Abstract
Vaccinia virus encodes an enzyme with DNA modifying activity that cleaves and inefficiently cross-links cruciformic DNA. This enzyme is contained within the virion, expressed at late times postinfection, and processes DNA in an energy-independent, Mg2+ ion-independent manner. Viral nuclease activity was measured in extracts from cells infected with well-defined viral mutants. Since some viral extracts lacked nuclease activity, the gene encoding the activity was postulated to be one of the open reading frames absent in the viruses lacking activity. Inducible expression of each candidate open reading frame revealed that only the gene VACWR035, or K4L, was required for nuclease activity. A recombinant virus missing only the open reading frame for K4L lacked nuclease activity. Extracts from a recombinant virus expressing K4L linked to a FLAG polypeptide were able to cleave and cross-link cruciformic DNA. There were no significant differences between the virus lacking K4L and wild-type vaccinia virus WR with respect to infectivity, growth characteristics, or processing of viral replicative intermediate DNA, including both telomeric and cross-linked forms. Purification of the K4L FLAG polypeptide expressed in bacteria yielded protein containing nicking-joining activity, implying that K4L is the only vaccinia virus protein required for the nicking-joining enzymatic activity.
Viruses have evolved numerous strategies for the selective replication of their genomes in the infected cell. Vaccinia virus contains a double-stranded DNA genome with covalently continuous hairpin termini (22). The replication of the poxvirus genome requires two distinct processes using different sets of viral genes (20). The initial stage, the synthetic phase, requires members of the early gene class, including a virus-encoded DNA polymerase complex, for the rapid replication of viral DNA in the cytoplasm. The mechanism for initiation of DNA synthesis remains unknown, as DNA replication occurs in the absence of a typical origin of replication (5, 21). The most likely mechanism for initiation of DNA replication takes advantage of the structure of the genomic termini by the introduction of a single-stranded nick near the terminus followed by strand displacement synthesis to copy the terminal hairpin. The resulting double-stranded copy of the hairpin can rearrange to form two self-complementary hairpins, one of which contains the necessary 3′ OH for the elongation strand displacement reaction to copy the genome. However, the synthesis phase of DNA replication does not generate simple catenated genomes. The replicative intermediates of DNA replication are complicated multiple-branched DNA molecules, with the hairpin represented as double-stranded concatemer junctions (2, 3, 20, 23). These structures may arise from a combination of strand elongation synthesis and strand invasion utilizing both DNA replication and recombination. The vaccinia virus DNA polymerase can promote strand annealing in vitro (30), and the gene products required for recombination in vaccinia virus are members of the early class of viral genes (16).
The second stage of poxvirus DNA replication, the processing phase, entails the conversion of the DNA replicative intermediates generated by the synthetic phase of DNA replication into unit-length genomes. This phase requires members of the late class of viral genes and/or late transcription (3, 20). Two types of problems must be solved to generate the unit-length genomes. The hairpin termini must be generated from the double-stranded concatemer junction. The regeneration of the genomic termini requires a pair of specific nucleotide sequences arranged as an inverted repeat in the concatemer junction and involves strand exchange (4, 17, 19). The concatemer junction can be organized as a Holliday junction and resolved into hairpin termini by strand exchange and branch migration. The vaccinia virus WR protein encoded by A22R can process Holliday junction in vitro and is required for efficient telomere processing in vaccinia virus infections (11, 12). In addition to the processing of the genomic termini, the branch points in the cross-linked replicative intermediate must be resolved by an activity which can cleave and rejoin DNA strands. Since these branch points are distributed across the genome, the processing of these intermediates most likely requires a process that is structure, but not sequence, specific.
An enzymatic activity that cleaves and rejoins DNA strands, the nicking-joining enzyme, was detected in vaccinia virus virions or infected cell extracts (14, 18, 25). The cleavage and rejoining of DNA strands in supercoiled plasmids was confined to areas with single-stranded regions of DNA. Some of the linear molecules were cross-linked at their termini, generating covalently continuous molecules with a hairpin end. This cleavage and rejoining activity is consistent with enzymatic properties necessary for the processing of concatemeric replicative to unit-length genomes.
In this report we describe the identification of the vaccinia virus WR open reading frame encoding the virion nicking-joining enzyme. Using a combination of extracts from cells infected with poxviruses and transfection with plasmids containing candidate open reading frames, the VACWR035, or K4L gene, was identified as the gene encoding the viral nuclease activity. The protein encoded by the K4L open reading frame expressed and purified from bacteria contained both nuclease and rejoining activities and, thus, is the only viral protein required for the nicking-closing activity. However, the nuclease activity encoded by K4L is not required for viral growth in tissue culture or in mice. Therefore, this gene is not associated with DNA replicative intermediate processing, unless the virus also encodes redundant processes that can substitute for its activity.
MATERIALS AND METHODS
Cells and viruses.
BS-C-1 cells (ATCC CCL6) were maintained in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, and 50 μg/ml gentamicin. BOS cells, a BS-C-1-derived cell line expressing the bacteriophage T7 RNA polymerase, were obtained from the laboratory of Bernard Moss, National Institute of Allergy and Infectious Diseases, NIH, and were maintained in complete MEM, 10% FBS, supplemented with 1 mg/ml G418 (Sigma). The thymidine kinase-deficient cells 143B (ATCC CRL8303) were maintained in MEM, 10% FBS, supplemented with 50 μg/ml bromodeoxyuridine (BrdU). Cells were incubated in a humidified air-5% CO2 atmosphere at 37°C. Viral titers were determined by standard plaque assay on BS-C-1 cells (9). Preparations of purified virus were made by centrifugation through a 36% sucrose cushion (8).
Virion extract.
The virion extract containing the viral nicking-joining enzyme was prepared as described by Gross and Shuman (13) with some modifications. Virus was purified by centrifugation over a 36% sucrose cushion (9), and 2.5 optical density units (at 260 nm) were resuspended in 800 μl of buffer A (50 mM Tris-HCl, pH 8.0, 10 mM dithiothreitol [DTT], 0.05% NP-40) and incubated on ice for 10 min. The sample was spun in a Beckman Microfuge at 13,000 rpm for 5 min. The pellet was resuspended in 150 μl of 300 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1 mM EDTA, 50 mM DTT, 0.1% sodium deoxycholate and incubated on ice for 30 min. The sample was centrifuged in a Beckman Microfuge at 13,000 rpm for 5 min, and the supernatant was applied to an Ultrafree-MC DEAE centrifugal filter device (Millipore) and centrifuged for 1 minute at 5,000 × g. The device was preequilibrated by the addition of 400 μl of 300 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1 mM EDTA, 50 mM DTT, 0.1% sodium deoxycholate and centrifugation at 5,000 × g for 1 minute. The eluant was stored at −20°C after the addition of a one-quarter volume (37.5 μl) of 200 mM Tris-HCl, pH 8.0, 8 mM DTT, 4 mM EDTA, 40% glycerol.
Nuclease assay.
The nuclease assay reaction follows the conversion of supercoiled plasmid to nicked circular and linear forms by agarose gel electrophoresis as described previously (18). The DNA substrate for the reactions was pECHC (19), a plasmid containing a copy of the duplex concatemer junction of vaccinia virus. This plasmid, when supercoiled, will extrude a cruciform. A reaction volume of 50 μl containing 1 μg of pECHC and 1 μl of extract in 10 mM 2-(N-morpholino)-ethanesulfonic acid, pH 6.5, 10 mM EDTA, 100 μg/ml bovine serum albumin was incubated at 55°C for 30 min. The reaction mixtures were cooled to room temperature and extracted three times with an equal volume of phenol, phenol-chloroform, and finally chloroform. Samples were mixed with DNA agarose dye buffer and separated by electrophoresis through neutral or alkaline agarose gels (18).
Plasmids and recombinant viruses.
The plasmid pECHC used as the substrate for the nuclease assay contains concatemer junction of vaccinia virus WR (19). The plasmid pT7K4LFLAG contains the open reading frame for K4L in the FLAG peptide expression plasmid pPG552. The plasmid pPG552 was generated by ligating the annealed oligonucleotides MM552 (CATGGACTACAAAGACGATGACGACAAGCATATGAGCT) and MM553 (CATATGCTTGTCGTCATCGTCTTTGTAGTC) to pPG174 digested with NcoI and SacI. The plasmid pPG174 was a gift from Paul Gershon (TAMHSC-IBT) and was derived from pTM-1 (10), a plasmid containing a T7 RNA polymerase promoter proximal to a polylinker and flanked by sequences from vaccinia virus WR. The RNA transcript encoded by the T7 RNA polymerase promoter in pTM-1 contains the encephalomyocarditis virus untranslated leader region to enhance translation in the absence of a cap. The pPG174 contains the same nucleotide sequence elements as pTM-1, but the T7 RNA polymerase promoter, the encephalomyocarditis virus untranslated leader region, and polylinker are inverted with regard to the flanking vaccinia virus DNA sequences. Additionally, the plasmid pPG552 encodes a FLAG tag peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) in frame with an ATG supplied by an NdeI site in the polylinker. The plasmid pT7K4LFLAG was constructed in two steps. First of all, the open reading frame corresponding to K4L was cloned into the TA TOPO cloning vector pCRII (Invitrogen) using the PCR product of the oligonucleotides MM528 (5′-GGGGGGCATATGAATCCGGATAATACAATCGCA) and MM529 (5′-GGGGGGCTCGAGGCATCTTGTTAACGGGCTCGT) and vaccinia WR DNA, generating pTAK4L. The NdeI and XhoI restriction endonuclease sites in oligonucleotides MM528 and MM529 are underlined, and the initiation ATG is represented in bold for oligonucleotide MM528. The DNA fragment product containing the open reading frame for K4L from pTAK4L after digestion with the restriction enzymes NdeI and XhoI was gel purified using Geneclean (Q-Biogene) and ligated to pPG552 digested with NdeI and XhoI. This plasmid, pT7K4LFLAG, encodes the product of the open reading frame K4L linked to a FLAG peptide at the N terminus under the transcriptional control of the T7 promoter. The nucleotide sequence for the insert in pT7K4LFLAG was determined and shown to correspond to the open reading frame K4L.
The plasmid pK4LKO was designed to eliminate the open reading frame for K4L when recombined into vaccinia virus WR. The plasmid contains a pair of vaccinia virus nucleotide sequences corresponding to each flanking region of the K4L open reading frame inserted into the cloning vector pZippy-NEO/GUS (7). The vector contains two clusters of multiple unique restriction enzyme sites flanking the selection marker Escherichia coli neomycin resistance (neo) and the detection marker β-glucuronidase (gusA) genes to facilitate isolation of recombinant viruses after recombination. The plasmid pK4LKO was constructed in four stages. A DNA fragment proximal to the 3′ end of K4L was generated from the PCR using MM516 (5′-GGGGGGACTAGTGTCCCTAGTCATATGTTTAAA) and MM517 (5′-GGGGGGAAGCTTTCCCAATTTACGAGCCCGTTA) with vaccinia virus WR DNA and ligated into the TA cloning vector pCRII, generating pTAK4Lleft. The SpeI and HindIII restriction endonuclease sites in MM516 and MM517 are shown underlined. The DNA fragment derived from digestion of pTAK4Lleft with SpeI and HindIII was purified using Geneclean after separation using agarose gel electrophoresis and ligated into pZippy-NEO/GUS digested with the restriction enzymes SpeI and HindIII to produce pK4LKOleft. The 5′-proximal flanking DNA fragment for K4L from PCR withMM518 (5′-GGGGGGGAGCTCTGAAGATGATGTTTGGCACCT) and MM519 (5′-GGGGGGGGCGGCCGCACGGACATTTCACCACCAGAT) and vaccinia virus WR DNA was cloned into the TA cloning vector pCRII, generating pTAK4Lright. The SacI and NotI restriction endonuclease sites in MM518 and MM519 are shown underlined The DNA fragment derived from digestion of pTAK4Lright with the restriction endonucleases SacI and NotI was purified after separation using agarose gel electrophoresis and Geneclean and inserted into pK4LKOleft digested with SacI and NotI, generating pK4LKO.
The recombinant virus vΔK4L was the recombination product of the plasmid pK4LKO and vaccinia virus WR. The plasmid pK4LKO was transfected into BS-C-1 cells infected with WR using DOTAP (Boehringer Mannheim). After 3 days the cells were collected, concentrated by centrifugation, resuspended in 1 ml of MEM, and broken open by three freeze-thaw cycles. The product of the transfection was used to infect BS-C-1 cells incubated with 0.64 mg/ml G418 (Sigma). The BS-C-1 cells were preincubated with G418 and 4 mM HEPES, pH 7.5, for at least 6 h before infection. After three rounds of infection in BS-C-1 monolayers in a 25-cm2 flask, the infectious stock was plaqued on monolayers of BS-C-1 cells overlaid with 0.5% methyl cellulose-MEM in the presence of G418 and 5-bromo-4-chloro-3-indolyl β-d-glucuronide at 0.2 mg/ml (Clontech). Plaques staining blue were isolated and used to infect BS-C-1 monolayers in 12-well dishes in the presence of G418 and HEPES. Infected cells were treated with DNAzol (Invitrogen), and the DNA was analyzed by PCR to discriminate between single and double crossover recombinants. A double crossover recombinant virus lacking the open reading frame for K4L was identified and denoted vK4LKO. The structure of vK4LKO was confirmed by Southern blot analysis.
The virus vK4Lrestore is a recombinant virus containing the open reading frame for K4L linked to sequences encoding the FLAG polypeptide inserted within the thymidine kinase gene of vK4LKO. The plasmid pT7K4LFLAG was recombined with vK4LKO in BS-C-1 cells using DOTAP (Boehringer Mannheim), and the infected cells were collected after 3 days. The infectious stock was carried through three rounds of plaque purification in thymidine kinase-deficient 143B cells overlaid with 0.5% methyl cellulose supplemented with 50 μg/ml BrdU. Plaques were isolated, and the viral stock was expanded in 143B monolayers in 12-well dishes in the presence of BrdU. Infected cells were treated with DNAzol (Invitrogen), and the DNA was analyzed by PCR to discriminate between single and double crossover recombinants. A recombinant virus was isolated containing the open reading frame for FLAG-K4L at the thymidine kinase locus and denoted vK4Lrestore. The structure of vK4Lrestore was confirmed by Southern blot analysis.
Expression of virion nuclease after transfection into eukaryotic cells.
Twelve large (100-mm-diameter) plates of confluent BOS cells were infected with vaccinia virus WR in the presence of 40 μg/ml cytosine arabinoside and transfected with pPG552 (four plates) or pT7K4LFLAG (eight plates) using Lipofectamine (Invitrogen). The cells were collected 50 h after infection by scraping, rinsed once with phosphate-buffered saline, and resuspended in 0.5 ml (pPG552) or 1 ml (pT7K4LFLAG) T-lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0) and carried through three freeze and thaw cycles. The samples were clarified by low-speed centrifugation and stored at −20°C.
Expression of virion nuclease in bacteria.
The protein encoded by the vaccinia virus open reading frame K4L linked to the FLAG polypeptide was expressed in bacteria. The plasmid pT7K4LFLAG was used to transform ED8439 cells, and a 5-ml culture was grown in Luria broth to saturation overnight at 37°C. A small aliquot was added to 500 ml of Luria broth with 0.2% maltose and shaken at 37°C. When the optical density at 600 nm reached approximately 1.0, the MgSO4 concentration was increased to 10 mM and 50 ml of bacteriophage CE6 (Novagen), a recombinant lambda phage that contains T7 RNA polymerase at 3.4 × 1010 PFU/ml, was added. After 4 h at 37°C the cells were concentrated by centrifugation and stored at −20°C. After thawing, the pellet was resuspended in 7 ml HEMGN (100 mM KCl, 25 mM HEPES, pH 7.6, 0.1 mM EDTA, 12.5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 10% glycerol) with 20 μl of 50 mg/ml lysozyme and 100 μl of protease inhibitor cocktail (Sigma). After 30 min on ice the sample was sonicated for two 30-second bursts in a cup sonicator. The material was centrifuged for 30 min at 10,000 rpm in a KA-21.50 rotor (Kompspin). The supernatant was passed over a 1-ml column composed of anti-FLAG M2 affinity gel (Sigma) 10 times. The column was rinsed with four 2-ml washes of TBS (50 mM Tris-HCl, pH 7.4, 150 mM NaCl), and the proteins were eluted with a 4-mg/ml solution of the FLAG peptide (Sigma). A parallel purification procedure was also undertaken using extract from CE6-infected ED8439 cells containing the pPG552 plasmid.
RESULTS
The gene encoding the virion nuclease is located on the left side of the genome.
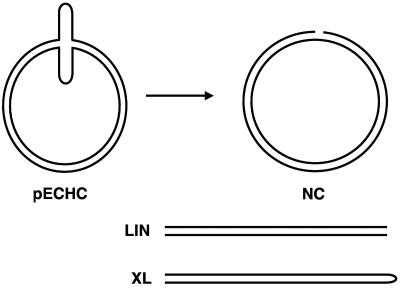
The single-stranded nuclease activity present in vaccinia virus virions (27) was shown to cleave plasmid DNA specifically at regions with single-stranded characteristics and most efficiently at regions that could be extruded as cruciforms (14, 18). Three reaction products were observed after incubation with the viral nuclease (Fig. 1): a singly cleaved nicked circle, a linear molecule, and in much smaller amounts, a cleaved and rejoined hairpin molecule. The cross-linked form has the same mobility as the linear form on neutral agarose gels. Evidence for the cross-linking is provided by electrophoresis through denaturing gels or by demonstration of a “snap-back” covalently continuous molecule after denaturation and quick cooling. The ability to cleave and rejoin DNA in a conservative manner was recognized as a necessary step in the processing of the replicative intermediates observed during poxvirus replication.
FIG. 1.
Reaction products after virion nuclease reaction. Plasmids containing single-stranded regions, such as an extruded cruciform (pECHC), were converted to nicked circular (NC), linear (LIN), or covalently continuous cross-linked (XL) forms after incubation with the nuclease.
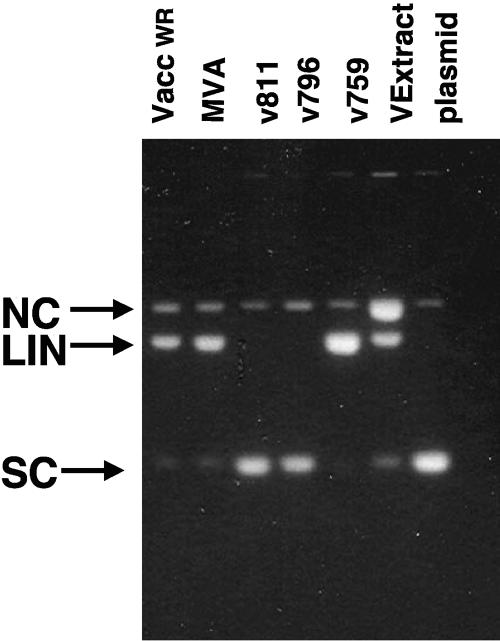
Previous efforts to characterize and identify the virion nicking-joining enzyme in vaccinia virus utilizing biochemical chromatographic separation demonstrated that the enzyme is a hydrophobic 50-kDa protein (14, 25). This work describes the use of a genetic screen to identify the open reading frame encoding the enzyme. Cytoplasmic extracts from cells infected with vaccinia virus strains missing portions of the genome in comparison to vaccinia virus WR were screened for nuclease activity. The four virus strains used were as follows: modified virus Ankara (MVA) was generated by serial passage of vaccinia virus Ankara through CEF cells (15) and is significantly smaller than vaccinia WR (1); the viral recombinants v759, v811, and v796 were constructed by making tailored deletions in vaccinia virus Copenhagen strain (24). v796 and v759 are missing a portion of the left and right of vaccinia virus Copenhagen, respectively. The virus v811 is missing both regions of Copenhagen absent in v759 and v796. Cytoplasmic extracts from BS-C-1 cells infected with these four viruses and vaccinia virus WR strain were tested for nuclease activity (Fig. 2). The nuclease activity was present in extracts from vaccinia WR, MVA, and v759 but absent in both v811 and v796. Since both of these viral constructs are missing a substantial portion of the left end of the genome, the gene expressing the viral nuclease must reside in the left side of the genome.
FIG. 2.
Nuclease activity is not present in all poxvirus recombinants. Cytoplasmic lysates from BS-C-1 cells infected at a multiplicity of infection of 1 with vaccinia virus WR, MVA, v811, v796, v759, and virion extract from vaccinia virus WR (left to right) were used in a nuclease assay at 55°C for 2 hours, and the DNA sample was resolved by electrophoresis through a 1.4% agarose gel. The rightmost lane contains the plasmid used as a substrate in the nuclease assays, pECHC. The positions of the pECHC nicked circle (NC), linear (LIN), and supercoiled (SC) forms are denoted by arrows.
To identify candidate genes that encode nuclease activity, a catalog of the genes missing in v796 and v811 but retained in MVA was made by comparing the nucleotide sequence of MVA to the genomes lacking nuclease activity (1). There were five open reading frames present in MVA but absent in v796 and v811 (Table 1). Since the nuclease activity is associated with a molecular mass of approximately 50 kDa (14, 18, 25, 27), the proteins encoded by K4L and F3L were considered likely candidates for the virion nuclease.
TABLE 1.
Characterization of open reading frames
| Open reading framea | Gene nameb | Predicted molecular mass | Gene function | Gene class |
|---|---|---|---|---|
| VACWR020 | C8L | 20,753 | Unknown | Late |
| VACWR028 | N1L | 13,961 | Virokine, host defense modifier | Early+late |
| VACWR035 | K4L | 48,872 | Phospholipase D-like | Late |
| VACWR039 | K7R | 17,467 | Unknown | Unknown |
| VACWR042 | F3L | 55,775 | Kelch-like | Unknown |
The open reading frame for vaccinia virus strain WR, as designated at www.poxvirus.org.
The homologous gene designation in vaccinia virus strain Copenhagen.
The open reading frame K4L encodes a protein with nuclease activity.
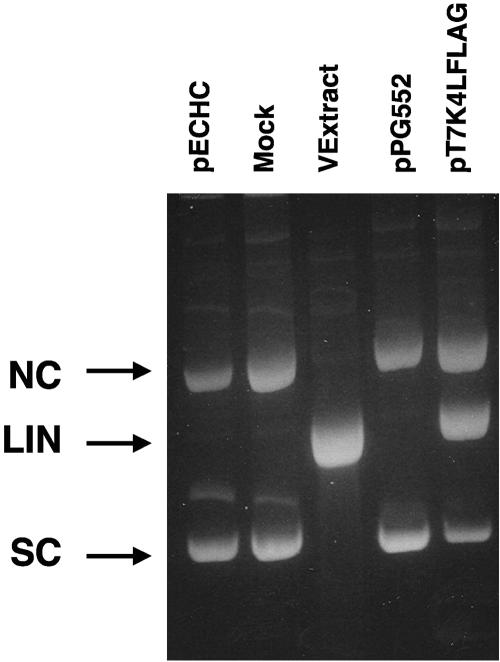
To identify the protein that encodes the vaccinia virus nicking-joining enzyme, plasmids were constructed that expressed the proteins K4L and F3L linked at the N terminus to a FLAG polypeptide under the transcriptional control of the bacteriophage T7 RNA polymerase. The expression of genes under the transcriptional control of the T7 promoter after transfection into mammalian cells expressing T7 RNA polymerase is enhanced greatly by coinfection with vaccinia virus. Since vaccinia virus nuclease activity occurs late after infection, cells were transfected with plasmids and infected with vaccinia virus WR in the presence of an inhibitor of DNA replication, cytosine arabinoside. Under these conditions, only vaccinia virus early class and genes under T7 transcription control will be expressed. A protein of approximately 50 kDa was observed to react with an anti-FLAG peptide in extracts from cells transfected with pT7K4LFLAG (data not shown). Nuclease activity was detected in cytoplasmic extracts from the cells transfected with pT7K4LFLAG (Fig. 3). Plasmids containing the open reading frame for F3L under the control of the T7 promoter did not display nuclease activity when transfected into BOS cells (data not shown).
FIG. 3.
Nuclease activity is present in cells transfected with K4L. Nuclease assays were run using mock-infected extract, virion extract derived from vaccinia virus WR, or cytoplasmic extracts prepared from cells transfected with pPG552 or pT7K4LFLAG and infected with vaccinia virus WR in the presence of cytosine arabinoside (second through fifth lanes). These samples, and the plasmid pECHC (left-most lane) used in the nuclease assays, were resolved by electrophoresis through a 1.4% agarose gel. The positions of the pECHC nicked circle (NC), linear (LIN), and supercoiled (SC) forms are denoted by arrows.
Expression of K4L is required for nuclease activity.
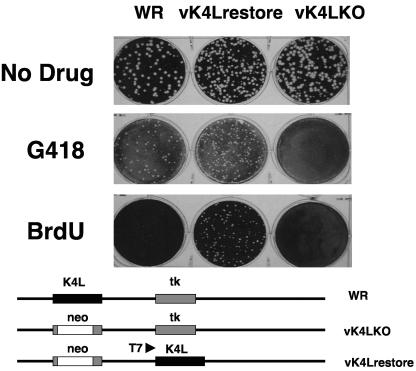
To investigate the supposition that K4L participates in virion nuclease activity, a recombinant virus lacking the open reading frame for K4L was constructed and screened for nuclease activity. The virus vK4LKO was constructed by isolating the recombination products of pK4LKO and vaccinia virus WR. This virus contains the entire WR genome with the K4L open reading frame replaced by the bacterial neomycin resistance and β-glucuronidase genes. The open reading frame for K4L was reinserted into the vK4LKO genome, replacing the viral thymidine kinase gene by isolating the recombination products of pT7K4LFLAG and vK4LKO. The structure of both recombinant viruses was confirmed by PCR and Southern blot analysis (data not shown). In addition, the viruses displayed the predicted resistance or sensitivity to drugs that interfere with wild-type vaccinia virus replication (Fig. 4). The recombinant vK4LKO was resistant to G418 and sensitive to incubation with BrdU. The recombinant vK4Lrestore is resistant to both G418 and BrdU. Expression of the K4L open reading frame was confirmed by Western blot analysis with the observation of an approximately 50-kDa protein that reacted with anti-FLAG antibody in vK4Lrestore virions.
FIG. 4.
Sensitivity of virus recombinants to inhibitors of viral replication. Duplicate monolayers of BS-C-1 cells were infected with the same dilution of vK4LKO, vK4Lrestore, or vaccinia virus WR and incubated with no drug (upper panel), 1 mg/ml G418 (middle panel), or 50 μg/ml bromodeoxyuridine (lower panel).
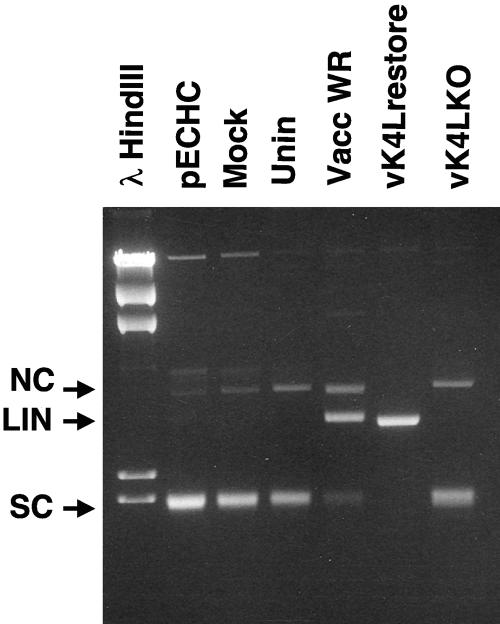
Cytoplasmic extracts from BOS cells infected with the recombinant viruses or vaccinia virus WR were used in a nuclease assay (Fig. 5). Extracts made from the vaccinia virus WR- and vK4Lrestore-infected cells contained nuclease activity; however, extracts made from vK4LKO did not contain nuclease activity. These results indicate that vaccinia virus nuclease activity requires the expression of the K4L protein.
FIG. 5.
The K4L open reading frame is required for nuclease activity. Cytoplasmic extracts from uninfected BOS cells or cells infected with vaccinia virus WR, vK4Lrestore, and vK4LKO and pECHC plasmid alone (mock lane) were used in a nuclease assay with pECHC plasmid, and the products were resolved by electrophoresis through a 1.4% agarose gel. The position of the pECHC nicked circle (NC), linear (LIN), and supercoiled (SC) forms are denoted by arrows.
The vaccinia virus K4L gene encodes the nicking-joining enzyme.
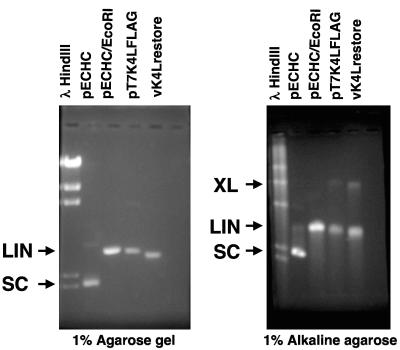
Vaccinia virus nuclease activity was only observed if the K4L open reading frame was expressed; however, it remains unclear if the product of K4L is the only viral protein required for nuclease activity. Initial characterization of the nuclease activity indicated that the activity was associated with a native molecular mass of approximately 100 kDa (14, 28), suggesting that K4L protein exists as a dimer or is associated with another protein. To determine if K4L encodes the only viral protein required for nicking-joining activity, K4L was expressed in bacteria in the absence of any other vaccinia virus proteins. Attempts to express the open reading frame encoding K4L under the control of the T7 promoter in bacteria that express T7 RNA polymerase were unsuccessful, suggesting that the expressed protein was detrimental to cell viability; however, the plasmid pT7K4LFLAG was stable in the absence of T7 RNA polymerase. Therefore, expression of K4L was accomplished by introducing T7 RNA polymerase into bacteria using the λ phage CE6. Infection of bacteria containing pT7K4LFLAG led to the production of K4L protein containing a FLAG peptide at the N terminus of the protein. The protein was purified from extracts of bacteriophage-infected bacterial cells using an anti-FLAG column. After purification, the predominant protein observed in a silver-stained polyacrylamide gel migrated with a molecular mass of approximately 50 kDa (data not shown). The nuclease activity of the purified bacterial extract was compared to that of a virion extract derived from the recombinant virus vK4Lrestore. The purified K4L protein extract from bacteria and the virion extract from vK4Lrestore were active in the nuclease assay. The linear products of the nuclease reaction were electrophoresed through an agarose gel and compared to pECHC plasmid and pECHC plasmid linearized after digestion with EcoRI on neutral agarose gels (Fig. 6, left panel). The linear band derived from the vK4Lrestore extract was slightly smaller than the band observed from the K4L bacterial-derived protein sample. The shorter linear band in the virion extract reactions may be due to increased exonuclease activity at high levels of K4L nuclease or due to an additional exonuclease present in the viral extract (also observed in Fig. 3 and 5). To determine if the extracts contained cross-linking activity, samples from the nuclease reaction were separated by electrophoresis through alkaline agarose gels (Fig. 6, right panel). A high-molecular-mass band with twice the mass of the linear fragment was observed in both the purified bacterial protein and virion-derived extracts, corresponding to a cross-linked pECHC molecule. The bacterial protein converted about 7% of the pECHC into the cross-linked form, while the virion extract derived from vK4Lrestore converted about 13% of the pECHC into the cross-linked form. No nuclease activity was detected in bacterial extracts derived from λ phage CE6-infected bacterial cell extracts containing pPG552 and passed over an anti-FLAG column (data not shown). Therefore, the vaccinia virus K4L protein is sufficient, requiring no other viral proteins, for nicking-joining activity.
FIG. 6.
The K4L open reading frame encodes the viral nicking-joining enzyme. An affinity-purified preparation of the bacterial K4L protein (pT7K4LFLAG) and the virion extract from the recombinant virus vK4Lrestore were used in a nuclease assay with pECHC, and the products were resolved by electrophoresis through a 1.4% agarose gel. The band corresponding to the linear pECHC molecule was purified and used for electrophoresis through a neutral agarose (left panel) or alkaline agarose (right panel) gel with untreated pECHC or pECHC linearized by digestion with the restriction endonuclease EcoRI. The positions of the pECHC cross-linked (XL), linear (LIN), and supercoiled (SC) forms are denoted by arrows. Band intensity was quantitated on a Fuji LAS-1000 after staining the gels with SYBR Gold (Molecular Probes).
DISCUSSION
Biochemical fractionation of virion extracts revealed the existence of an endonuclease activity which efficiently cleaved single-strand DNA at low salt concentrations in the absence of Mg2+ (14, 27, 28). Incubation of the nuclease with plasmid DNA revealed site-specific cleavage associated with regions of known partially base-paired secondary structure, and incubation at elevated temperatures resulted in the conversion of some of the linear molecules into terminally cross-linked DNA molecules (14, 18). Since poxviruses contain covalently continuous double-stranded DNA genomes with cross-links at the termini, this combination of strand scission and rejoining activity associated with this enzyme fueled the speculation that this activity is associated with the processing of poxvirus genomes.
As a first step in determining the role of the nicking-joining enzyme in the poxvirus life cycle, we decided to unambiguously identify the open reading frame responsible for the nuclease activity. Extracts were made from cells infected with a series of vaccinia viruses missing defined portions of the genome. This genetic screen for nuclease activity reduced the number of candidate genes for the nicking-joining enzyme to two, K4L and F3L. Further assays using extracts from the individually expressed proteins revealed that only the protein encoded by K4L possessed nuclease activity. To demonstrate that the K4L protein was sufficient for nicking-joining activity, it was expressed in prokaryotes, purified over an antibody column, and tested for activity. The bacteria-derived K4L protein was able to cleave and covalently join the DNA, demonstrating that K4L is the vaccinia virus gene encoding the viral nicking-joining enzyme.
The rate of conversion of plasmid into cross-linked molecules using bacterial protein or virion extract was similar to the reported value of 15% conversion observed previously (14, 25). The bacterially expressed form of the K4L open reading frame was able to carry out all of the reactions ascribed to the viral nicking-joining enzyme. However, it is unclear if the specific activity of the bacterial protein is similar to that of the eukaryotic-derived viral K4L protein. The amount of the K4L gene product in the virion extract reaction mixture could not be determined, since the extract is comprised of many proteins. The rate of cross-link formation was lower in the reactions using the K4L protein derived from bacteria, even though highly enriched protein was used, implying that the viral extract may include proteins that enhance the conversion into the cross-linked form. Additionally, the virus-derived protein may be different, due to posttranslation modifications, from the bacterial protein.
The gene encoding K4L is not essential for growth in tissue culture, as a viral recombinant lacking the K4L gene was facilely isolated. The role of K4L in virus growth was investigated by comparing the growth of wild-type vaccinia virus, strain WR, the viral recombinant missing K4L, vK4LKO, and the recombinant virus containing the open reading frame encoding K4L placed back into vK4LKO under the transcriptional control of the T7 promoter, vK4Lrestore. The infectivities of the purified viruses in tissue culture, as measured by the PFU per gram of protein, were nearly identical (data not shown). The extent of telomere processing was measured by Southern blot analysis of DNA from infected BS-C-1 or BOS cells separated by agarose gel electrophoresis after digestion with the restriction endonuclease BstEII. The amounts of unprocessed concatemer junctions (2.6-kb BstEII fragment) and resolved telomeres (1.3-kb BstEII fragment) were identical in cells infected with each of the three viruses (data not shown). The extent of branch resolution, as measured by the distribution of DNA among unit-length genomes and well-retained material in CHEF agarose gel electrophoresis, was the same for BS-C-1 or BOS cells infected with the three viruses (data not shown). This indicates that if the K4L protein participates in DNA processing, vaccinia virus must encode redundant activities. Previously, it has been demonstrated that efficient processing of vaccinia virus telomeres requires expression of the open reading frame for the A22R protein (11, 12).
Although the K4L gene is not required for growth of vaccinia virus in tissue culture, the role of K4L in host virulence was investigated by comparing the 50% lethal dose (LD50) in mice for vK4LKO to vaccinia virus WR. Groups of five BALB/c mice were injected by the intraperitoneal route with graded doses of vaccinia virus WR or vK4LKO, and the LD50 for each virus was computed using the method of Reed and Muench (26). The LD50 values for the two viruses were almost identical, suggesting that K4L is not involved in host pathogenesis in mice. The K4L open reading frame is designated VACWR035 at www.poxvirus.org and is identified as possessing homology to eukaryotic phospholipase D. Assays designed to demonstrate phospholipase D activity (Molecular Probes) failed to show activity significantly higher in extracts from BOS cells infected with vK4Lrestore compared to uninfected cells or cells infected with vaccinia virus WR or vK4LKO (data not shown). The protein does not contain any motifs that correlate with nucleic acid activities or binding, and BLAST searches against the protein database did not reveal any strong homologies outside of proteins from other poxvirus strains and phospholipase D proteins from mammalian sources.
The results presented here do not provide strong evidence for the role of K4L in the vaccinia virus life cycle. The ability of the protein encoded by K4L to nick and join DNA in vitro does not guarantee that these activities are germane to the role of K4L in the growth of vaccinia virus. The K4L protein is expressed late and, as demonstrated by its fractionation during purification on chromatographic resins (25), is hydrophobic. The majority of the K4L protein is found in the insoluble fraction with detergent extraction (data not shown), suggesting the protein may be part of the viral core. As part of the viral core, K4L may participate in the binding and condensation of the viral DNA. The demonstration that K4L binds and nicks vaccinia virus hairpin termini in vitro (6) is consistent with a role in the packaging of viral DNA. The ability of K4L to nick hairpin DNA suggests a possible role in the initiation of DNA replication. Since K4L is not essential for virus growth, the participation of K4L in the initiation of replication of DNA is unlikely unless vaccinia virus encodes redundant proteins for the process. Nevertheless, the importance of the K4L gene is underscored by the observation that a homologue for the open reading frame for VACWR053, or K4L, is present in all poxviruses (www.poxvirus.org), although most appear to retain a shorter version of the open reading frame.
Although the role of K4L in the vaccinia virus life cycle is unclear, the gene encodes a vaccinia virus nicking-joining enzyme. Previous efforts to generate in vitro systems that mimic telomere resolution were limited by the presence of nuclease activity interfering with the ability to detect sequence-specific telomere resolution (29). The demonstration that the nicking-joining activity is dispensable for viral growth provides an opportunity to revisit in vitro approaches for the study of telomere resolution.
Acknowledgments
The BOS cells and MVA virus were a kind gift of Bernard Moss (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.). v759, v796, and v811 were provided by Enzo Paoletti (Virogenetics), and the plasmid pPG174 was provided by Paul Gershon (TAMHSC-IBT).
REFERENCES
- 1.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 2.Baroudy, B. M., S. Venkatesan, and B. Moss. 1983. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47:723-729. [DOI] [PubMed] [Google Scholar]
- 3.DeLange, A. M. 1989. Identification of temperature-sensitive mutants of vaccinia virus that are defective in conversion of concatemeric replicative intermediates to the mature linear DNA genome. J. Virol. 63:2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLange, A. M., and G. McFadden. 1987. Efficient resolution of replicated poxvirus telomeres to native hairpin structures requires two inverted symmetrical copies of a core target DNA sequence. J. Virol. 61:1957-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLange, A. M., and G. McFadden. 1986. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc. Natl. Acad. Sci. USA 83:614-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMasi, J., S. Du, D. Lennon, and P. Traktman. 2001. Vaccinia virus telomeres: interaction with the viral I1, I6, and K4 proteins. J. Virol. 75:10090-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dvoracek, B., and T. Shors. 2003. Construction of a novel set of transfer vectors to study vaccinia virus replication and foreign gene expression. Plasmid 49:9-17. [DOI] [PubMed] [Google Scholar]
- 8.Earl, P., B. Moss, L. Wyatt, and M. W. Carroll. 1998. Generation of recombinant vaccinia viruses, p. 16.17.1-16.17.19. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. Greene Publishing Associates & Wiley Interscience, New York, N.Y. [Google Scholar]
- 9.Earl, P. L., N. Cooper, L. Wyatt, B. Moss, and M. W. Carroll. 1998. Preparation of cell cultures and vaccinia virus stocks, p. 16.16.1-16.16.13. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. Greene Publishing Associates & Wiley Interscience, New York, N.Y. [Google Scholar]
- 10.Elroy-Stein, O., and B. Moss. 1998. Gene expression using the vaccinia virus/T7 RNA polymerase hybrid system, p. 16.19.1-16.19.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 3. Greene Publishing Associates & Wiley Interscience, New York, N.Y. [Google Scholar]
- 11.Garcia, A. D., L. Aravind, E. V. Koonin, and B. Moss. 2000. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl. Acad. Sci. USA 97:8926-8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia, A. D., and B. Moss. 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 75:6460-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, C. H., and S. Shuman. 1996. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J. Virol. 70:8549-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakritz, N., P. D. Foglesong, M. Reddy, S. Baum, J. Hurwitz, and W. R. Bauer. 1985. A vaccinia DNase preparation which cross-links superhelical DNA. J. Virol. 53:935-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayr, A., V. Hochstein-Mintzel, and H. Stickl. 1975. Abstammung, eigenshaften und verwendung des attenuierten vaccinia-stammes MVA. Infection 3:6-14. [Google Scholar]
- 16.Merchlinsky, M. 1989. Intramolecular homologous recombination in cells infected with temperature-sensitive mutants of vaccinia virus. J. Virol. 63:2030-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merchlinsky, M. 1990. Resolution of poxvirus telomeres: processing of vaccinia virus concatemer junctions by conservative strand exchange. J. Virol. 64:3437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchlinsky, M., C. F. Garon, and B. Moss. 1988. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J. Mol. Biol. 199:399-413. [DOI] [PubMed] [Google Scholar]
- 19.Merchlinsky, M., and B. Moss. 1989. Nucleotide sequence required for resolution of the concatemer junction of vaccinia virus. J. Virol. 63:4354-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merchlinsky, M., and B. Moss. 1989. Resolution of vaccinia virus DNA concatemer junctions requires late gene expression. J. Virol. 63:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merchlinsky, M., and B. Moss. 1988. Sequence-independent replication and sequence-specific resolution of plasmids containing the vaccinia virus concatemer junction: requirements for early and late trans-acting factors. Cancer Cells 6:87-94. [Google Scholar]
- 22.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2672. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 23.Moyer, R. W., and R. L. Graves. 1981. The method of cytoplasmic orthopox replication. Cell 27:391-401. [DOI] [PubMed] [Google Scholar]
- 24.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, E. K. Norton, and E. Paoletti. 1991. Deletion of 55 open reading frames from the termini of vaccinia virus. Virology 180:406-410. [DOI] [PubMed] [Google Scholar]
- 25.Reddy, M. K., and W. R. Bauer. 1989. Activation of the vaccinia virus nicking-joining enzyme by trypsinization. J. Biol. Chem. 264:443-449. [PubMed] [Google Scholar]
- 26.Reed, L., and H. Muench. 1938. Simple method of determining fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 27.Rosemond-Hornbeak, H., and B. Moss. 1974. Single-strand deoxyribonucleic acid-specific nuclease from vaccinia virus. Endonucleolytic and exonucleolytic activities. J. Biol. Chem. 249:3292-3296. [PubMed] [Google Scholar]
- 28.Rosemond-Hornbeak, H., E. Paoletti, and B. Moss. 1974. Single-strand deoxyribonucleic acid-specific nuclease from vaccinia virus. Purification and characterization. J. Biol. Chem. 249:3287-3291. [PubMed] [Google Scholar]
- 29.Stuart, D., K. Ellison, K. Graham, and G. McFadden. 1992. In vitro resolution of poxvirus replicative intermediates into linear minichromosomes with hairpin termini by a virally induced Holliday junction endonuclease. J. Virol. 66:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer, D. O., X. D. Yao, M. J. Mann, and D. H. Evans. 2000. In vitro concatemer formation catalyzed by vaccinia virus DNA polymerase. Virology 278:562-569. [DOI] [PubMed] [Google Scholar]