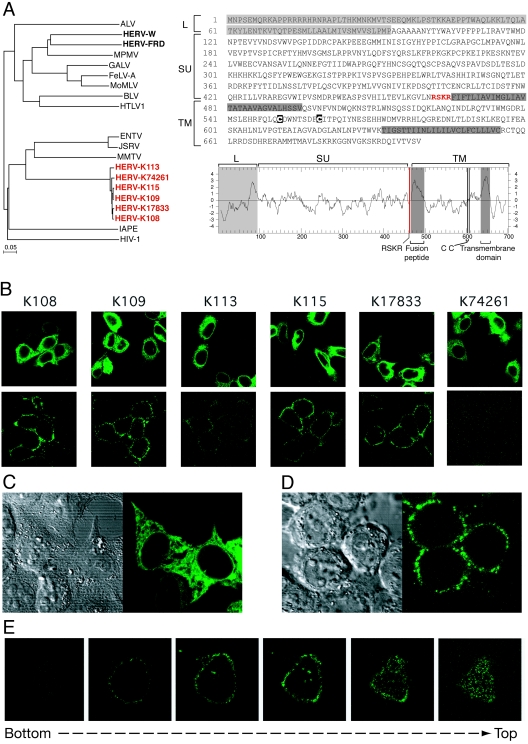
The proteins encoded by the six complete HERV-K env genes are highly conserved, with more than 97% identity at the amino acid level, consistent with their recent integration into the human genome. A phylogenetic tree based on the retroviral transmembrane (TM) subunit (i.e., the most conserved ENV domain) shows that the HERV-K copies cluster together with the betaretrovirus group, i.e., the Jaagsiekte sheep retrovirus, the enzootic nasal tumor virus, and the mouse mammary tumor virus, and are clearly distinct from the group of the murine leukemia virus (MLV)-like retroviruses, which includes HERV-W and HERV-FRD (Fig. 1A, left panel). As illustrated in Fig. 1A (right panel), the structural organization of the HERV-K ENVs is canonical, with a signal peptide at the N-terminal end (albeit longer than usual), an RX(K/R)R consensus cleavage site for the cellular furin protease that splits the surface (SU) and TM subunits, a hydrophobic fusion domain at the N-terminal end of the TM subunit, the two conserved cysteine residues in the ectodomain of the TM subunit, and a hydrophobic transmembrane anchor domain.
FIG. 1.
Structure and expression of the HERV-K ENV proteins. (A, left) Phylogenetic tree based on the ENV TM domain showing the clustering of the six HERV-K copies with the betaretrovirus group. Protein sequences were aligned using ClustalW, and the resulting alignment was manually refined before the tree was calculated using the neighbor-joining method (PHYLIP software). Nucleotide sequences of the six env genes are as follows (accession number, position in the sequence entry, orientation): for K108, AC072054, 30,365 to 32,464 (minus); for K109, AF164615, 6,412 to 8,508 (plus); for K113, AY037928, 6,451 to 8,550 (plus); for K115, AY037929, 6,442 to 8,541 (plus); for K17833, Y17833, 5,581 to 7,680 (plus); and for K74261, AC074261, 93,508 to 95,604 (plus). (Right) Sequence and hydrophobicity profile of the HERV-K108 ENV highlighting the canonical ENV features (see the text). ALV, avian leukemia virus; MPMV, Mason-Pfizer monkey virus; GALV, gibbon ape leukemia virus; FeLV-A, feline leukemia virus A; MoMLV, Moloney MLV; BLV, bovine leukemia virus; HTLV1, human T-cell leukemia virus type 1; ENTV, enzootic nasal tumor virus; JSRV, Jaagsiekte sheep retrovirus; MMTV, mouse mammary tumor virus; IAPE, intracisternal A-particle-related envelope-encoding element. (B-E) Immunofluorescence analysis of HeLa cells transiently transfected with expression vectors of each of the six fully coding HERV-K env genes. (B) In the upper panels, cells were fixed, permeabilized, and stained for HERV-K env expression (whole-cell staining); in the lower panels, living cells were observed directly after staining without prior fixation or permeabilization (cell surface staining). (C, D) Higher magnification of representative images of fixed and permeabilized cells (C) or of living cells (D) transfected previously with the HERV-K108 env gene. (Left) Image of the cells under phase-contrast microscopy; (right) image of the same cells stained for the HERV-K ENV by immunofluorescence. (E) Successive confocal images of a living cell stained for the HERV-K108 ENV, demonstrating cell surface localization. In all experiments, HERV-K ENV detection was performed on HeLa cells grown on glass coverslips approximately 24 h posttransfection. HERV-K ENVs were detected using a mouse monoclonal antibody (raised by us against a recombinant His-tagged protein corresponding to amino acids 223 to 437 of K109 ENV, using the pET-28 vector from Novagen), and an Alexa Fluor 488-conjugated anti-mouse secondary antibody (Molecular Probes). No signal was detected with cells transfected with an irrelevant expression vector (not shown). Observations were made under a Zeiss LSM 510 laser scanning confocal microscope.
To experimentally characterize the six complete HERV-K envelope proteins, we first cloned the corresponding open reading frames into a human cytomegalovirus (CMV) promoter-driven expression vector, starting from the env initiation codon down to the R domain of the provirus 3′ long terminal repeat, as described in reference 7. In a first assay, we checked that all six HERV-K env genes were correctly expressed. To do so, we used a monoclonal antibody that we raised against a recombinant protein corresponding to part of the SU subunit and analyzed by immunofluorescence confocal microscopy the cellular distribution of the ENVs in transiently transfected HeLa cells (Fig. 1B to E). As can be clearly observed in the figure, the ENV proteins have a broad cytoplasmic distribution—as expected for a protein synthesized in the endoplasmic reticulum—with approximately the same level of expression for the six HERV-K env genes (Fig. 1B, upper panels, and C). When the staining was performed on living, nonpermeabilized cells, the HERV-K proteins could be detected—in all cases except K74261—at the cell surface (albeit at a reduced level for the HERV-K113 protein), a result consistent with the expected localization of a functional ENV (Fig. 1B, lower panels, D, and E).
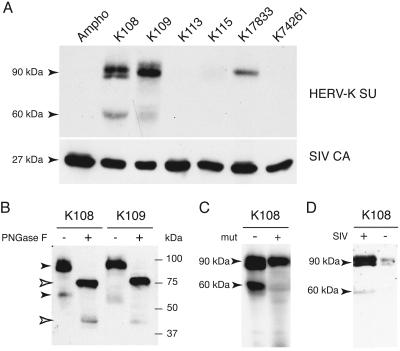
We then asked whether these ENV proteins could be incorporated into retroviral particles. Human embryonic kidney 293T cells were transfected with an expression vector for the simian immunodeficiency virus (SIV) lentiviral core proteins, a corresponding lacZ gene-marked defective retroviral vector, and an expression vector for the env gene to be tested (or a vector for the amphotropic MLV ENV as a control). Cell supernatants were collected 48 h posttransfection, virions were concentrated by ultracentrifugation, and the pellets were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As shown in the Western blot presented in Fig. 2A, three bands can be detected specifically (not observed with the MLV ENV control) with the HERV-K108 and -K109 ENV pseudotypes, with apparent molecular masses of approximately 90, 80, and 60 kDa, consistent with the sizes of the full-length unprocessed precursor (with and without its signal peptide) and of the SU subunit, respectively (theoretical values of 80, 68, and 42 kDa for nonglycosylated proteins). The level of the putative processed SU subunit was reproducibly observed to be higher for the K108 than for the K109 ENV in this assay, whereas a faint signal of the expected size for the unprocessed precursor was also detected for the HERV-K17833 ENV, indicating that this protein can also be incorporated into the SIV particles but with a much reduced efficiency compared to that of the K108 and K109 proteins.
FIG. 2.
Western blot analysis of the incorporation of HERV-K ENVs into SIV pseudotypes. (A) Virions contained in the supernatant of human 293T cells cotransfected with an expression vector for the SIV core proteins, a corresponding lacZ gene-marked defective retroviral vector, and an expression vector for the env gene to be tested (or a vector for the amphotropic MLV envelope as a control [Ampho]) were assayed for the presence of the HERV-K ENV protein. Forty-eight hours posttransfection, supernatants of the 293T cells were harvested, pelleted by ultracentrifugation onto a 20% sucrose cushion, and recovered for SDS-PAGE analysis. (B) SIV particles pseudotyped with K108 and K109 ENVs were purified as described for panel A and treated or not treated with peptide-N-glycosidase F (PNGase F; NEB Biolabs) as recommended by the manufacturer before SDS-PAGE. (C) SIV particles were pseudotyped with the K108 ENV protein or a K108 ENV protein mutated at the furin cleavage site (RSKR to ASAR) (mut) and analyzed as described for panel A. As the K108 mutant protein is less expressed than its wild-type counterpart, twice the volume of cell medium was used for the purification step. (D) The supernatants of 293T cells transfected with the K108 ENV expression vector and either a control plasmid (CMV β, without SIV) or the SIV plasmids (see panel A; with SIV) were concentrated and analyzed by SDS-PAGE as described for panel A. All blots were first used for the detection of HERV-K ENV proteins, using the same monoclonal antibody as in Fig. 1. The membrane in panel A was thereafter tested for SIV p27 capsid using a mouse monoclonal antibody (2F12; National Institutes of Health AIDS Research and Reference Reagent Program), to check for equal loadings of the lanes (lower panel). The membrane in panel D was also incubated with an antibody against actin (Santa Cruz) to exclude the possibility of contamination by cell proteins in the course of virion purification (not shown).
To ascertain the nature of the observed protein bands and obtain the “exact” molecular weight of the ENV-derived products, we performed the same SDS-PAGE analysis on viral pellets treated, or not treated, with peptide-N-glycosidase F so as to remove all N-linked carbohydrates. As shown in Fig. 2B, the ENV precursors (not separated as a doublet on this gel) have a molecular mass of approximately 75 kDa after treatment, and the SU subunits migrate as 40-kDa proteins, which match exactly the expected values. Also consistent with the predicted nature of the smaller protein, point mutations that we introduced to disrupt the furin cleavage site of K108 ENV (RSKR → ASAR) led to its disappearance from purified SIV particles (Fig. 2C), without loss of the larger proteins. Last, we analyzed the concentrated supernatants of 293T cells transfected with the K108 env expression vector together with a control plasmid (CMV β) or the SIV vectors. We detected only trace amounts of K108 ENV protein in the absence of the SIV vectors, definitely demonstrating that the bands that are detected result from the specific incorporation of ENV proteins into SIV particles (Fig. 2C).
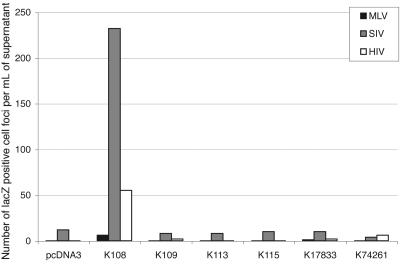
Finally, the infectivity of the pseudotyped particles was assayed as previously described (8). In this assay, human 293T cells are cotransfected with an expression vector for the retroviral proteins—except ENV—from a gammaretrovirus (MLV) or lentiviruses (human immunodeficiency virus type 1 [HIV-1] and SIV), a corresponding lacZ gene-marked defective retroviral vector, and an expression vector for the env gene to be tested (or an empty vector as a negative control and a vector for the amphotropic MLV ENV as a positive control). Then, the pseudotyped virions are assayed for infectivity; after recovery of the transfected cell supernatant 36 h posttransfection, transfer of the supernatant onto target cells, centrifugation of the plates (i.e., spinoculation [see reference 12]), and an additional 60-h incubation period, lacZ-positive cell colonies are counted following in situ histochemical staining for β-galactosidase activity. The results of such an assay are given in Fig. 3 for the six env genes and the various viral cores (MLV, SIV, and HIV-1) tested, using feline astrocytic cells as a target (see below). It can be clearly observed that one—and only one—of the HERV-K envelopes, namely, K108, confers infectivity to the virions when it is expressed on SIV-derived particles, with a viral titer of 100 to 300 CFU/ml. This result is consistent with the HERV-K108 ENV being efficiently integrated and processed within the SIV particles and with the HERV-K109 protein disclosing an altered processing profile (Fig. 2) (but other defects might as well be responsible for its lack of infectivity). It can also be noted that the MLV core is much less efficient than the lentiviral cores, if it is efficient at all, as was similarly observed previously for the HERV-W and HERV-FRD ENVs (1, 8). It is noteworthy that the viral titer measured for the HERV-K108 ENV is low compared to that of the amphotropic MLV ENV, as was previously observed for other HERV ENVs, most probably because they do not perfectly fit in with the core particles available for pseudotyping.
FIG. 3.
Assay for infectivity of the viral particles pseudotyped with the proteins encoded by the six fully coding HERV-K env genes. Human 293T cells were cotransfected with an expression vector for the retroviral proteins—except ENV—from a gammaretrovirus (MLV) or lentiviruses (HIV-1 and SIV), a corresponding lacZ gene-marked defective retroviral vector, and an expression vector for the env gene to be tested (or an empty vector [pcDNA3] as a negative control and a vector for the amphotropic MLV ENV [not shown] as a positive control). Viral supernatants were collected 36 h after transfection of 293T cells, filtered through 0.45-μm-pore-size membranes, and added to the target feline G355.5 cells in 24-well plates that were then subjected to spinoculation (12). After an additional 60 h of incubation, the lacZ-positive cell colonies were counted following in situ histochemical staining for β-galactosidase activity. A positive control performed with the amphotropic MLV ENV gave titers of 8 × 105, 4 × 105, and 6 × 104 infectious particles per ml for the MLV, SIV, and HIV cores, respectively. This graph gives the results obtained in an experiment representative of two to five independent assays performed under the same conditions (the mean viral titer observed for K108 ENV with the SIV core is 150, with variations ranging from 60 to 400 depending on the overall efficiency of each infection assay).
Further characterization of the cell tropism of the HERV-K108 ENV was achieved using a large series of mammalian cells (Table 1). As can be observed, under conditions where the amphotropic MLV ENV used as a positive control demonstrated infectivity for all—except BHK21—cells tested, the HERV-K108 ENV-pseudotyped virions were infectious for a restricted set of cells (i.e., the human 293T and SH-SY5Y, rodent BHK21 and LOK, and feline G355.5 cell lines). Lack of infection of other target cells might be due to the absence of the appropriate receptor on these cells, or alternatively to receptor interference if the cells express small amounts of endogenous envelope proteins (e.g., 2102Ep is known to express HERV-K envelope proteins [6]).
TABLE 1.
Infection host ranges of the HERV-K108 and control envelope proteinsa
| Target cells | Titerb
|
||
|---|---|---|---|
| ENV K108 | ENV FRD | ENV W | |
| Human | |||
| HeLa (cervix adenocarcinoma) | − | − | ++ |
| TE671 (rhabdomyosarcoma) | − | ++ | + |
| 293T (embryonic kidney) | ++ | ++ | ++ |
| SH-SY5Y (neuroblastoma) | + | +++ | ++ |
| NTera2D1 (teratocarcinoma) | − | ++ | ND |
| 2102Ep (teratocarcinoma) | − | ND | ND |
| NCCIT (teratocarcinoma) | − | ND | ND |
| Primate | |||
| Cos-7 (kidney) | − | − | ++ |
| Vero (kidney) | − | + | + |
| Mouse | |||
| WOP (embryo fibroblast) | − | − | ND |
| MCA (fibrosarcoma) | − | − | ND |
| LOK (kidney) | + | − | − |
| Rat | |||
| 208F (embryo fibroblast) | − | − | − |
| Hamster | |||
| BHK21 (kidney) | + | + | − |
| CHO (ovary) | − | − | − |
| Cat | |||
| G355.5 (brain) | ++ | ++ | − |
| Dog | |||
| MDCK (kidney) | − | + | − |
Infection range was determined using SIV lentiviral pseudotypes.
Titers (CFU/ml) were as follows: −, <10; +, 10 to 100; ++, 100 to 1,000; and +++, >1,000. ND, not determined.
In conclusion, the present results demonstrate that one of the HERV-K(HML2) family members, i.e., K108, has conserved a functional env gene. The encoded ENV protein can be exported to the cell surface and incorporated into lentiviral pseudotypes with cleavage into SU and TM subunits, finally generating infectious pseudotyped particles. The infection range of this envelope protein is restricted, being limited to 5 out of 15 cell types tested, and is distinct from that of the two other infectious HERV ENVs, thus suggesting usage of a different receptor for cell entry. Interestingly, not all the env genes of the HERV-K family tested are positive in the infection assay, and indeed the corresponding proteins seem to be defective at various steps of pseudotype formation, despite a rather high sequence conservation among the various copies (more than 97% identity in amino acid sequences). This could simply account for the nonfunctionality of previously described HERV-K env genes (15, 21). Sequence alignment of natural mutants could now be useful in identifying key residues critical for function by generating hybrids or even point mutations within the identified genes and alleles. Yet, identification of a functional env is an important issue since HERV-K(HML2) is one of the youngest HERV families, with evidence for a recent burst in the human lineage (17, 20, 22). Such amplifications could a priori originate from strictly intracellular retrotransposition events, not requiring an intact env gene, but could as well be the result of a bona fide infection of germ line cells (see reference 4). In this respect, the occurrence of a still-functional ENV among the recent HERV-K copies of the human genome could be a hint for the second scenario, although the direct demonstration of primary germ line cell infection is still lacking. In addition, among all the HERV families present in the human genome, HERV-K(HML2) is the only one with potentially fully coding gag and pol genes (16, 22). Along this line, it is noteworthy that HERV-K-related particles have been detected in culture supernatants from several cell lines derived from tumors, including teratocarcinomas and melanomas (6, 10, 13, 19). It is therefore conceivable that infectious retroviruses are produced, provided that they incorporate the HERV-K108 ENV protein. The present results lend credence to this hypothesis, and the search for HERV-K copies with active gag-pol genes (possibly within K108 alleles) could lead to the unraveling of a fully competent, infectious, endogenous human retrovirus.
Acknowledgments
We thank M. Huesca for generating the mouse monoclonal anti-SU-K antibody, A. Jalil for assistance with confocal microscopy and helpful advice for the staining experiments, A. Richaud and J. F. Casella for technical assistance, F. L. Cosset for providing the 2F12 anti-SIV capsid mouse monoclonal antibody and the complete set of retroviral plasmids used for pseudotyping experiments, S. Fichelson for the gift of LOK cells, and C. Lavialle for critical reading of the manuscript.
This work was supported by the CNRS and by grants from the Ligue Nationale contre Le Cancer (Equipe Labellisée).
REFERENCES
- 1.An, D. S., Y. Xie, and I. S. Chen. 2001. Envelope gene of the human endogenous retrovirus HERV-W encodes a functional retrovirus envelope. J. Virol. 75:3488-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannert, N., and R. Kurth. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc. Natl. Acad. Sci. USA 13:14572-14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 4.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 25:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benit, L., P. Dessen, and T. Heidmann. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709-11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieda, K., A. Hoffmann, and K. Boller. 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J. Gen. Virol. 82:591-596. [DOI] [PubMed] [Google Scholar]
- 7.Blaise, S., N. de Parseval, L. Bénit, and T. Heidmann. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 100:13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaise, S., A. Ruggieri, M. Dewannieux, F.-L. Cosset, and T. Heidmann. 2004. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity on retroviral particles and functional conservation among simians. J. Virol. 78:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boller, K., H. König, M. Sauter, N. Mueller-Lantzsch, R. Löwer, J. Löwer, and R. Kurth. 1993. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology 196:349-353. [DOI] [PubMed] [Google Scholar]
- 11.de Parseval, N., V. Lazar, L. Bénit, J. Casella, and T. Heidmann. 2003. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 77:10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavillette, D., M. Marin, A. Ruggieri, F. Mallet, F. L. Cosset, and D. Kabat. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76:6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löwer, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Mueller-Lantzsch, J. Löwer, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 90:4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lower, R., R. R. Tonjes, C. Korbmacher, R. Kurth, and J. Lower. 1995. Identification of a rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayer, J., M. Sauter, A. Racz, D. Scherer, N. Mueller-Lantzsch, and E. Meese. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21:257-258. [DOI] [PubMed] [Google Scholar]
- 17.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi, S., X. Lee, X. Li, G. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Tang, P. Edouard, S. Howes, J. J. Keith, and J. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 17:785-789. [DOI] [PubMed] [Google Scholar]
- 19.Muster, T., A. Waltenberger, A. Grassauer, S. Hirschl, P. Caucig, I. Romirer, D. Fodinger, H. Seppele, O. Schanab, C. Magin-Lachmann, R. Lower, B. Jansen, H. Pehamberger, and K. Wolff. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 63:8735-8741. [PubMed] [Google Scholar]
- 20.Steinhuber, S., M. Brack, G. Hunsmann, H. Schwelberger, M. P. Dierich, and W. Vogetseder. 1995. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum. Genet. 96:188-192. [DOI] [PubMed] [Google Scholar]
- 21.Tonjes, R. R., C. Limbach, R. Lower, and R. Kurth. 1997. Expression of human endogenous retrovirus type K envelope glycoprotein in insect and mammalian cells. J. Virol. 71:2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 23.Villesen, P., L. Aagaard, C. Wiuf, and F. S. Pedersen. 2004. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]