Abstract
Simian immunodeficiency virus (SIV) has been shown to progress through a number of changes that lead to the emergence of pathogenic viral variants in macaques initially infected with a mildly cytopathic variant, SIVMneCL8. One of these late-stage isolates, SIVMne170, replicates to high levels in vivo and causes a rapid disease course when reintroduced into naïve macaques, resulting in a viral set point up to 3,000-fold higher than the set point of the parental virus, SIVMneCL8. However, in cell culture both viruses replicate with similar kinetics. One major difference between in vivo and in vitro cultures is the life span of the infected cells. Here, we manipulated the life span of infected cells in vitro, and we show that the fitness of SIVMne170 in cultures with a limited cell life span dramatically increased compared to its fitness in cultures with a nonlimited life span of cells. The increase in fitness was at least partially due to the fact that the rapid turnover system eliminates the negative influence of the cytopathic effects associated with replication of SIVMne170. Because the relative fitness of SIVMneCL8 and SIVMne170 observed in the rapid turnover system more accurately reflects their fitness in vivo, the system represents an improved approach to comparing relative fitness of viruses.
Human and simian immunodeficiency viruses (HIV and SIV) are known to continuously evolve and adapt to specific selective pressures found within the host, leading to the emergence of variants that may differ from the earlier variants in their replication properties. The fitness of such evolving variants is directly linked to the precise conditions under which viral replication occurs. For example, an inhibitor-resistant variant demonstrates higher fitness than the wild-type virus in the presence of the inhibitor but may have lower fitness when the drug is removed (5, 9, 26). In general, the conditions found in tissue culture do not adequately reflect the selective pressures acting on the virus within a host. To define culture conditions that predict in vivo fitness would require studying variants with clear fitness differences in vivo. However, it is difficult to identify HIV type 1 (HIV-1) variants that show a clear difference in replication fitness in vivo, because in any one individual, the replication fitness of different viruses may be as much host dependent as virus dependent. Variants with different fitness can be identified in a SIV animal model system because the replication fitness of a given virus can be examined experimentally in multiple hosts.
In a study of evolution of a cloned SIV variant in macaques, we have shown that newly emerging viral variants replicated to much higher levels and were more pathogenic than their parent virus, SIVMneCL8, when transmitted to a new host (11). Some of the increase in replication levels could be attributed to changes in the glycosylation of the envelope protein, which led to low immunogenicity (3, 22). Measurements of viral RNA in plasma of infected animals showed that a virus with only these specific glycosylation changes replicated to ∼100-fold-higher levels than the parental virus. However, infections of virus-naïve macaques with SIVMne170, a late-stage derivative of SIVMneCL8, resulted in 1,000- to 3,000-fold-higher viral RNA levels in plasma, compared to virus-naïve macaques infected with SIVMneCL8. This suggests that some factor other than sensitivity to envelope-directed antibodies may be responsible for an additional 10- to 30-fold increase in viral replication ability (11). Therefore, the early and late isolates of SIVMne present a convenient in vitro system to dissect the features important for increased viral pathogenesis in vivo. Interestingly, the replication kinetics of SIVMneCL8 and SIVMne170 were similar under standard tissue culture conditions and thus did not correlate to their replicative ability in vivo (12).
In general, immune escape plays an important role in HIV and SIV selection in the host (recently reviewed in reference 2). In addition, HIV and SIV have been observed to become increasingly pathogenic in a way that is independent of immune system selection/pressure (4, 11, 17, 25). The mechanism of increased virulence is not clear, but it seems to correlate with viral ability to form syncytia in T-cell lines, its in vitro replicative fitness, and the ability to use a different receptor for entry (1, 14, 21, 23, 24). While generally predictive of increased virulence, these correlations are not absolute and, therefore, other yet-undiscovered features may be important.
Most in vitro experiments measuring viral fitness are performed under conditions where cells turn over at a rate much slower than observed in vivo. It has been estimated that within an infected human, the life span of infected cells is ∼2 days (19, 27). This turnover rate is a combined effect of cytotoxic T-cell responses directed to HIV-1-infected cells, cytopathic effects of the virus, and normal death rates of activated cells in the body. Hence, the in vitro conditions that are commonly used are unlikely to lead to accurate insights into the selective pressure imposed on viral replication by rapid clearance of infected cells by the immune system. Previously, we have shown that this selective pressure can be reproduced in tissue culture using a rapid turnover system (8). In such a system, the life span of cells is limited to 2 to 3 days by either killing or removing a large proportion of cells in the infected culture and adding uninfected cells, which selects for virus that preferentially spreads via cell-cell interactions rather than through cell-free infections (7, 8) Therefore, the rapid turnover system may better represent at least some of the in vivo conditions of viral replication.
In this study we sought to define tissue culture conditions that would better represent some of the conditions encountered by SIV within a host. We expected that relative fitness of the late-stage virus SIVMne170 would increase when conditions better mimicked the in vivo conditions that shaped its evolution from SIVMneCL8. We hypothesized that the short life span of infected cells observed in vivo (10, 19, 27) may provide selective pressure that affects the relative fitness of SIVMne170. Using competition experiments, we measured the relative fitness of SIVMneCL8 and SIVMne170 in cultures with both nonlimited life span of cells and with rapid cell turnover, limiting the life span to 2 to 3 days. SIVMne170 demonstrated increased fitness only when the life span of cells was limited. This indicates that during viral evolution within the host, SIVMne170 acquired features that allow it to replicate better in the face of rapid turnover of infected cells, whether caused by the immune system of the host or by viral cytopathic effects.
MATERIALS AND METHODS
Cells and viruses.
CEMx174 cells, expressing secreted alkaline phosphatase (SEAP), were a kind gift of R. Desrosiers (16). The cells were maintained in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin.
To generate stocks of infectious virus, 293T cells were transiently transfected with each proviral clone using TransIT reagent (Mirus, Madison, WI), and the produced virus was used to infect CEMx174 cells. The virus was collected as soon as cytopathic effects were evident, filtered through a 0.22-μm filter, and treated with 30 U/ml of DNase I in the presence of 10 mM MgCl2. Virus aliquots of 1 ml were frozen at −80°C. Viral titer was determined as the 50% tissue culture infective dose on 2 × 105 CEMx174 cells using 100 μl of virus serially diluted fourfold in replicates of six. Viral replication was observed as an increase in SEAP activity in culture medium using a PhosphaLight detection kit according to the manufacturer's instructions (Applied Biosystems, Foster City, CA).
Competition experiments.
To establish infected cultures, 2 × 105 CEMx174 cells were infected separately with approximately 104 50% tissue culture infective doses of competing viruses. Two days later infected cells were washed and mixed in various ratios. The relative levels of infected cells were normalized by SEAP activity in the medium. For competitions with nonlimited life span of cells, 104 cells from infected cultures (corresponding to approximately 500 infected cells) were mixed with 4 × 105 noninfected cells in a final volume of 1 ml (giving an effective multiplicity of infection [MOI] of 0.0025). On each consecutive day the culture medium was completely replaced with fresh medium. A 50-μl aliquot of culture medium was frozen for subsequent GeneScan analysis, and 20 μl was used to measure SEAP activity. The viability of cells was assessed by staining with trypan blue and counting the number of live and dead cells. The cultures were split threefold as necessary to maintain the cell density below 2 × 106 cells/ml. The experiment was continued for 12 days or until all cells were dead.
For competitions in the rapid turnover system, 2 × 104 cells from infected cultures (corresponding to approximately 1,000 infected cells) were mixed with 2 × 105 noninfected cells in a final volume of 1 ml (for an MOI of 0.01). The virus was transferred to an excess of noninfected cells every 2 days. For cell-based passage, 10% of the culture was taken out and the rest was discarded. Infected cells were washed twice with complete medium and mixed with 2 × 105 noninfected cells. For DC-SIGN-based passage, 90% of the infected culture was discarded and the remaining 10% was cleared from infected cells by centrifugation. The cells were discarded, and the virus-containing supernatant was incubated with 4 × 104 DC-SIGN-expressing Raji B cells for 40 min. After incubation the cells were washed twice with complete medium and mixed with 2 × 105 noninfected CEMx174 cells. A 20-μl aliquot of culture medium was used to monitor the spread of virus by SEAP activity, and 50 μl of medium was frozen for consecutive GeneScan analysis.
GeneScan assay.
In order to measure the ratio of SIVMneCL8 viral RNA to that of SIVMne170, virus-containing medium was lysed by the addition of 0.04% Triton X-100 (Sigma, St. Louis, MO) and subjected to reverse transcription-PCR (RT-PCR), followed by PCR product purification and GeneScan analysis. The lysis of virions and RT-PCR were performed in a single step using a modification of the OneStep RT-PCR kit (QIAGEN, Valencia, CA). The sample (2 μl of culture medium) was put in a reaction volume of 20 μl with final concentrations of reagents as follows: 1× reaction buffer, 0.4% Triton X-100, 0.1 pmol/μl primers, 0.4 mM deoxynucleoside triphosphate, 1× enzyme mix, 5 U/ml RNaseOUT inhibitor (Invitrogen, Carlsbad, CA). The RT step was performed at 50°C for 30 min followed by an RT inactivation step (95°C for 15 min) and two-step PCR amplification (1 min at 61°C and 15 seconds at 95°C) for 30 cycles. The PCR product was purified using Sephadex G-50 fine Sepharose columns (Amersham Biosciences, Piscataway, NJ) prepared in MultiScreen-HV plates (Millipore, Bedford, MA).
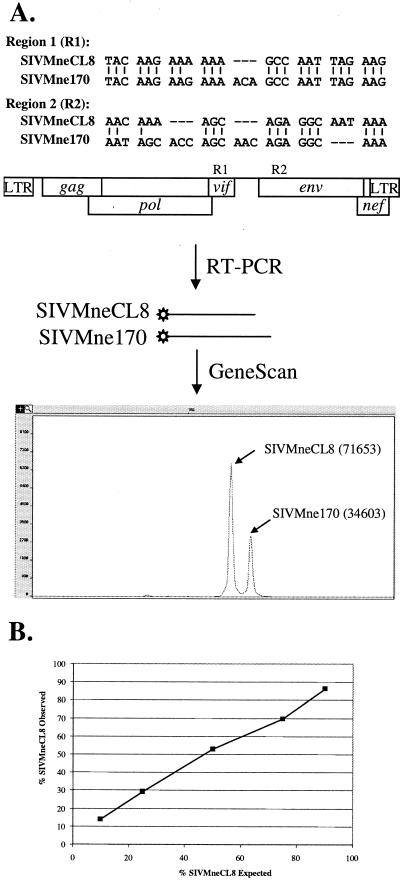
The primers for PCR were designed to flank the region with a 3-bp insertion in the SIVMne170 sequence. For region 1 (Fig. 1A) the primers were 5′-GCAGAGTAATCTTCCCACTA-3′ and 5′-carboxyfluorescein (FAM)-ATTCCAATACCCTTGTACTTCTAA-3′. For region 2 (Fig. 1A) the primers were 5′-FAM-CCCATTATGCATTACTATGAAATGCAACAAAAG-3′ and 5′-GTTCCAAGCCTGTGCAATTA-3′. As a result, products formed by amplification of SIVMneCL8 and SIVMne170 RNA differed by 3 nucleotides and could be distinguished on a polyacrylamide gel. The relative amount of each product is proportional to the area of the corresponding peak.
FIG. 1.
The GeneScan-based assay to measure relative levels of SIVMneCL8 and SIVMne170 RNA. The assay was developed for two regions in the viral genome, vif (R1) and env (R2), which each contain a 3-bp insertion in SIVMne170 (A, top). Viral RNA regions amplified by RT-PCR with a 5′-labeled 6-FAM primer (asterisk in the figure) could be distinguished by GeneScan because the product from SIVMne170 is 3 bp longer than the product from SIVMneCL8 (A, middle). The relative amount of RNA of each variant was determined from relative areas of corresponding peaks. In the example shown (numbers in parentheses), the proportion of SIVMneCL8 was 67.4% (A, bottom). (B) The GeneScan assay was validated using plasmids encoding SIVMneCL8 and SIVMne170, mixed in known proportions (1:9, 1:3, 1:1, 3:1, and 9:1). The graph shows the correlation between the expected and the observed proportions of SIVMneCL8.
Relative viral fitness.
The relative viral fitness (w) was calculated using the equation w(p) = log[(p1/q1) × (p0/q0)], where p and q are the proportions of competing viruses and 0 and 1 refer to consecutive time points. Thus, when the relative amounts of viruses do not change over time, their fitness w is equal to 0. When the proportion of one of the viruses increases, its fitness becomes positive, while the fitness of the out-competed virus becomes negative, and absolute values, w , are equal for both viruses. For experiments with nonlimited life span of cells the time points were 1 day apart, while for the rapid turnover system the time points coincided with transfers and were 2 days apart.
Cell doubling time and half-life time of cell death.
The doubling time and half-life time of cell death rates were measured by fitting experimental data with exponential curves using Microsoft Office Excel 2003. The exponential rates were divided by ln(2) to obtain doubling time and half-life time of decay.
RESULTS
SIVMneCL8 is more fit than SIVMne170 in cultures with nonlimited life span of cells.
Often, the replication properties of two viruses are compared in parallel cultures, where minor differences in conditions may be present. However, a better measure of the replicative capacity of two viral variants in culture is a competition experiment, which provides a measure of relative viral fitness under identical culture conditions (9, 15, 20). In this setting, one variant dominates and displaces the other variant at a rate that is proportional to the difference in fitness of the two viruses. In order to monitor the relative proportion of SIVMneCL8 (an early variant) and SIVMne170 (a late variant) in cell-free culture supernatant over time, we developed a GeneScan assay that distinguishes the two viruses by the presence of 3-bp insertions that SIVMne170 has acquired in its genome relative to SIVMneCL8 (Fig. 1A). One of the insertions is located immediately after the pol gene, and the other is in the first third of the env gene. The linear range of the GeneScan assay was validated by using mixtures of plasmids encoding SIVMneCL8 and SIVMne170 proviruses (Fig. 1B) as well as by site-specific real-time PCR analysis (data not shown). Two separate markers were used to control for outgrowth of a recombinant virus.
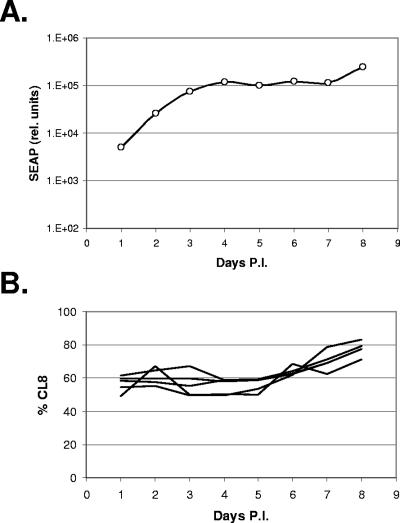
Previously, we reported that SIVMneCL8 and SIVMne170 have similar replication rates in CEMx174 cells (CD4+ human T-B hybrid cell line) (12) when they are compared in parallel cultures. Therefore, we predicted that, under these same conditions, these two variants would also show similar fitness in a competition experiment. To test this, we infected CEMx174 cells that contain an LTR-SEAP reporter gene (16) with SIVMneCL8 and SIVMne170 and monitored the relative levels of the two variants over time. The medium was completely replaced each day to more accurately measure the spread of virus and to limit the GeneScan measurements to the virions released only in the previous 24 h. At day 4 postinfection, SEAP activity reached an initial plateau, indicating that most of the cells in the culture were infected (Fig. 2A). As expected, during this 4-day period the relative levels of SIVMneCL8 and SIVMne170 did not change significantly, indicating that these two variants had similar fitness under these conditions (Fig. 2B). The average fitness of SIVMneCL8, measured from five replicates during the first 4 days of infection, was −0.002 ± 0.118 (see Materials and Methods).
FIG. 2.
Results of competition in a system with nonlimited life span of cells. (A) The increase in the number of infected cells was observed by measuring SEAP activity in media of cultures infected with mixes of SIVMneCL8 and SIVMne170 viruses. Initial spread of virus through the culture (increase in SEAP activity on days 1 through 4) is followed by exhaustion of available cells (plateau in SEAP activity on days 4 through 7) and expansion of infected cells (additional increase in SEAP activity on day 8). (B) Percentage of SIVMneCL8 RNA in culture medium at different times postinfection measured by GeneScan R1 probe. The relative amount of SIVMneCL8 remained approximately the same during the first 5 days and increased thereafter.
After all of the cells in a culture are infected, the relative amounts of viruses could be predicted to change due to differences in survival rates of the infected cells. A highly cytopathic virus, such as SIVMne170, kills infected cells and should lead to their eventual disappearance from the culture, while cells infected with mildly cytopathic variant SIVMneCL8 should survive for longer periods of time and even continue to divide (persistent infection). Indeed, the proportion of SIVMneCL8 increased at 7 days postinfection. This increase correlated with an increase in cell numbers (data not shown) and in SEAP activity in the medium (Fig. 2A), suggesting an outgrowth of SIVMneCL8-infected cells at this time. The relative fitness of SIVMneCL8 during this period was +0.170 ± 0.028, significantly higher than its fitness during the initial 4 days of infection (P < 0.005, two-tailed equal variance t test). In an independent repeat of the experiment using cultures maintained for a longer period of time, SIVMneCL8 replaced the SIVMne170 variant in all five independent cultures by day 11 (data not shown) and SIVMneCL8 fitness measured +0.151 ± 0.054. To examine the possibility that we were observing an outgrowth of a recombinant virus, we measured the relative levels of SIVMneCL8 and SIVMne170 using GeneScan probes targeted to both regions R1 and R2 (Fig. 1A). These two probes produced almost identical results (data not shown), suggesting that recombinant viruses were not preferentially selected in our experiments and that we were observing an increase in the number of cells infected with full-length SIVMneCL8.
In summary, these data suggest that SIVMne170 was as fit as the parental clone SIVMneCL8 during the first 4 days after inoculation and less fit thereafter. Therefore, the relative fitness of SIVMneCL8 and SIVMne170 observed in these experiments after prolonged cultures was opposite to their fitness in vivo.
SIVMne170 is more fit than SIVMneCL8 in rapid turnover systems.
To reproduce in vitro the short half-life of infected cells observed in vivo, we performed viral transfer experiments in cultures containing SIVMneCL8- and SIVMne170-infected cells (Fig. 3B). Every 2 days, 90% of the cells were discarded and replaced with noninfected cells. Therefore, the life span of cells in these cultures was less than 2 days for the majority of both SIVMneCL8- and SIVMne170-infected cells. This rate of cell reduction is equivalent to the reduction caused by exponential decay with a half-life of ∼14 h, which is similar to the rate of disappearance of infected cells observed in vivo (19, 27). The ratio between SIVMneCL8 and SIVMne170 was measured immediately before each transfer step. Due to continuous viral replication, the probability of recombination in these experiments was much higher than in cultures with nonlimited cell life span. Therefore, we limited our analysis to the first three passages, when the effects of recombination were minor (data not shown).
FIG. 3.
Overview of different culture conditions tested for effects on viral fitness. (A) Cultures with nonlimited life span of infected cells. Cultures were inoculated with virus at a low MOI (0.005), and the virus was allowed to spread. Cells were maintained at density below 2 × 106 and the medium was replaced every day, but no fresh cells were added. (B) Cell-based rapid turnover system. Cultures were inoculated at a 0.01 MOI. Virus was passaged to an excess of fresh cells every 2 days by transferring 1/10 of cells from infected cultures. (C) Virus-based rapid turnover system. Cultures were inoculated at a 0.01 MOI. Virus was passaged to fresh cells every 2 to 3 days by transferring 1/10 of cell-free virus-containing medium. (D) DC-SIGN-based rapid turnover system. Cultures were inoculated at a 0.01 MOI. Virus was passaged to fresh cells every 2 days by prebinding of cell-free virus to Raji cells expressing DC-SIGN, washing off unbound virus, and mixing these cells with noninfected target cells.
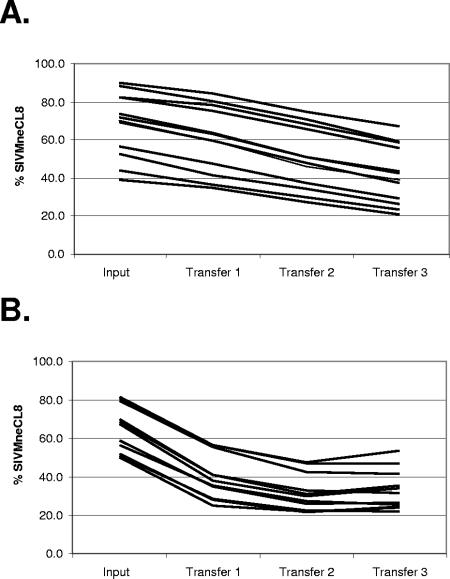
In 12 independent experiments, the starting proportions of SIVMneCL8 to SIVMne170 in mixes were between 40% and 90%. Regardless of the starting ratio, the proportion of SIVMneCL8 in each of these 12 cultures decreased in each of three consecutive transfers (Fig. 4A). The relative viral fitness was calculated separately for each transfer step and for each replicate. There were no statistically significant differences in relative viral fitness either between different transfer steps (t test, P > 0.15) or between different starting proportions (t test, P > 0.25). Therefore, the relative fitness of SIVMne170 was averaged for all three transfers and all 12 replicates, resulting in a value of +0.180 ± 0.043. SIVMne170 was therefore more fit than SIVMneCL8 in cultures with a limited life span of infected cells.
FIG. 4.
High relative fitness of SIVMne170 in rapid turnover systems. Cultures were inoculated with SIVMneCL8- and SIVMne170-infected cells mixed in different ratios. The virus was transferred from infected cultures to uninfected cells by transferring 1/10 of cells (A) or incubating cell-free virus with DC-SIGN-expressing Raji B cells (B) and mixing these cells with uninfected CEMx174 cells. The proportion of SIVMneCL8 RNA in culture medium was determined before the first transfer and after each consecutive transfer. Each line represents an independent competition and shows the reduction in proportion of SIVMneCL8 after each transfer, indicating higher fitness of SIVMne170 in the rapid turnover assays.
We attempted to reduce the half-life of infected cells even further by discarding all cells from the infected cultures and transferring only the cell-free virus to fresh cells (Fig. 3C). However, viral replication in such cultures could not be sustained, even when the transfers were performed every 3 days (data not shown). Therefore, we used a modified version of the rapid turnover assay developed by us previously (8) that included DC-SIGN-expressing Raji B cells (Fig. 3D) (28). The cells loaded with viral particles were washed to remove unbound virus and then mixed with fresh target cells. As expected, such DC-SIGN-mediated infection was more efficient than infection via cell-free virus and allowed sustained viral replication over multiple 3-day passages. The SIVMne170 fitness relative to SIVMneCL8 in this system was +0.470 ± 0.051 (Fig. 4B). Similar results were obtained in four more independent repeats of this experiment, with an average SIVMne170 fitness of +0.382. Thus, the higher fitness of SIVMne170 in a rapid turnover system was shown to be independent of the method of viral transfer.
The rapid turnover system limits cytopathic effects of SIVMne170.
Our previous studies have shown that in cultures infected with the SIVMne170 variant, the viability of cells was significantly lower than in cultures infected with SIVMneCL8 (12). We hypothesized that the increase in SIVMne170 fitness observed in cultures with a limited life span of infected cells was at least in part due to the alleviation of cytopathic effects of this virus. The cytopathic effects of virus replication in cell lines are most likely caused by direct killing of infected cells. However, these effects must occur within the time scale of viral transfers (2 to 3 days) in order to affect viral fitness in the rapid turnover system. Because previous measurements were done in nonsynchronized cultures (12), the time scale and the magnitude of cytopathic effects of SIVMneCL8 and SIVMne170 could not be determined from those data. In order to better understand and directly compare the cytopathic properties of SIVMneCL8 and SIVMne170, we separately infected CEMx174 cells with high doses of each virus (MOI, ∼0.5) in triplicate. After 2 days all of the cells in culture were infected, and we started monitoring the viability of cells (Fig. 5). Compared to uninfected cells, whose viability remained above 90%, SIVMneCL8-infected cultures showed a temporary decrease in viability to 70% and eventually recovered to 90% of viable cells, suggesting that the cytopathic effects of SIVMneCL8 replication were relatively modest. In contrast, infection with SIVMne170 resulted in decreased cell numbers and eventually led to the death of infected cultures. The cytopathic effects of clone SIVMne170 first became evident after 2 days, when the viability of cells began decreasing. In the following days, the number of live cells decreased exponentially, with a half-life of approximately 1 day until most cells in the culture were dead. These findings indicate that limiting the life span of cells to 2 days in a rapid turnover system eliminates the majority of cytopathic effects of SIVMne170 replication, which could therefore increase its relative fitness compared to conditions where cytopathic effects occur.
FIG. 5.
Measurement of cytopathic effects of SIVMneCL8 and SIVMne170 viruses. Cells were infected with either SIVMneCL8 (black triangles) or SIVMne170 (open circles), and the viability of cells in the cultures was compared to that of noninfected cultures (black squares). The synchronized infection allows direct measurements of cytopathic effects of each virus. All data points are averages and standard deviations of three independent replicates.
Syncytium formation is not important for increased fitness of SIVMne170 in the rapid turnover system.
Our studies indicate that under conditions of rapid cell turnover, where difference in cytopathicity does not impact fitness, SIVMne170 is more fit than SIVMneCL8. To examine the possibility that some of the known properties that distinguish SIVMne170 from SIVMneCL8 are responsible for its increased replicative ability in the rapid turnover system, we tested chimeric viruses of SIVMneCL8 and SIVMne170.
SIVMne170, unlike SIVMneCL8, causes a large amount of syncytia when replicating in CEMx174 cells (12). This raised the question of whether this ability increases SIVMne170's fitness in the rapid turnover system. We tested a previously described chimeric virus, SIVMne8/170xr-su, which contains a portion of the SIVMne170 sequence, including the vpx/vpr genes and the SU portion of the env gene, in the background of SIVMneCL8. This chimera has an in vitro replication rate similar to that of SIVMneCL8 but produces a large number of syncytia, as SIVMne170 does (12). We performed competition experiments between this virus and the two parental viruses, both when the life span of cells was not limited as well as in the rapid turnover system. In competition with SIVMneCL8, the starting ratio did not change over time in either system, indicating that the fitness of SIVMne8/170xr-su was very similar to that of SIVMneCL8 (data not shown). When we compared the chimera to the SIVMne170 virus, we observed that the chimera out-competed SIVMne170 when the life span of cells was not limited and did the converse in the rapid turnover system. Thus, the overall phenotype of this chimera was similar to that of SIVMneCL8 (Table 1). Therefore, we concluded that syncytium formation is not important for increased fitness of SIVMne170 in a rapid turnover system.
TABLE 1.
Relative fitness of chimeric viruses measured in competitions with parental viruses
| Virus | Nonlimited lifespan
|
Rapid turnover
|
||||
|---|---|---|---|---|---|---|
| SIVMneCL8 | SIVMne170 | Fitnessa | SIVMneCL8 | SIVMne170 | Fitness | |
| SIVMneCL8 | Win | High | Lose | Low | ||
| SIVMne170 | Lose | Low | Win | High | ||
| 8/170xr-sub | Equal | Win | High | Equal | Lose | Low |
| 8/170 | Lose | Equal | Low | Equal | Lose | Low |
| 170/8 | Equal | Win | High | Win | Equal | High |
“High” and “low” fitness phenotypes were defined from the relative fitness of SIVMneCL8 and SIVMne170 (in bold). For chimeric viruses, the phenotype was determined from competitions with each of the parental variants. The outcome of the competition was labeled according to the relative fitness value of the chimera (w). For 0.1 < |w| < 0.1, the outcome was judged as a tie (labeled as equal); for w > 0.1, the outcome was judged as “lose” (chimera is less fit); for w > 0.1, the outcome was judged as “win” (chimera is more fit). The chimera's phenotype was assigned as “high” when the chimera had equal fitness with the high-fitness parental strain and won competition with the low-fitness parental strain (w > 0.1). The chimera's phenotype was assigned as “low” when the chimera had equal fitness with the low-fitness parental strain and lost the competition with the high-fitness parental strain.
8/170xr-su chimera contains a portion of the SIVMne170 sequence responsible for syncytium formation, including the vpx/vpr genes and the SU portion of the env gene, in the background of SIVMneCL8; 8/170 chimera contains the 5′ half of SIVMneCL8 and the 3′ half of SIVMne170; 170/8 chimera contains the 5′ half of SIVMne170 and the 3′ half of SIVMneCL8.
The determinant of higher fitness of SIVMne170 in the rapid turnover system is located in the 5′ half of its genome.
To further examine the region responsible for increased fitness of SIVMne170 in the rapid turnover system, we tested two reciprocal chimeras containing different regions of SIVMneCL8 and SIVMne170. The 8/170 chimera contains the 5′ half of SIVMneCL8 and the 3′ half of SIVMne170. When the life span of cells was not limited, this chimera had low relative fitness, losing competition with SIVMneCL8 and remaining in equilibrium with SIVMne170 (Table 1). In the rapid turnover system, the 8/170 chimera showed fitness similar to that of SIVMneCL8 but lower than the fitness of SIVMne170. The fact that the 8/170 chimera exhibited low fitness in both systems, when the life span of cells was limited as well as when it was not, suggested that determinants of increased fitness were not encoded in this chimera.
The 170/8 chimera contained the 5′ half of SIVMne170 and the 3′ half of SIVMneCL8. This chimera showed high fitness in the system with nonlimited life span of cells, maintaining the input proportion in competition with SIVMneCL8 and increasing its proportion in competition with SIVMne170 (Table 1). In the rapid turnover system, the 170/8 chimera also had high fitness. It was able to out-compete SIVMneCL8 and remained in equilibrium with SIVMne170 (Table 1). The combined results of competition experiments with chimeric viruses indicated that the determinant of higher fitness of SIVMneCL8 in systems with nonlimited life span of cells was located in the 3′ half of the virus, while the determinant of higher fitness of SIVMne170 in a rapid turnover system was located in the 5′ half of the virus.
DISCUSSION
In this study, we measured the in vitro fitness of two variants of SIVMne that have very different replicative capacities and pathogenesis in vivo. We found that the relative fitness of the two variants depended on the life span of cells in the culture. The early-stage, less pathogenic variant, SIVMneCL8, was more fit in cultures with a nonlimited life span of cells. The late-stage, highly pathogenic variant, SIVMne170, was more fit in a rapid turnover system, when the life span of cells was limited to 2 days. This indicates that the SIVMne170 variant has an increased capacity to replicate under conditions with a high turnover of infected cells, such as occurs in vivo. We believe that this increase in fitness is caused by two separate phenomena. First, the rapid turnover system eliminates the negative effect of SIVMne170's cytopathic properties, which lead to low fitness of this virus in the system with nonlimited life span of cells. Second, the 5′ portion of SIVMne170 contains a specific determinant responsible for the increased fitness of this variant in the rapid turnover system.
Several studies have found that the life span of HIV-infected (or SIV-infected) cells in vivo is very short, with half-life estimates ranging from 1.6 to 3 days (19, 27). We therefore predicted that a rapid turnover system represents conditions that the virus encounters within a host better than do standard culture conditions, where the life span of infected cells is not limited. The studies presented here support that prediction and suggest that the rapid turnover system may provide a useful tool for characterizing the replication properties of HIV and SIV isolates. This assay may be particularly useful in cases where the variants differ in cytopathic effects and rate of spread. In the case of SIVMne170, which naturally evolved from SIVMneCL8 within the host, both its cytopathic properties and its infectivity appeared to be different from the parental isolate. The changes, however, were not apparently beneficial when tested under regular tissue culture conditions, and only the rapid turnover system was able to recapitulate the higher fitness of SIVMne170 observed in vivo.
Culture conditions influence viral fitness.
In the cultures with rapid cell turnover, the long life span of infected cells does not provide an advantage, and other factors, such as replication rate, become more important. The fact that the fitness of SIVMne170 in the rapid turnover system was higher than the fitness of SIVMneCL8 indicated that SIVMne170 acquired some property or properties that increased its replication rate. Previous studies from our group comparing parallel infected cultures, as well as competition studies presented here, failed to demonstrate the existence of any advantageous property of SIVMne170 in cell cultures with a nonlimited life span (12), because its positive effect was counter-balanced by the negative effect of increased cytopathicity of this virus. In fact, the disadvantages of SIVMne170's cytopathic effects were approximately balanced out by the advantages of its higher rate of spread, resulting in a replication rate that was very similar to that of SIVMneCL8 under standard culture conditions. Our conclusion that a standard tissue culture system poorly reflects in vivo SIV fitness is in agreement with a study of another late-stage SIVMne isolate, SIVMne027, which showed that the increased replication capacity of this variant was evident in dendritic cell-T-cell cocultures but not in regular cultures of CEMx174 cells or activated monocytes (13).
Cultures with a nonlimited life span of infected cells favor viruses that are able to fully exploit the long life span of cells by establishing persistent infections. This was exemplified by the relatively noncytopathic SIVMneCL8 virus, which has characteristics of viruses present early in HIV and SIV infection (22). It is tempting to speculate that less cytopathic viruses have their greatest advantage very early in infection, because a longer life span of the infected cell may increase the chances of local viral spread at the site of infection and/or viral amplification. At these early stages of infection, cell killing by cytotoxic T lymphocytes is not important, allowing infected cells to persist if the virus that infects them is not directly cytopathic to the cell. Such noncytopathic viruses would thus be selectively amplified over cytopathic viruses under these conditions, but not at later times when replication under conditions of rapid cell turnover is more important.
Determinants of higher fitness of SIVMneCL8 in a system with nonlimited life span of cells.
SIVMneCL8 showed an increased fitness versus SIVMne170 in cultures where the majority of cells were infected for several days. Our experiments with chimeric viruses point to the 3′ half of SIVMneCL8 as the main determinant of increased fitness in tissue culture systems with nonlimited life span of cells (Table 1). We believe that the main effect of this region is reduced cytopathic effects, resulting in persistently infected cells, which survive longer than cells infected with a highly cytopathic virus. In our earlier studies, the cytopathic properties of the 170/8 chimera and the SIVMne170 variant could not be distinguished when they were examined in parallel cultures (12). The competition experiments described here revealed that the 170/8 chimera had a higher fitness than SIVMne170 when the life span of cells was not limited, suggesting that the chimera may be less cytopathic than SIVMne170, which also encodes an additional cytopathic determinant(s) in the envelope. Alternatively, cytopathic properties may not be the only determinant of increased viral fitness in this system. Future experiments with additional chimeras or with more accurate measurements of cytopathic effects may allow us to distinguish these two hypotheses.
Determinants of higher fitness of SIVMne170 in a rapid turnover system.
The reason for increased viral fitness of SIVMne170 compared to SIVMneCL8 in a rapid turnover system is not clear and may have multiple explanations. SIVMne170 acquired multiple changes throughout the genome compared to the parent, SIVMneCL8. Thus, the advantageous change(s) may affect almost any stage of the virus life cycle, from generation of more stable virions to faster kinetics of reverse transcription and integration. Our analysis of chimeric viruses 8/170 and 170/8 indicated that the genetic determinant(s) of this increased fitness is localized in the 5′ half of SIVMne170's genome, including gag, pol, and the 5′ portion of the vif gene. Thus, we can exclude several determinants previously implicated in increased fitness of SIVs in vitro and in vivo, such as vpr, env, and nef genes, as well as the U3 region (6, 13, 17, 18), because these are not present in the 5′ region of SIVMne170. The 5′ region identified here has 32 amino acid changes relative to SIVMneCL8, including nine nonsynonymous mutations in the gag gene. It is possible that that the adaptation to rapid turnover of infected cells will map to mutations that affect late stages of the viral life cycle, such as virion formation and release. Regardless of the exact nature of the adaptation, it provides a specific advantage for virus during rapid cell turnover because both SIVMne170 and 170/8 chimera had a higher fitness than SIVMneCL8 in that system, but not in a system with a nonlimited life span of cells.
Pathogenesis and in vitro fitness.
It is tempting to speculate that increased fitness of SIVMne170 in the rapid turnover system is responsible for the increased pathogenesis of the variant in vivo. Resistance to neutralizing antibodies, faster replication, and increased cytopathicity of the virus may lead to a more profound impact on the state of the immune system and, as a result, to a higher steady-state viral load. Interestingly, the increased direct killing of infected cells, which can reduce the apparent replicative fitness of the virus in vitro (Fig. 2 and 4), can also be responsible for selective destruction of HIV-specific T cells and, thus, for the increase in overall fitness and pathogenicity of the virus. However, it should be noted that other factors, unaddressed so far, may play an important role in pathogenesis of this virus. For example, changes in the envelope (as well as in other regions) of SIVMne170 may not only render it resistant to antibodies but also affect the cytotoxic T-cell response of the host and, correspondingly, the in vivo fitness of the virus. Our approach does not allow us to investigate this possibility.
In conclusion, we were able to demonstrate that a limited life span of infected cells is an important factor for viral replication, influencing viral fitness. Rapid turnover systems appear to favor variants with a higher rate of viral spread and reduce the negative effects of viral cytopathic properties. While the less-fit virus was observed to quickly disappear from the competition, the relative contribution of higher fitness in the rapid turnover system to the overall viral fitness in vivo was not assessed in this study. We propose that the rapid turnover system could be further enhanced to include other selection conditions that may help uncover adaptive pressures imposed on viruses by other factors, such as neutralizing antibody responses, presence of target cells expressing different receptors, and antiretroviral drug treatments.
Acknowledgments
We acknowledge the assistance of the FHCRC Genomics Shared Resource in running the GeneScan analysis. We thank Masahiro Yamashita, Shari Kaiser, and Mario J. Pineda for critical reading of the manuscript, Gretchen Strauch for expert editorial assistance, and Jason Kimata, who constructed the chimeric viruses used here.
This work was supported by NIH grants AI 34251 (to J.O.) and R37 AI30827 (to M.E.). Y.V. was partially supported by grant number 106594-36-RFGN from the American Foundation for AIDS Research.
REFERENCES
- 1.Arien, K. K., A. Abraha, M. E. Quinones-Mateu, L. Kestens, G. Vanham, and E. J. Arts. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, J., J. N. Blankson, M. Wind-Rotolo, and R. F. Siliciano. 2004. Mechanisms of HIV-1 escape from immune responses and antiretroviral drugs. Curr. Opin. Immunol. 16:470-476. [DOI] [PubMed] [Google Scholar]
- 3.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edmonson, P., M. Murphey-Corb, L. N. Martin, C. Delahunty, J. Heeney, H. Kornfeld, P. R. Donahue, G. H. Learn, L. Hood, and J. I. Mullins. 1998. Evolution of a simian immunodeficiency virus pathogen. J. Virol. 72:405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao, Y., E. Paxinos, J. Galovich, R. Troyer, H. Baird, M. Abreha, C. Kityo, P. Mugyenyi, C. Petropoulos, and E. J. Arts. 2004. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J. Virol. 78:5390-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 7.Gummuluru, S., V. N. KewalRamani, and M. Emerman. 2002. Dendritic cell-mediated viral transfer to T cells is required for human immunodeficiency virus type 1 persistence in the face of rapid cell turnover. J. Virol. 76:10692-10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gummuluru, S., C. M. Kinsey, and M. Emerman. 2000. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J. Virol. 74:10882-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrigan, P. R., S. Bloor, and B. A. Larder. 1998. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. J. Virol. 72:3773-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 11.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 12.Kimata, J. T., and J. Overbaugh. 1997. The cytopathicity of a simian immunodeficiency virus Mne variant is determined by mutations in Gag and Env. J. Virol. 71:7629-7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimata, J. T., J. M. Wilson, and P. G. Patel. 2004. The increased replicative capacity of a late-stage simian immunodeficiency virus mne variant is evident in macrophage- or dendritic cell-T-cell cocultures. Virology 327:307-317. [DOI] [PubMed] [Google Scholar]
- 14.LaBonte, J. A., T. Patel, W. Hofmann, and J. Sodroski. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J. Virol. 74:10690-10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maree, A. F., W. Keulen, C. A. Boucher, and R. J. De Boer. 2000. Estimating relative fitness in viral competition experiments. J. Virol. 74:11067-11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Means, R. E., T. Greenough, and R. C. Desrosiers. 1997. Neutralization sensitivity of cell culture-passaged simian immunodeficiency virus. J. Virol. 71:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen, C., C. Pedersen, J. D. Lundgren, and J. Gerstoft. 1993. Biological properties of HIV isolates in primary HIV infection: consequences for the subsequent course of infection. AIDS 7:1035-1040. [DOI] [PubMed] [Google Scholar]
- 18.Patel, P. G., M. T. Yu Kimata, J. E. Biggins, J. M. Wilson, and J. T. Kimata. 2002. Highly pathogenic simian immunodeficiency virus mne variants that emerge during the course of infection evolve enhanced infectivity and the ability to downregulate CD4 but not class I major histocompatibility complex antigens. J. Virol. 76:6425-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 20.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rangel, H. R., J. Weber, B. Chakraborty, A. Gutierrez, M. L. Marotta, M. Mirza, P. Kiser, M. A. Martinez, J. A. Este, and M. E. Quinones-Mateu. 2003. Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J. Virol. 77:9069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudensey, L. M., J. T. Kimata, E. M. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schramm, B., M. L. Penn, R. F. Speck, S. Y. Chan, E. De Clercq, D. Schols, R. I. Connor, and M. A. Goldsmith. 2000. Viral entry through CXCR4 is a pathogenic factor and therapeutic target in human immunodeficiency virus type 1 disease. J. Virol. 74:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trkola, A., H. Kuster, C. Leemann, C. Ruprecht, B. Joos, A. Telenti, B. Hirschel, R. Weber, S. Bonhoeffer, and H. F. Gunthard. 2003. Human immunodeficiency virus type 1 fitness is a determining factor in viral rebound and set point in chronic infection. J. Virol. 77:13146-13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuttle, D. L., C. B. Anders, M. J. Aquino-De Jesus, P. P. Poole, S. L. Lamers, D. R. Briggs, S. M. Pomeroy, L. Alexander, K. W. Peden, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2002. Increased replication of non-syncytium-inducing HIV type 1 isolates in monocyte-derived macrophages is linked to advanced disease in infected children. AIDS Res. Hum. Retrovir. 18:353-362. [DOI] [PubMed] [Google Scholar]
- 26.Weber, J., H. R. Rangel, B. Chakraborty, M. Tadele, M. A. Martinez, J. Martinez-Picado, M. L. Marotta, M. Mirza, L. Ruiz, B. Clotet, T. Wrin, C. J. Petropoulos, and M. E. Quinones-Mateu. 2003. A novel TaqMan real-time PCR assay to estimate ex vivo human immunodeficiency virus type 1 fitness in the era of multi-target (pol and env) antiretroviral therapy. J. Gen. Virol. 84:2217-2228. [DOI] [PubMed] [Google Scholar]
- 27.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 28.Wu, L., T. D. Martin, M. Carrington, and V. N. KewalRamani. 2004. Raji B cells, misidentified as THP-1 cells, stimulate DC-SIGN-mediated HIV transmission. Virology 318:17-23. [DOI] [PubMed] [Google Scholar]