Abstract
Cytotoxic T lymphocytes (CTLs) are the most powerful weapon of the immune system to eliminate cells infected by intracellular parasites or tumors. However, very often, escape mechanisms overcome CTL immune surveillance by impairing the classical HLA class I antigen-processing pathway. Here, we describe a strategy for CTL activation based on the ability of Tat to mediate transcellular delivery of viral proteins encompassing HLA class I-restricted epitopes. In this system, the recombinant protein TAT-NpFlu containing the transduction domain of Tat of human immunodeficiency virus type 1 fused to the amino acid region 301 to 498 of the nucleoprotein of influenza A virus is proven to sensitize different human cells to lysis by HLA-B27-restricted, Flu 383-391-specific CTL lines. The fusion protein is processed very effectively, since a comparable biological effect is obtained with an amount of protein between 1 and 2 orders of magnitude lower than that of the synthetic peptide. Interestingly, while part of TAT-NpFlu undergoes fast and productive cleavage, a large amount of it remains intact for up to 24 h. Confocal microscopy shows that TAT-NpFlu accumulates in the trans-Golgi network (TGN), where it starts to be detectable 1 h after transduction. Using TAT-NpFlu mutants and hybrid constructs, we demonstrate that enrichment in the TGN occurs only when the carboxy-terminal region of NpFlu (amino acids 400 to 498) is present. These data disclose an unconventional route for presentation of epitopes restricted for HLA class I molecules.
Cytotoxic T lymphocytes (CTLs) are key cells in immune defense against virus-infected or -transformed cells. However, very often, escape mechanisms overcome CTL immune surveillance by impairing the classical HLA class I antigen-processing pathway (10, 15). There is therefore a great interest in identifying novel routes for presentation on HLA class I molecules to be targeted for immunotherapy.
For a long time it was believed that the strict division of HLA class I and class II processing was necessary to identify different physiological responses of the immune system to endogenous (HLA class I) or internalized (HLA class II) proteins. However, although exogenous proteins are normally excluded from presentation by HLA class I molecules, exceptions to this rule have been described in the last 10 years that indicate the possibility, in given circumstances, of engagement and presentation of exogenous antigens on HLA class I molecules either in vivo or in vitro (16, 26). In this respect, the immunological meaning of exogenous and endogenous processing should be reevaluated, since several examples of CTLs primed in vivo by proteins present in extracellular fluids have been provided previously (27), and a different pathway for exogenous proteins could be a possible explanation. Moreover, this pathway could be the initiating step for CTL activation in antitumoral immunity and, eventually, parasite infections through cross-priming. Two mechanisms have been suggested to explain exogenous peptide loading on HLA class I molecules: the first one links to the classical HLA class I pathway via TAP-endoplasmic reticulum-Golgi, and the second proposes cleavage and encounter with HLA class I molecules in the endosomes (1, 14, 23).
Many reports have described the potentiality of Tat of HIV-1 as a vehicle to introduce macromolecules inside cells (18, 29, 39, 40), and several groups have recently used protein transduction domains to successfully treat subclinical models of human diseases (3, 6, 9). Here, we characterize a novel pathway that leads to an efficient production and presentation of HLA class I epitopes. Our strategy is based on the use of a recombinant protein (TAT-NpFlu) in which the nucleoprotein of influenza A virus (amino acids [aa] 301 to 498), containing the HLA-B*2705-restricted, immunodominant epitope Flu 383-391, was fused to the protein transduction domain of Tat. We show here that this chimeric protein crosses cell membranes and accumulates inside trans-Golgi network (TGN) vesicles, where it is likely to represent a readily available source of antigen for HLA class I-mediated presentation to specific cytotoxic T lymphocytes.
MATERIALS AND METHODS
Cloning and sequencing.
To produce genetic in-frame Tat fusion proteins, DNA sequences encoding viral proteins were inserted into the expression vector pTAT-HA (21). In this vector, the ATG start codon is followed by a six-His tag, the Tat protein transduction domain (YGRKKRRQRRR) flanked by glycine residues, a hemagglutinin (HA) tag, and a multicloning site.
The cDNAs encoding for the nucleoprotein of influenza A virus (NpFlu) and for Ebna3C of Epstein-Barr virus were used as a template to amplify the DNA sequence coding for the amino acid region 301 to 498 of NpFlu and for the amino acid region 204 to 396 of Ebna3C. The following specific primers containing the KpnI (forward primer) and the EcoRI (reverse primer) sites (underlined) were used for PCR amplification: NpFlu forward (5′-CGGGGTACCATAGACCCTTTCAAACTACTT-3′), NpFlu reverse (5′-CCGGAATTCACATTGTCGTACTCTTCTGCATT-3′), Ebna3C forward (5′-CGGGGTACCACTATGTTAACTGCCACATTTG-3′), and Ebna3C reverse (5′-CCGGAATTCATATTGGGATCGATATATGG-3′).
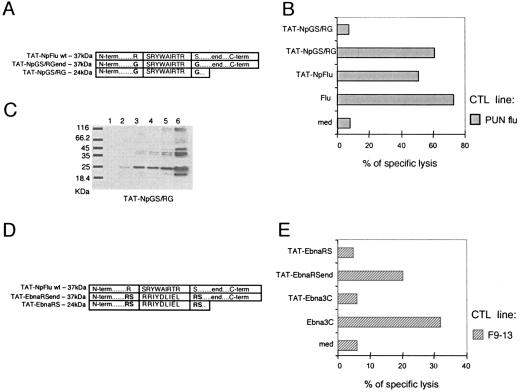
After double digestion with KpnI/EcoRI and purification from an agarose gel, PCR fragments were inserted into pTAT-HA vector. To generate the mutated TAT-NpGS/RGend and TAT-EbnaRSend constructs, a restriction site for XhoI was inserted immediately upstream of the Flu383-391 epitope in TAT-NPFlu vector. Afterward, this construct was double digested with XhoI and Eco0109I located downstream of the epitope (partial digestion), and the mutated sequences (approximately 80 bp) were inserted instead of NpFlu wild-type (wt) sequence. The mutated sequences have been amplified with the following primers, adding XhoI and Eco0109I restriction enzyme sites at the N and C termini, respectively: NPGS/RGend forward (5′-AGTACTCGAGAACTGGGAAGCAGG-3′), NPGS/RSend reverse (5′-CGGAGGCCCTCTGTTGATTAGTGTTTCCTCCACCCCTGG-3′), Ebna RS forward (5′-AGTACTCGAGAACGGTCCCGAAGAATCTATGATTTGATA-3′), and Ebna RS reverse (5′-CGGAGGCCCTCAGAGAGCTACGCAGTTCTATCAAATCATA-3′).
The truncated mutants TATNpGS/RG and TATEbnaRS were obtained from TAT-NpGS/RGend and TAT-EbnaRSend by deleting the C-terminal region of 98 aa (end) through digestion with Eco0109I and religation. TAT-Npshort was obtained by the same procedure starting from TAT-NpFlu. The sequence of all recombinant constructs has been confirmed by using a BigDye Terminator cycle sequencing kit (Applied Biosystems) and an ABI 377 DNA sequencer (Applied Biosystems).
Production, purification, and detection of Tat fusion proteins.
Tat fusion proteins have been produced according to Nagahara et al. (21). The engineered pTAT-HA vectors were used to transform the highly expressing bacterial strain BL21(DE3)pLysS, and different isolates were inoculated overnight in LB/Amp. The following day, 1 liter of LB/Amp was inoculated with overnight cultures and grown for 10 h in presence of 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside). To purify the Tat recombinant proteins, Ni-nitrilotriacetic acid (Ni-NTA) resin has been used according to the protocol provided by QIAGEN. After 24 h of dialysis with 1× phosphate-buffered saline (PBS), the samples were concentrated with an Ultrafree-CL 10 or 30 system (Millipore). The proteins were loaded on a gel for 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by Coomassie blue staining. For Western blotting analysis, after SDS-PAGE, the samples were transferred to nitrocellulose filter and marked with mouse monoclonal immunoglobulin G (IgG) anti-HA (probe F7; Santa Cruz) diluted 1:200 in 1× PBS-0.1% Tween 20-5% dry milk for 1 h. The goat anti-mouse IgG conjugated to horseradish peroxidase (Pierce) was diluted 1:8 × 104 as suggested.
Synthetic peptides.
The synthetic peptides SRYWAIRTR (Flu 383-391) and RRIYDLIEL (Ebna3C 258-266) were prepared using 9-fluorenylmethyoxy carbonyl chemistry. The purity of peptides was always higher than 90%, as estimated by high-pressure liquid chromatography analysis. Peptides were dissolved in dimethyl sulfoxide. Concentrations were assayed by bicinchoninic acid assay (Pierce).
Generation of CTL lines.
Four HLA-B*2705 (Bar, Cuc, Pet, and Pun)-positive donors and one HLA-B*2702 (F9)-positive donor were bled after giving their informed consent. Analysis of the HLA-B27 subtype was performed using an HLA-B27 high-resolution kit (Dynal). Peripheral blood mononuclear cells (PBMCs) from these subjects were isolated on a gradient of Lymphoprep and incubated with Flu 383-391 or with Ebna3C 258-266 (7.5 μM) or in medium alone and screened by use of a gamma interferon enzyme-linked immunospot assay to identify the responders (7). To generate Flu-specific CTL lines, cell suspensions from gamma interferon enzyme-linked immunospot assay-positive wells were recovered and stimulated with antigen-pulsed, γ-irradiated autologous PBMCs and cultured in RPMI 1640 supplemented with 10% heat-inactivated human serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 20 U/ml of human recombinant interleukin-2 (rIL-2) (Roche Applied Science). The phenotype of the T-cell suspension was characterized by indirect immunofluorescence using OKT4 and OKT8 monoclonal antibodies (MAbs) and analyzed with a FACSCaliber instrument (Becton Dickinson). T cells were totally CD8 positive. To obtain CD8+ CTL lines specific for Ebna3C 258-266 from donor F9, PBMCs were stimulated with autologous mature dendritic cells (DCs) and cultured in 10% human serum-RPMI medium with 20 U/ml of human rIL-2. DCs were differentiated from monocyte cells isolated from PBMCs by use of Macs Microbeads CD14+ (Mylteni) and maintained in heat-inactivated 10% fetal calf serum (FCS)-RPMI medium supplemented with 20 ng/ml human recombinant granulocyte-macrophage colony-stimulating factor and 2 ng/ml IL-4 (Eurobio). For maturation, DCs were treated with 100 ng/ml of lipopolysaccharide (Sigma).
Cell lines.
The following cell lines have been used in this study: Epstein-Barr virus-transformed B-lymphoblastoid cell lines (B-LCLs), TAP-deficient T2 cells, and HeLa and HT29 cells, all stably transfected with cDNA encoding B*2705. B-LCLs and T2 and HT29 cells were cultured in 10% FCS-RPMI medium and HeLa cells in 10% FCS-Dulbecco's minimal essential medium. All B*2705 transfectants were maintained in medium containing hygromycin B (200 μg/ml).
Detection of Tat recombinant proteins.
To detect the fusion proteins inside the transduced B-LCLs or HeLa B*2705, 1 × 105 cells were incubated for from 30 min to 24 h with Tat recombinant proteins (1.4 μM), washed extensively with 1× PBS, and lysed in 50 μl of loading buffer (100 mM Tris, 200 mM dithiothreitol, 4% SDS, 20% glycerol) for 15 min at 100°C. Each sample was then subjected to electrophoresis on a 12% SDS-PAGE gel and analyzed by Western blotting using the anti-HA MAb.
Cytotoxicity assays.
The cytotoxic activity of T-cell lines was assessed according to the standard 51Cr release assay. CTL were mixed in 96-well U-bottom plates with 51Cr-labeled target cells for 4 h at 37°C. The effector/target cell ratio was always 20:1. Target cells had been previously incubated overnight (or for 2 h in the case of inhibition assays) with Flu 383-391 (30 μM) or Tat fusion proteins (0.1 μM when not differently indicated). At 1 h before adding the synthetic peptide or the recombinant proteins, brefeldin A (Sigma) was supplied to target cells at 36 μM and it was constantly present at 11 μM during testing (24).
In cell membrane blocking experiments, target cells were fixed with 0.05% glutaraldehyde (Sigma) for 45 s at room temperature and washed with 0.2 M lysine. For HLA-B27 inhibition studies, MAb ME1 (6 μg/ml) was added to peptide-pulsed or Tat-transduced 51Cr-labeled target cells 1 h prior to mixing with effector T cells.
Fluorescence microscopy.
HeLa B*2705 transfectants were grown on coverslips for 30 h and incubated with 0.4 μM Tat fusion proteins. At established times, cells were fixed in 4% paraformaldehyde in 1× PBS for 20 min at 25°C, followed by treatment with 0.1 M glycine in 1× PBS for 15 min at 25°C and with 0.1% Triton X-100 in 1× PBS for an additional 5 min at 25°C to allow permeabilization. For double-staining experiments, cells were incubated for 1 h at 25°C with the following primary antibodies: anti-HA MAb (F7; Santa Cruz Biotechnology) (1:200 in PBS); anti-CD63 (BD Bioscences) (1:200 in PBS), a multivesicular-body marker; anti-giantin (Covance) (1:200 in PBS) for cis and medium Golgi; and anti-Cav-1 (Santa Cruz) (1:200 in PBS) specific for caveolin 1 resident in caveolae. Primary antibodies were visualized, after appropriate washing in 1× PBS, using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Cappel) (1:50 in PBS) or Texas Red-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories) (1:200 in PBS). HLA class I molecules were stained using W6/32-FITC (Sigma) (1:50 in PBS). Specific markers for early (Transferrin-Texas red; Molecular Probes) and late (Lysotracker red; Molecular Probes) endosomes were added to culture medium 10 min and 30 min, respectively, prior to fixation. Rabbit anti-furin polyclonal serum has been kindly provided by Wolfgang Garten (Philipps-Universität Marburg, Germany) (28). Colocalization of fluorescence signals was evaluated by use of confocal vertical (x and z) sections (interval, 0.5 μm) and a Zeiss confocal laser scan microscope (Zeiss, Oberkochen, Germany). The multitrack function has been used to prevent cross-talk between the two signals. Quantitative analysis of the extent of colocalization was performed using a Zeiss KS 300 3.0 image processing system: 50 cells for each condition, randomly taken from three independent experiments, were analyzed. Data are expressed as mean percentage ± standard deviation of colocalization.
RESULTS
TAT-NpFlu fusion protein is correctly processed by cell lines of different origins.
To produce a genetic in-frame Tat fusion protein, we used the bacterial expression vector pTAT-HA (21, 37), in which we cloned the cDNA encoding the amino acidic region 301 to 498 of the nucleoprotein of influenza A virus (NpFlu). This region contains the immunodominant epitope Flu 383-391 (SRYWAIRTR), which is restricted for HLA-B*2705 molecules. The recombinant protein TAT-NpFlu was produced in the BL21(DE3)pLysS bacterial strain and purified by affinity chromatography using the Ni-NTA resin to which the six-His tag is bound.
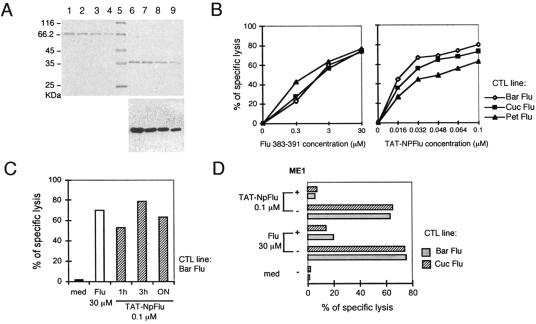
Fig. 1A shows SDS-PAGE results for the purified protein and Western blotting results obtained using the antibody anti-HA that confirmed the predicted molecular mass of 37 kDa (lanes 6 to 9). This recombinant protein was firstly employed to transduce B-lymphoblastoid cell lines. Fig. 1B shows the results obtained with one representative B-LCL (Bar) incubated with increasing doses of TAT-NpFlu recombinant protein (right panel) or with Flu 383-391 peptide (left panel) and used as a target in a standard 51Cr release assay. Three different CTL lines (Bar, Pet, Cuc) generated from unrelated HLA-B27-positive donors were used. The biological effect, measured as a percentage of specific lysis, occurred in a dose-dependent manner and was highly efficient, since less than 0.03 μM (Bar), 0.032 μM (Cuc), and about 0.1 μM (Pet) of TAT-NpFlu evoked a cytotoxic response comparable to that obtained with a concentration of 3 μM of the Flu 383-391 synthetic peptide.
FIG. 1.
Processing of TAT-NpFlu. (A) TAT-NpFlu recombinant protein purified by Ni-NTA resin was loaded on 12% SDS-PAGE and visualized by Coomassie blue staining. Lanes 1 to 4: curve of BSA (0.8, 0.6, 0.4, and 0.2 μg of protein, respectively). Lane 5: protein marker. Lanes 6 to 9: decreasing amounts of TAT-NpFlu. Bottom panel: Western blotting of TAT-NpFlu protein by use of the anti-HA MAb. (B) Lysis of B-LCL (Bar) sensitized with increasing doses of either Flu 383-391 peptide or TAT-NpFlu fusion protein by three different Flu 383-391-specific, HLA-B27-restricted CTL lines. Target cells were incubated overnight with the recombinant protein or with the synthetic peptide. The experiment shown is representative of four. (C) Target cells (B-LCL Bar) were preincubated for 1 h, 3 h, and overnight with TAT-NpFlu recombinant protein before cytotoxicity test was run for 4 h. The results for one of three separate experiments are shown. med, medium. (D) The anti-B27 ME1 MAb inhibits the lysis of B-LCL target cells (Bar) by Flu 383-391-specific CTLs both in cells pulsed with the synthetic peptide and in cells transduced with TAT-NPFlu fusion protein. 51Cr-labeled target cells were incubated for 1 h with ME1 MAb (6 μg/ml) before mixing with effector cells for further 4 h. One of two independent experiments is reported here.
The protein TAT-NpFlu was processed in short time; 1 h of incubation prior to testing was enough to sensitize target cells to an efficient lysis (Fig. 1C). It must, however, be considered that the test proceeds for 4 h, during which processing and expression of HLA class I-peptide complexes may still proceed.
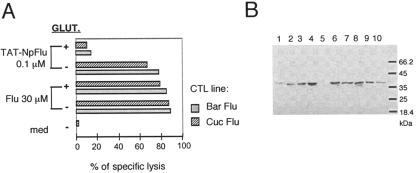
To assure the involvement of HLA-B27 molecules in the presentation of the immunogenic epitope derived from TAT-NpFlu, we used an antibody (ME1) specific for HLA-B27, routinely employed in our laboratory to assay the HLA-B27 restriction since it inhibits the recognition of the complex HLA-B27/Flu by T-cell receptor (5). As shown in Fig. 1D, ME1 antibody was able to block the presentation of the epitope derived from either intracellular processing (TAT-NpFlu) or external binding (synthetic peptide Flu). To formally demonstrate the strict requirement of TAT-NpFlu to enter the target cells to be correctly processed, the fluidity of cell membrane was blocked using 0.05% glutaraldehyde, and subsequently a cytotoxicity assay was performed. As shown in Fig. 2A, glutaraldehyde only prevented the presentation of the epitope derived from TAT-NpFlu, whereas it did not affect presentation of the synthetic peptide Flu 383-391.
FIG. 2.
Uptake and presentation of TAT-NpFlu recombinant protein. (A) 51Chromium release assay using, as targets, the B-LCL Bar, pretreated or not, for 45 min at room temperature, with 0.05% glutaraldehyde (GLUT.), prior to transduction overnight with TAT-NPFlu (0.1 μM) or incubation with Flu 383-391 synthetic peptide (30 μM). The results for one of two experiments are shown. med, medium. (B) Kinetics of uptake of TAT-NpFlu chimera by B-LCL Bar cells. The results of Western blotting with anti-HA MAb of the total protein extract from 1 × 105 B-LCL Bar treated with TAT-NpFlu (1.4 μM) for different times are shown. Lane 1: 30 min; lane 2: 60 min; lane 3: 90 min; lane 4: 2 h; lane 5: 2 h with 0.05% glutaraldehyde added; lane 6: 4 h; lane: 7: 6 h; lane 8: 8 h; lane 9: 12 h; lane10: 24 h. Protein concentration peaked at 2 h. The sample run in lane 5 had been treated with 0.05% glutaraldehyde 1 h prior to addition of TAT-NpFlu.
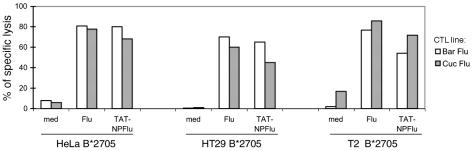
The kinetics of entry of TAT-NpFlu was assessed in B-LCL Bar transduced with the recombinant protein, harvested and lysed at different times, and loaded on an SDS-polyacrylamide gel (Fig. 2B). Western blot analysis indicated that TAT-NpFlu is detectable as early as 30 min and that it is not completely degraded until 24 h. As expected, treatment with glutaraldehyde prevented the entry of the recombinant protein and therefore the presentation of the epitope (lane 5, Fig. 2B) while not affecting that of the synthetic peptide Flu 383-391. To verify whether other cells besides B-LCL were able to produce the epitope Flu 383-391, HeLa, HT29, and T2 cells stably transfected with HLA-B*2705 cDNA were transduced with TAT-NpFlu. The first two cell lines are cancer cells of epithelial origin from cervix and colon, respectively, and T2 cells are hybridoma cells derived from a fusion of B and T lymphocytes with a deletion in the HLA region, therefore lacking components of the antigen presentation machinery, among which are peptide transporters (Tap) (22). Fig. 3 shows that all B*2705-transfected cells are equally sensitized to lysis by nucleoprotein (NP)-Flu-specific CTL lines after TAT-NpFlu protein transduction, indicating that they are all equipped with components necessary for epitope processing and that TAP molecules are not required for this processing route.
FIG. 3.
TAT-NPFlu recombinant protein is productively processed by cells of different origin. The results from a cytotoxicity test in which HLA-B*2705 transfectants (T2, HeLa, and HT29) incubated overnight with TAT-NpFlu (0.1 μM), with Flu 383-391 peptide (30 μM), or in medium (med) alone were used as targets of two Flu 383-391-specific CTL lines (Bar Flu; Cuc Flu) are shown. Results for one experiment representative of several separate experiments are reported here.
Processing of TAT-Ebna3C and mutagenesis of TAT-NpFlu.
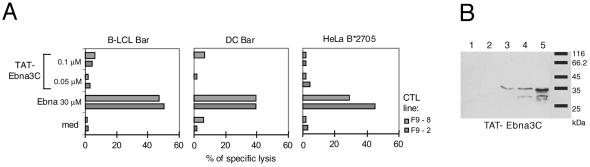
To test the versatility of our system, other Tat chimeric proteins such as the fusion protein TAT-Ebna3C including the amino acid region 204 to 396 of Ebna-3C from Epstein-Barr virus encompassing the HLA-B27-restricted epitope 258-266 (RRIYDLIEL) were produced. The protein was isolated and purified as described above for TAT-NpFlu. Neither B-LCLs nor HeLa B*2705 transduced with ∼0.05 μM or ∼0.1 μM TAT-Ebna3C for one night were able to cleave and present the epitope. We also produced dendritic cells from an HLA-B*2705 donor, but even these cells transduced with TAT-NpFlu did not perform any better, although they were good targets for EBNA3C-specific CTL lines (lines F9-8 and F9-2) when pulsed with the synthetic peptide (Fig. 4A). This failure was not due to defects in the entry of this protein inside the cells; indeed, the transduced protein was detectable after 90 min, reaching a maximum concentration after 4 h (Fig. 4B).
FIG. 4.
Failure of TAT-EBNA3C fusion protein to sensitize transduced cells to cytolysis. (A) B-LCL Bar, dendritic cells from the same donor Bar, and HeLa B*2705 cells were incubated overnight at two concentrations (0.05 μM and 0.1 μM) of the recombinant protein TAT-Ebna3C and used as targets for Ebna3C-specific CTL lines. (B) Western blot with anti-HA MAb of the total cell lysate from HeLa B*2705 incubated with TAT-Ebna3C (1.4 μM) and harvested at 30 min (lane 1); 60 min (lane 2); 90 min (lane 3); 2 h (lane 4); and 4 h (lane 5) after protein transduction.
To investigate why the recombinant protein TAT-NpFlu was productively processed while TAT-Ebna3C was not, we changed the two twin sequences Arg/Ser flanking the epitope Flu 383-391 (Fig. 5A) on the supposition that they could be the target motif for a protease releasing the epitope Flu 383-391. Therefore, the Arg382 and the Ser392 were both mutated to Gly to create the protein TAT-NpGS/RGend (Fig. 5A). This protein was processed by HeLa B*2705 cells with the same efficiency as the wt (TAT-NpFlu), as shown by the results of the 51Cr release assay presented in Fig. 5B. Therefore, these data excluded the involvement of a putative protease, recognizing the Arg/Ser sequence, as responsible for the correct processing of the protein TAT-NpFlu. Starting from this chimeric protein, we made a deletion mutant by taking away the C terminus to generate the truncated protein (TAT-NpGS/RG; 24 kDa) spanning aa 301 to 400 (Fig. 5A). Target cells transduced with TAT-NpGS/RG were not lysed by Flu-specific CTLs (Fig. 5B) even when the deletion mutant protein was supplied at higher doses (data not shown). However, this truncated form, the only one among the TAT-NpFlu chimeras not productively cleaved, does transduce cells and, like TAT-NpFlu wt (Fig. 2B), reaches the highest concentration after 90 min (Fig. 5C). These experiments demonstrated that the presence of the C-terminal region of these TAT-NpFlu constructs is an essential requisite for a correct release of the Flu 383-391 epitope. To further support these data, we created hybrid constructs between NpFlu and Ebna3C. In particular, we replaced the sequence of the Flu 383-391 epitope with that of Ebna3C 258-266 in the context of the protein TAT-NpFlu, and from this hybrid we produced both the long and the C-terminal deletion forms (TAT-EbnaRSend and TAT-EbnaRS, respectively; see Fig. 5D). The ability of target cells to correctly process these proteins and to present the epitope Ebna3C 258-266 was tested in a standard cytotoxicity assay. The results presented in Fig. 5E confirm the requirement of the C-terminal end, since among the three constructs, TAT-Ebna3C and the two hybrid forms, TAT-EbnaRS and TAT-EbnaRSend, only the last was able to trigger the effector functions of a Ebna3C 258-266-specific CTL line.
FIG. 5.
Analysis of TAT-NpFlu mutants and hybrids. (A) Schematic representation of TAT-NpGS/RGend (37 kDa) obtained from TAT-NpFlu wt by mutation of Arg382 and Ser392 to Gly and TAT-NpGS/RGend (24 kDa) generated from the latter by deleting 98 amino acids from the carboxyl terminus (end). (B) 51Cr release assay in which HeLa B*2705 cells, transduced overnight with TAT-NpFlu or with the mutant proteins TAT-NpGS/RGend and TAT-NpGS/RG at 0.1 μM, were used as targets for the Pun Flu CTL line. One representative experiment of two is shown. med, medium. (C) Western Blot analysis to assess the entry of TAT-NpGS/RG into HeLa B*2705 cells. Cells were incubated with 1.4 μM of the mutant protein, and the cell lysate was harvested after 30 min (lane 1); 60 min (lane 2); 90 min (lane 3); 2 h (lane 4); and 4 h (lane 5). Lane 6: 0.3 μM of TAT-NpGS/RG protein. The mutant protein is detectable inside the cells 60 min after transduction. (D) Schematic representation of the NpFlu/Ebna3C hybrid proteins. TAT-EbnaRSend derives from TAT-NpFlu wt by replacement of the sequence encoding Flu 383-391 with that of Ebna3C 258-266. The double motif Arg/Ser was maintained at the N and C terminus of the epitope as described for the TAT-NpFlu wt. TAT-EbnaRS is identical to TAT-EbnaRSend but lacks the C-terminal region (end). (E) Cytotoxicity assay with an Ebna3C-specific CTL line (F9-13) as effector and HeLa B*2705 cells as target cells incubated overnight with three different TAT fusion proteins at 0.1 μM concentration.
Confocal microscopy analysis.
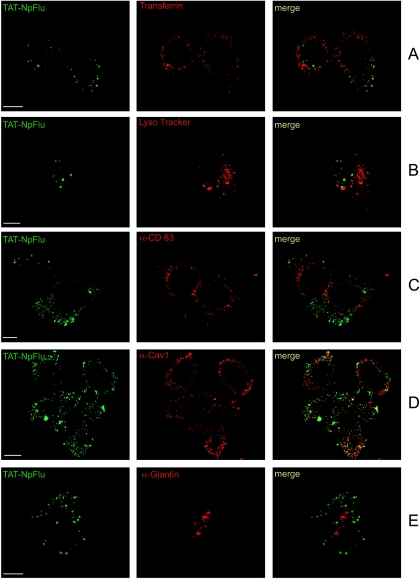
The intracellular localization of TAT-NpFlu was assessed in HeLa B*2705 cells. To enter the cell, Tat must first bind, on the cell surface, to anionic charges like heparin sulfate proteoglycans (34) and then flip inside the cell. Accordingly, treatment of cells with heparin (50 μg/ml) 1 h prior addition of TAT-NpFlu completely prevented TAT-NpFlu cell entry (data not shown). To identify the intracellular compartments where the TAT-NpFlu protein was localized, HeLa B*2705 cells were treated with the fusion protein and assayed at different times from 15 min to 3 h. However, at the 15-min and 30-min time points we were not able to trace the protein inside the cells, which is consistent with the biochemical data previously shown (Fig. 2B). Therefore, all confocal microscopy studies were performed in cells incubated with TAT-NpFlu for 1 h up to 3 h, which gave the same results. Fig. 6 and 7 present the results for the time point at 2 h for each experiment. We used transferrin as a marker for the early endosomes and Lyso Tracker for the late endosomes, and the lysosomes were added 10 min and 30 min, respectively, before fixing the cells. In both cases, the protein TAT-NpFlu did not colocalize with these markers at up to 3 h (Fig. 6A and B). Likewise, the chimeric TAT protein did not colocalize with the tetraspanin CD63, a marker for multivesicular bodies and lysosomes (Fig. 6C). Analysis of the caveolae (α-Cav-1; Fig. 6D) and of the cis and medium Golgi (α-Giantin; Fig. 6E) shows that TAT-NpFlu was not detectable inside these compartments. A widespread colocalization of the recombinant protein was found with the protease furin (Fig. 7). This protease, a human subtilisin-related proprotein convertase, has ubiquitous cell expression, with a net prevalence in the TGN (32).
FIG. 6.
Confocal analysis of the distribution of TAT-NpFlu chimeric protein. TAT-NpFlu fusion protein is visualized in green (left panels) and cellular compartments are depicted in red (middle panels); the overlays are shown on the right. (A) Transferrin, the marker for early endosomes. (B) Lysotracker, marker for late endosomes and lysosomes. (C) Anti-CD-63 for lysosomes and multivesicular bodies. (D) Anti-caveolin-1 (Cav-1) for caveolar compartments. (E) Anti-Giantin for cis and medium Golgi. TAT-NpFlu is marked with FITC-conjugated secondary antibody. Bars, 10 μm.
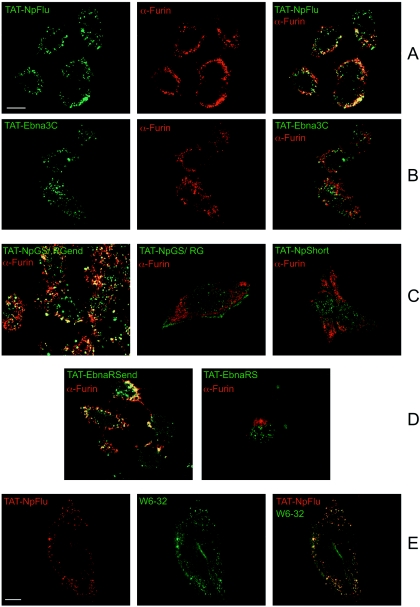
FIG. 7.
Tat fusion proteins and localization in the TGN. (A) Full colocalization, visible as yellow (right panel), of the TAT-NpFlu protein depicted in green (left panel) with the protease Furin mainly resident in TGN and visualized in red (middle panel). (B) TAT-Ebna3C shows only a partial colocalization with furin (right panel). (C) Merge of the furin with TAT-NpGS/RGend (left panel), with TAT-NpGS/RG (middle panel), and with TAT-Np-short (right panel) fusion proteins. (D) Merge of the furin with TAT-EbnaRSend (left panel) and with its truncated counterpart TAT-EbnaRS (right panel). Panels C and D show that C-terminal deletion mutants do not accumulate in the TGN. (E) TAT-NpFlu colocalizes with HLA class I molecules (marked with W6/32-FITC). Bars, 10 μm.
The experiment was then repeated with the other constructs. Fig. 7 shows the merging of the furin with these chimeric proteins. Clearly, functional data correlate with the localization of the proteins: only those constructs possessing the C terminus (TAT-Np-Flu; TAT-GS/RG-end; TAT-EbnaRS-end) accumulate in the TGN, whereas for the others (TAT-GS/RG; TAT-EbnaRS; TAT-NpShort) no colocalization was detectable. TAT-EBNA3C, which was not productively processed, was present in the TGN, although at a lower amount (21.42 ± 5.5%, P < 0.001, versus 74.13 ± 5.3%, P < 0.001, for TAT-NpFlu). As expected, a clear colocalization was also found with W6/32 monoclonal antibody, which recognizes HLA class I molecules (25).
Brefeldin A inhibits epitope presentation.
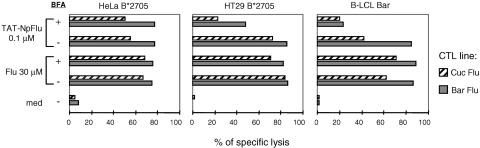
Since the TAT-NpFlu protein seems to accumulate in the TGN, where it is likely to encounter the HLA class I molecules, we asked whether this process could be inhibited by brefeldin A, which has been reported to be a very potent Golgi disassembler: it disrupts the egress of immune complexes from the endoplasmic reticulum to the Golgi, preventing access to the plasma membrane and thus inhibiting HLA class I-mediated presentation (30). Interestingly, the processing of TAT-NpFlu was highly inhibited by the use of this drug in B-LCLs and HT29B*2705 (inhibition ranging from 40% to 75%), whereas in HeLa cells expressing B*2705, processing was not affected (Fig. 8) even at higher doses (72 μM instead of 36 μM; data not shown). These data suggest that the epitope NP-Flu 383-391, derived from the processing of the chimeric protein in lymphocytes and epithelial cells, is mainly loaded on de novo-synthesized HLA molecules. The lack of inhibition by brefeldin A in HeLa cells suggests, however, that recycling HLA class I molecules can also take part in this process.
FIG. 8.
Effect of brefeldin on TAT-NPFlu processing. 51Chromium-release assay using as target cells HeLa B*2705, HT29 B*2705, and B-LCL Bar not pretreated or pretreated for 1 h with brefeldin A (37 μM) prior to incubation for 2 h with TAT-NpFlu (0.1 μM) or Flu 383-391 peptide (30 μM). Two Flu 383-391-specific CTL lines (Bar Flu and Cuc Flu) have been employed as effector cells. The experiment reported here is representative of three.
DISCUSSION
In this paper we describe an unconventional cellular route followed by proteins containing HLA class I-restricted epitopes by use of the Tat transduction domain and containing the C terminus of the nucleoprotein of influenza A virus. These proteins are transported into furin-rich TGN vesicles, where the epitope is presumably cleaved and encounters the HLA class I molecules. Although this pathway is artificially induced, it has a natural counterpart since the sequence that makes this process highly effective derives from a viral protein, the nucleoprotein of influenza A virus, which, however, has been depleted of the major nuclear localization signal (4), thus probably subverting or forcing its intracellular trafficking. Accordingly, the contribution of this pathway to the immunological response against this specific immunodominant epitope during a viral infection cannot be assessed here. We demonstrate, however, that the exogenously introduced nucleoprotein can be processed and that epitopes from its own or other protein sequences, which are not processed this way in their natural context, could be successfully presented to the immune system.
The use of the Tat transduction domain allows the introduction of the exogenous proteins in a very efficient way. In particular, a chimeric protein, TAT-NpFlu, in which a region of the C terminus of the nucleoprotein of influenza A virus (aa 301 to 498), containing the HLA B*2705-restricted epitope Flu 383-391 (SRYWAIRTR) has been fused with the transduction domain of TAT, turns out to be an excellent activator for specific CTLs. Interestingly, CTLs are able to lyse the transduced cells as early as few hours after protein entry, suggesting that the chimeric protein is processed very effectively. An apparent contradiction is the fact that this same protein is stored within the cell for at least 24 h. In agreement with our findings, others have reported the presence of TAT-transduced proteins inside the cells as long as for 20 h (8). These data may be explained by considering that few HLA-peptide complexes need to be engaged by T-cell receptors to trigger cytolysis and that they may be stable for several hours (35, 36). Therefore, it is possible that the amount of the protein which is processed is little in any case, either because part of the protein forms aggregates that hampers the cleavage or because of a low efficiency of the specific proteases. Nevertheless, the protein stored in the TGN could represent a reservoir of antigen that can be processed through time. In any case, the transduced protein was by no means toxic (cells were healthy and duplicated regularly).
The entrance of the protein requires a fluid membrane, as shown by the inhibition induced by treatment with glutaraldehyde, ruling out the possibility that the epitope is generated by serum proteases. Interestingly, HeLa and HT29 tumor cells transfected with HLA-B*2705 were as competent as HLA-B27-positive B-LCL to process the recombinant TAT protein, showing that cells of different origin are equipped with the proteases necessary for antigen trimming through this route. The process is inhibited by brefeldin A at least in B-LCL and HT29 but not in HeLa cells. This may indicate that the first two cell lines make use of nascent rather than recycling HLA class I molecules to present the epitope whereas HeLa cells do the opposite. HeLa cells are highly selected tumor cells which may have some defects in HLA class I trafficking. In this context, a recent report has shown that HeLa cells are refractory to the Nef-mediated disruption of HLA transport to the cell surface of newly synthesized HLA class I molecules but not of the recycling molecules (17). It is therefore conceivable that epitope presentation by HeLa cells is mainly dependent on recycling of HLA class I molecules. These molecules, either de novo synthesized or internalized from cell membrane, are supposed to gain access to the TGN already complexed with peptides: therefore, the efficiency of presentation may depend on the extent to which lower-affinity peptides can be replaced by Flu epitope (20). Imaging experiments indicate that after 1 h TAT-NpFlu accumulates in the TGN. Before this time, we were not able to detect the protein inside the cells; therefore, we do not know whether the protein gains access to the TGN after trafficking through the endosomes or other vesicles or directly from the cytoplasm. In the TGN the epitope is likely to be released from the protein by unknown specific enzymes. Interestingly, a recent work exploiting a Tat transducing system to introduce multiple epitopes inside target cells has shown that a more efficient processing was obtained when furin-sensitive linkers were introduced in the region flanking the epitopes (19). Therefore, as probably in our case, antigen trimming occurs in the TGN where furin is mainly resident (11, 12, 33). However, Flu epitope is not surrounded by furin consensus motifs; therefore, a direct role of this enzyme can be excluded even though an indirect action, i.e., activating the proper enzyme, is still possible.
A leading role in this process appears to be played by the C terminus of the NP, certainly in the localization of the protein in the TGN, since those constructs lacking the C terminus do not colocalize with furin although entering the cell. TAT-EBNA3C which enters the cell appears to be present in TGN in a smaller amount compared with TAT-NpFlu; however, it is not processed, even when target cells are incubated overnight. The EBNA3C epitope becomes readily available for CTL recognition when inserted into the context of the nucleoprotein, thus suggesting the presence in the C terminus of sequences relevant for localization and/or processing by resident proteases. It has been already reported that the epitope 383-391 from the nucleoprotein of influenza A virus can be processed and presented in TAP-deficient cells (38). Moreover, other epitopes carried by this protein appear to follow different routes for processing (13). The delivery of internalized HLA class I molecules to the TGN is also well documented, and this route is targeted by some viral proteins to escape immune surveillance (2). Therefore, our observations are in agreement with evidence that points to the TGN as a site of intense processing.
Published data have already underlined the potentiality of Tat as a vehicle for tumor-associated antigens and in vaccine design against tumors (31, 41). Here, we provide an example of how an alternative delivery route may be exploited by fusing Tat transduction domain to the C terminus of the influenza virus nucleoprotein into which HLA class I immunodominant epitopes of other proteins can be inserted and processed. Although some details of this route, i.e., which proteases are responsible for the generation of the correct epitopes, still need to be elucidated, nevertheless, the system is highly efficient and specific, lending itself to exploitation in vaccines.
Acknowledgments
This work was supported by grants from Associazione Italiana Ricerca Cancro (AIRC) to R. Sorrentino and to M. R. Torrisi, from Ministero Istruzione, Università e Ricerca (MIUR) to M. R. Torrisi, and from MIUR, Fondo Investimenti Ricerca di Base (FIRB) and Associazione Italiana Sclerosi Multipla to R. Sorrentino.
We thank Luigi Finocchi at the University of Rome “La Sapienza” for the synthesis of peptides and W. Garten at the Institut fur Virologie der Philipps-Universitat Marburg, Marburg, Germany, for the generous provision of anti-furin serum. Acknowledgments are also given to all members of the laboratory for fruitful discussions and to Federica Lucantoni for technical assistance.
REFERENCES
- 1.Ackerman, A. L., and P. Cresswell. 2004. Cellular mechanisms governing cross-presentation of exogenous antigens. Nat. Immunol. 5:678-684. [DOI] [PubMed] [Google Scholar]
- 2.Blagoveshchenskaya, A. D., L. Thomas, S. F. Feliciangeli, C. H. Hung, and G. Thomas. 2002. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111:853-866. [DOI] [PubMed] [Google Scholar]
- 3.Cao, G., W. Pei, H. Ge, Q. Liang, Y. Luo, F. R. Sharp, A. Lu, R. Ran, S. H. Graham, and J. Chen. 2002. In vivo delivery of a Bcl-xL fusion protein containing the TAT protein transduction domain protects against ischemic brain injury and neuronal apoptosis. J. Neurosci. 22:5423-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cros, J. F., A. Garcìa-Sastre, and P. Palese. 2005. An unconventional NLS is critical for the nuclear import of the influenza A virus nucleoprotein and ribonucleoprotein. Traffic 6:205-213. [DOI] [PubMed] [Google Scholar]
- 5.Del Porto, P., M. D'Amato, M. T. Fiorillo, L. Tuosto, E. Piccolella, and R. Sorrentino. 1994. Identification of a novel HLA-B27 subtype by restriction analysis of a cytotoxic γδ T cell clone. J. Immunol. 153:3093-3100. [PubMed] [Google Scholar]
- 6.Dietz, G. P., E. Kilic, and M. Bahr. 2002. Inhibition of neuronal apoptosis in vitro and in vivo using TAT-mediated protein transduction. Mol. Cell. Neurosci. 21:29-37. [DOI] [PubMed] [Google Scholar]
- 7.Fiorillo, M. T., M. Maragno, R. Butler, M. L. Dupuis, and R. Sorrentino. 2000. CD8+ T cell autoreactivity to an HLA-B27-restricted self epitope correlates with ankylosing spondylitis. J. Clin. Investig. 106:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fittipaldi, A., A. Ferrari, M. Zoppé, C. Arcangeli, V. Pellegrini, F. Beltram, and M. Giacca. 2003. Cell membrane lipid rafts mediate caveolar endocytosis of HIV-1 Tat fusion proteins. J. Biol. Chem. 278:34141-34149. [DOI] [PubMed] [Google Scholar]
- 9.Fu, A. L., Q. Li, Z. H. Dong, S. J. Huang, Y. X. Wang, and M. J. Sun. 2004. Alternative therapy of Alzheimer's disease via supplementation with colin acetyltransferase. Neurosci. Lett. 368:258-262. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Lora, A., I. Algarra, A. Collado, and F. Garrido. 2003. Tumour immunology, vaccination and escape strategies. Eur. J. Immunogenet. 30:177-183. [DOI] [PubMed] [Google Scholar]
- 11.Gil-Torregosa, B. C., A. R. Castano, and M. Del Val. 1998. Major histocompatibility complex class I viral antigen processing in the secretory pathway defined by the trans-Golgi network protease furin. J. Exp. Med. 188:1105-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil-Torregosa, B. C., A. R. Castano, D. Lopez, and M. Del Val. 2000. Generation of MHC class I peptide antigens by protein processing in the secretory route by furin. Traffic 1:641-651. [DOI] [PubMed] [Google Scholar]
- 13.Golovina, T. N., E. J. Wherry, T. N. J. Bullock, and L. C. Eisenlohr. 2002. Efficient and qualitatively distinct MHC class I-restricted presentation of antigen targeted to the endoplasmic reticulum. J. Immunol. 168:2667-2675. [DOI] [PubMed] [Google Scholar]
- 14.Gromme, M., and J. Neefjes. 2002. Antigen degradation or presentation by MHC class I molecules via classical and non-classical pathways. Mol. Immun. 39:181-202. [DOI] [PubMed] [Google Scholar]
- 15.Hewitt, E. 2003. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jondal, M., R. Schirmbeck, and J. Reimann. 1996. MHC class I-restricted CTL responses to exogenous antigens. Immunity 5:295-302. [DOI] [PubMed] [Google Scholar]
- 17.Kasper, M. R., and K. L. Collins. 2003. Nef-mediated disruption of HLA-A2 transport to the cell surface in T cells. J. Virol. 77:3041-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsay, M. A. 2002. Peptide mediated cell delivery: application in protein target validation. Curr. Opin. Pharmacol. 2:587-594. [DOI] [PubMed] [Google Scholar]
- 19.Lu, J., Y. Higashimoto, E. Appella, and E. Celis. 2004. Multiepitope Trojan antigen peptide vaccines for the induction of antitumor CTL and Th immune responses. J. Immunol. 172:4575-4582. [DOI] [PubMed] [Google Scholar]
- 20.Luft, T., M. Rizkalla, T. Y Tai, Q. Chen, R. I. MacFarlan, I. D. Davis, E. Maraskovsky, and J. Cebon. 2001. Exogenous peptides presented by Transporter Associated with Antigen Processing (TAP)-deficient and TAP-competent cells: intracellular loading and kinetics of presentation. J. Immunol. 167:2529-2537. [DOI] [PubMed] [Google Scholar]
- 21.Nagahara, H., A. M. Vocero-Akbani, E. L. Snyder, A. Ho, D. G. Latham, N. A. Lissy, M. Becker-Hapak, S. A. Ezhevsky, and S. F. Dowdy. 1998. Transduction of full-length TAT-p27Kip1 induces cell migration. Nat. Med. 4:1449-1452. [DOI] [PubMed] [Google Scholar]
- 22.Nijman, H. W., J. G. Houbiers, M. P. Vierboom, S. H. van der Burg, J. W. Drijfhout, J. D'Amaro, P. Kenemans, C. J. Melief, and W. M. Kast. 1993. Identification of peptide sequences that potentially trigger HLA-A2.1-restricted cytotoxic T lymphocytes. Eur. J. Immunol. 23:1215-1219. [DOI] [PubMed] [Google Scholar]
- 23.Norbury, C. C., S. Basta, K. B. Donohue, D. C. Tscharke, M. F. Princiotta, P. Berglund, J. Gibbs, J. R. Bennink, and J. W. Yewdell. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates Science 304:1318-1321. [DOI] [PubMed] [Google Scholar]
- 24.Nuchtern, J. G., J. S. Bonifacino, W. E. Biddison, and R. D. Klausner. 1989. Brefeldin A implicates egress from endoplasmic reticulum in class I restricted antigen presentation. Nature 339:223-226. [DOI] [PubMed] [Google Scholar]
- 25.Parham, P., C. J. Barnstable, and W. F. Bodmer. 1979. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C, antigens. J. Immunol. 123:342-349. [PubMed] [Google Scholar]
- 26.Rock, K. L. 1996. A new foreign policy: MHC class I molecules monitor the outside world. Immunol. Today 17:131-137. [DOI] [PubMed] [Google Scholar]
- 27.Rock, K. L., and A. L. Goldberg. 1999. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 17:739-779. [DOI] [PubMed] [Google Scholar]
- 28.Schafer, W., A. Stroh, S. Berghofer, J. Seiler, M. Vey, M. L. Kruse, H. F. Kern, H. D. Klenk, and W. Garten. 1995. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 14:2424-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarze, S. R., A. Ho, A. N. Vocero-Akbani, and S. F. Dowdy. 1999. In vivo protein transduction: delivery of a biological active protein into the mouse. Science 285:1569-1572. [DOI] [PubMed] [Google Scholar]
- 30.SenGupta, D., P. J. Norris, T. J. Suscovich, M. Hassan-Zahraee, H. F. Moffett, A. Trocha, R. Draenert, P. J. Goulder, R. J. Binder, D. L. Levey, B. D. Walker, P. K. Srivastava, and C. Brander. 2004. Heat shock protein-mediated cross-presentation of exogenous HIV antigen on HLA class I and class II. J. Immunol. 173:1987-1993. [DOI] [PubMed] [Google Scholar]
- 31.Shibagaki, N., and M. C. Udey. 2003. Dendritic cells transduced with Tat protein transduction domain-containing tyrosinase-related protein 2 vaccinate against murine melanoma. Eur. J. Immunol. 33:850-860. [DOI] [PubMed] [Google Scholar]
- 32.Teuchert, M., S. Berghofer, H. D. Klenk, and W. Garten. 1999. Recycling of furin from the plasma membrane. Functional importance of the cytoplasmic tail sorting signals and interaction with the AP-2 adaptor medium chain subunit. J. Biol. Chem. 274:36781-36789. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyagi, M., M. Rusnati, M. Presta, and M. Giacca. 2000. Internalization of Hiv-1 Tat requires cell surface heparan sulfate proteoglicans. J. Biol. Chem. 276:3254-3261. [DOI] [PubMed] [Google Scholar]
- 35.Valitutti, S., S. Muller, M. Dessing, and A. Lanzavecchia. 1996. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J. Exp. Med. 183:1917-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Burg, S. H., M. J. W. Visseren, R. M. Brandt, W. M. Kast, and C. J. M. Melief. 1996. Immunogenicity of peptides bound to MHC class I molecules depends on the MHC-peptide complex stability. J. Immunol. 156:3308-3314. [PubMed] [Google Scholar]
- 37.Vocero-Akbani, A., N. Lissy, and S. F. Dowdy. 2000. Transduction of full-length Tat fusion proteins directly into mammalian cells: analysis of the T cell receptor activation-induced cell death. Methods Enzymol. 322:508-521. [DOI] [PubMed] [Google Scholar]
- 38.Voeten, J. T., G. F. Rimmelzwaan, N. J. Nieuwkoop, R. A. Fouchier, and A. D. Osterhaus. 2001. Antigen processing for MHC class I restricted presentation of exogenous influenza A virus nucleoprotein by B-lymphoblastoid cells. Clin. Exp. Immunol. 125:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wadia, J. S., and S. F. Dowdy. 2002. Protein transduction technology. Curr. Opin. Biotechnol. 13:52-56. [DOI] [PubMed] [Google Scholar]
- 40.Wadia, J. S., R. V. Stan, and S. F. Dowdy. 2004. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 10:310-315. [DOI] [PubMed] [Google Scholar]
- 41.Wang, H. Y., T. Fu, G. Wang, G. Zeng, D. M. Perry-Lalley, J. C. Yang, N. P. Restifo, P. Hwu, and R. F. Wang. 2002. Induction of CD4(+) T cell-dependent antitumor immunity by TAT-mediated tumor antigen delivery into dendritic cells. J. Clin. Investig. 109:1463-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]