Abstract
Group 2 coronaviruses encode an accessory envelope glycoprotein species, the hemagglutinin esterase (HE), which possesses sialate-O-acetylesterase activity and which, presumably, promotes virus spread and entry in vivo by facilitating reversible virion attachment to O-acetylated sialic acids. While HE may provide a strong selective advantage during natural infection, many laboratory strains of mouse hepatitis virus (MHV) fail to produce the protein. Apparently, their HE genes were inactivated during cell culture adaptation. For this report, we have studied the molecular basis of this phenomenon. By using targeted RNA recombination, we generated isogenic recombinant MHVs which differ exclusively in their expression of HE and produce either the wild-type protein (HE+), an enzymatically inactive HE protein (HE0), or no HE at all. HE expression or the lack thereof did not lead to gross differences in in vitro growth properties. Yet the expression of HE was rapidly lost during serial cell culture passaging. Competition experiments with mixed infections revealed that this was not due to the enzymatic activity: MHVs expressing HE+ or HE0 propagated with equal efficiencies. During the propagation of recombinant MHV-HE+, two types of spontaneous mutants accumulated. One produced an anchorless HE, while the other had a Gly-to-Trp substitution at the predicted C-terminal residue of the HE signal peptide. Neither mutant incorporated HE into virion particles, suggesting that wild-type HE reduces the in vitro propagation efficiency, either at the assembly stage or at a postassembly level. Our findings demonstrate that the expression of “luxury” proteins may come at a fitness penalty. Apparently, under natural conditions the costs of maintaining HE are outweighed by the benefits.
Coronaviruses are large enveloped positive-strand RNA viruses of mammals and birds (for a review, see reference 45). Their virions, as visualized by negative-staining electron microscopy, are roughly spherical particles measuring 80 to 120 nm in diameter. They contain a helical nucleocapsid comprised of the ∼30-kb genome and multiple copies of a single nucleocapsid protein species, N. The nucleocapsid is surrounded by a lipid-containing envelope, which is derived from the endoplasmic reticulum-Golgi intermediate compartment and into which are embedded at least three other structural proteins. Of these, the triple-spanning membrane glycoprotein M and the small envelope protein E are pivotal for virion morphogenesis (3, 15, 25, 57). The spike protein S, though not required for assembly, is crucial for infectivity as it mediates virus adsorption to specific host cell receptors (2, 5, 24, 28, 56, 69) and subsequent fusion of the viral envelope and the plasma membrane (for a review, see reference 10).
An additional, accessory structural protein, the hemagglutinin esterase (HE), occurs only in a subset of closely related coronaviruses; these viruses, designated group 2 coronaviruses, comprise, among others, mouse hepatitis virus (MHV), bovine coronavirus (BCoV), and human coronavirus OC43 (HCoV-OC43) (for a review, see reference 6). HE is an N-glycosylated class I membrane protein of approximately 65 kDa which forms disulfide-bonded homodimers (8, 16, 17, 20, 21, 44, 46). In electron micrographs of virions, these homodimers—or multimeric complexes thereof—can be discerned as a fringe of small 5- to 7-nm spikes (7, 53).
Intriguingly, the HE protein is not unique to coronaviruses: it displays 30% sequence identity to subunit 1 of the HE fusion protein of influenza C virus (6, 30). Moreover, an equally related HE homologue also occurs as a structural protein in toroviruses (11, 49). Apparently, the HE gene was acquired by a coronavirus group 2 predecessor via heterologous RNA recombination (30), an event which must have taken place relatively recently, after the group 2/severe acute respiratory syndrome coronavirus (SARS-CoV) split-off (47, 48).
The mere fact that this ancestral recombinant virus established itself in the field implies that the expression of HE must provide a strong selective advantage during natural infection. Presumably, the HE protein promotes virus spread and entry into host cells by facilitating virion attachment to O-acetylated sialic acids, i.e., established (co)receptors for several group 2 viruses (41, 42, 58, 59). Like the influenza virus C HE fusion protein, coronavirus HEs function as sialic acid (Sia)-binding proteins (8, 21, 35, 43, 54), although to what extent is still subject to debate. For instance, in the case of BCoV (40), HCoV-OC43 (23), and MHV strains S and JHM (63), virus attachment to Sia seems to be mediated primarily by the S protein. All coronavirus HEs, however, do display sialate-O-acetylesterase activity (22, 34-36, 43, 47, 52, 54, 58, 67). The HEs thus serve as virion-associated receptor-destroying enzymes, and in analogy to the O-acetylesterases and sialidases of ortho- and paramyxoviruses (32, 62), ensure that binding of viruses to cell-associated and cell-free Sias is reversible.
The HE protein is clearly not essential for the replication of murine coronaviruses in cultured cells: in various tissue culture-adapted murine coronavirus strains, the HE gene is inactivated (66). For example, MHV-A59 fails to produce the mRNA for HE, RNA2a, because of a nucleotide substitution within the transcription-regulating sequence (TRS); in addition, the open reading frame for the A59 HE protein is interrupted by a nonsense mutation at codon 15 (30, 44).
HE also seems to be dispensable for infection in vivo. Mice can be experimentally infected with HE-deficient MHV strains via both artificial and natural inoculation routes, resulting in hepatitis and acute or chronic diseases of the central nervous system (1, 27, 38). Moreover, upon intracranial inoculation of mice and rats with HE-expressing MHV strains, HE-defective variants are apparently selected for and can be isolated from the brain and spinal cord, in particular during prolonged infection (26, 68). Although one might interpret these findings as indicating that in murine coronaviruses, HE has become obsolete, there is good reason to assume that this protein actually contributes to viral fitness during natural infections. Phylogenetic studies indicate that in field populations of rodent coronaviruses, the HE gene is strictly maintained (47).
One possible explanation for the loss of HE expression in MHV laboratory strains departs from the assumption that in vitro mutations in HE are neutral. In the absence of positive selection for expression, mutations in the HE gene might accumulate during multiple rounds of replication and by chance may have become fixed upon plaque purification. Alternatively, however, under in vitro conditions the loss of HE might actually provide a gain of fitness. Either way, to further our understanding of coronavirus replication and evolution, it is important to explore the molecular basis of this phenomenon.
Previous attempts to define the role of the HE protein during infection involved the use of spontaneous MHV strain JHM variants, which exhibit different HE expression levels (64), and of defective interfering RNA-based HE expression vectors in combination with MHV-A59 (29, 70). Evidently, the benefits and possible consequences of HE expression during in vitro and in vivo replication would be best studied using isogenic recombinant viruses which differ solely in their expression of this protein. For this study, we have used targeted recombination (24) to generate MHV-A59 derivatives which express either the wild-type HE protein, an enzymatically inactive HE protein, or no HE at all. A detailed comparison of the in vitro growth properties of these viruses is presented, including mixed propagation-competition experiments and the biochemical analysis of acetylesterase-deficient variants, which arise spontaneously during the propagation of HE-expressing recombinant MHVs (rMHVs). We demonstrate that during propagation in cultured cells, rMHVs that express HE have a disadvantage compared to isogenic HE-deficient viruses. Mechanisms by which HE might decrease viral fitness in vitro are discussed.
MATERIALS AND METHODS
Viruses, cells, and antibodies.
All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), penicillin (100 IU/ml), and streptomycin (100 μg/ml) (DMEM-10).
The Utrecht derivative of MHV-A59 (A59U), MHV-S (American Type Culture Collection), and the MHV recombinants MHV-HE+, -HE0, and -HE− were propagated in mouse 17 clone 1 (17Cl1) cells, Sac(−) cells, or LR7 cells, an L-2 murine fibroblast cell line stably expressing the MHV receptor (24). fMHV (24), fMHVΔ2aHE, and feline infectious peritonitis virus (FIPV) strain 79-1146 were propagated in feline fcwf-D cells (American Type Culture Collection). Cells were routinely infected with virus suspensions diluted in phosphate-buffered saline containing 50 μg/ml DEAE dextran (PBS-DEAE). Prior to inoculation, monolayers were rinsed once with the same buffer.
Monoclonal antibodies (MAbs) J1.3 and J7.6, directed against the MHV M and S proteins, respectively (55), were provided by J. Fleming (University of Wisconsin, Madison, Wis.), and MAb 23F4.5, directed against FIPV S, was provided by Rhône Mérieux (Lyon, France). MAb 4G12-2F9, directed against MHV HE (65), and polyclonal antiserum UP3, specific for the MHV 2a protein (71), were kind gifts from S. Baker (Loyola University of Chicago, Maywood, Il.) and S. Weiss (University of Pennsylvania, Philadelphia, Pa.), respectively. The production of rabbit polyclonal antiserum K135 to MHV-A59 was described previously (37).
Construction of plasmids.
The transfer vector pFMΔ2aHE was constructed from pXH2aHE (12) by replacing the MHV-A59 S gene, located on an AvrII/SseI fragment, with a chimeric FIPV-MHV S gene, located on a corresponding AvrII/SseI fragment from pGTFMS (24).
The HE gene of MHV strain S was reverse transcription-PCR (RT-PCR) amplified from total intracellular RNA isolated from infected cells as a template. The gene was cloned into vector pGEM-T. Transfer vectors pMH54HE+, -HE0, and -HE− were constructed from pXH2aHE, using conventional cloning and mutagenesis techniques. They contained the 3′-most 10,838 residues of the MHV-A59 genome (residues 20497 through 31334; NC 001846), in which the A59 HE pseudogene (residues 22602 through 23921) was replaced by the HE gene of MHV strain S (residues 1 to 1330; AY771997) and in which the mutated TRS of RNA2b of A59 was restored to the wild-type sequence through a G22,584→A substitution (AATAAGC to AATAAAC). The HE0 mutant gene was created by splice extension overlap PCR, during which residues G134 and T135 were replaced by C and G, respectively; this resulted in a substitution of active-site Ser45 with Thr. The HE mutant gene was created by performing SwaI digestion at 37°C, followed by religation. The enzyme preparation fortuitously contained exonuclease activity, which in the selected construct generated a four-nucleotide deletion (residues 101 to 104) at the SwaI site. Translation of the mutated HE gene should yield a 58-residue peptide in which only residues 1 to 33 are HE derived.
Targeted recombination.
To introduce the chimeric FIPV-MHV S gene into the MHV genome and concomitantly delete the genes for 2a and HE, targeted RNA recombination was performed with pFMΔ2aHE-generated donor RNA and MHV-A59U as the recipient virus essentially as described previously (19). Recombinant virus fMHVΔ2aHE was isolated by three consecutive rounds of plaque purification in fcwf-D cells.
Recombinant MHV-A59 derivatives with either an intact or mutated HE gene were constructed via targeted RNA recombination as described above, but with fMHVΔ2aHE as the acceptor virus and with donor RNAs transcribed from transfer vectors pMH54HE+, -HE0, and -HE−. Candidate recombinant viruses were isolated by two consecutive rounds of plaque purification in LR7 cells. The procedure was performed in duplicate to allow the isolation of at least two completely independent recombinants for each of the envisaged viral mutants.
Metabolic labeling and analysis of viral mRNAs.
Metabolic labeling of intracellular viral RNAs was performed as described previously (39). LR7 cells grown in 35-mm wells were infected at a multiplicity of infection (MOI) of 10 PFU/cell. After a 1-h adsorption period, the inoculum was removed, and the monolayers were washed once with PBS. Then 2 ml of DMEM containing 2% FCS was added, and incubation at 37°C was continued. At 5 h postinfection (p.i.), the tissue culture supernatant was replaced with 1 ml DMEM-2% FCS supplemented with 20 μg/ml actinomycin D and 50 μCi/ml [3H]uridine (25 to 30 Ci/mmol; Amersham). Cells were labeled until 6 h p.i. The monolayers were washed twice with ice-cold PBS, and total cytoplasmic RNA was isolated as described previously (50). RNA samples, to the equivalent of 105 cells, were subjected to electrophoresis in 0.8% agarose-MOPS (morpholinepropanesulfonic acid)-formaldehyde gels and visualized by fluorography as described previously (39).
Analysis of intracellular viral proteins.
LR7 cells were grown in 35-mm dishes and infected with MHV-A59, MHV-S, or each of the MHV-A59 recombinant viruses at an MOI of 10 PFU per cell. Cells were depleted of cysteine and methionine by incubation with cysteine- and methionine-free minimal essential medium (MEM) containing 10 mM HEPES (pH 7.2) from 4 to 5 h p.i. The cells were metabolically labeled from 5 to 6 h p.i. by adding 35S in vitro cell-labeling mix (Amersham) to a final concentration of 170 μCi/ml. Subsequently, the cells were washed with ice-cold PBS and solubilized in 600 μl of lysis buffer, consisting of TES (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA) containing 1% Triton X-100 and 1 μg leupeptin, 40 μg aprotinin, and 1 μg pepstatin A per ml. Nuclei and cell debris were pelleted by centrifugation at 12,000 × g for 2 min at 4°C. From the supernatant, 50-μl aliquots were diluted with 1 ml of detergent solution (50 mM Tris-HCl [pH 8.0], 62.5 mM EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and 30 μl of 10% sodium dodecyl sulfate (SDS) was added. Antibodies were then added as follows: 3 μl of antiserum K135, 30 μl of MAb J1.3, 10 μl of MAb J7.6, 1 μl of MAb 4G12-2F9, or 3 μl of serum UP3. After 16 h of incubation at 4°C, immune complexes were adsorbed for 30 min to formalin-fixed Staphylococcus aureus cells (Pansorbin; Calbiochem) added as 50 μl of a 10% (wt/vol) suspension. In the case of MAb 4G12-2F9, an immunoglobulin G1 antibody, formalin-fixed streptococcus type G cells (Omnisorb; Calbiochem) were used. Immune complexes were collected by centrifugation at 12,000 × g for 1 min and washed three times with RIPA buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate). Pellets were resuspended in 30 μl of modified Laemmli sample buffer (9) and then heated for 5 min at 95°C or, where indicated, kept at room temperature. Samples were analyzed by electrophoresis in SDS-polyacrylamide gels followed by fluorography.
Analysis of virion proteins by virus immunopurification and sucrose gradient centrifugation.
LR7 cells were infected with MHV and depleted of methionine and cysteine from 1 to 2 h p.i., as described above. Cells were then washed twice with PBS, and incubation was continued until 10 h p.i. with cysteine- and methionine-free MEM-10 mM HEPES (pH 7.2) supplemented with 125 μg/ml Cys, 12.5 μg/ml Met, 1 μM HR2 fusion inhibitor (4), and 10 μCi/μl 35S in vitro cell-labeling mix (Amersham). Tissue culture supernatants (1 ml) were harvested and cleared by sequential low-speed (15 min, 1,500 × g) and high-speed (5 min, 14,000 × g) centrifugation at 4°C. Virus particles were affinity purified from 200 μl clarified supernatant supplemented with 300 μl MEM containing 1 μg/ml leupeptin, 40 μg/ml aprotinin, 1 μg/ml pepstatin A, and 0.2 mg/ml bovine serum albumin, to which was added 30 μl of MAb J1.3, 10 μl of MAb J7.6, or 2.5 μl of K135. Samples were processed and analyzed as described above, except that the S. aureus-bound immune complexes were washed three times with TM (10 mM Tris-HCl [pH 7.0], 10 mM MgCl2) instead of RIPA buffer. Alternatively, labeled virus particles present in the cleared tissue culture supernatants were purified by equilibrium centrifugation in 20 to 50% (wt/vol) linear sucrose gradients in TM, which were run in an SW50.1 rotor at 50,000 rpm at 4°C for 72 h; the gradient was fractionated from the bottom up into 25 fractions.
Detection of O-acetylesterase activity in SDS-PAGE gels.
Esterase staining in nondenaturing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels with α-naphthyl acetate was performed essentially as described previously (63). Samples (20 μl) from tissue culture supernatants of MHV-infected cells or from sucrose gradient-purified MHV were mixed with 10 μl of 3× modified Laemmli sample buffer without β-mercaptoethanol, incubated for 10 min at room temperature, and separated in 7.5% SDS-PAGE gels. Gels were soaked in PBS three times for 20 min each time at room temperature and stained for acetylesterase with an α-naphthyl acetate esterase detection kit (Sigma) according to the manufacturer's instructions.
Propagation-competition assays.
Monolayers of 106 LR7, 17Cl1, or Sac(−) cells grown in 35-mm wells were infected with pure or mixed virus preparations at an MOI of 0.01 PFU/cell in 750 μl PBS-DEAE at 37°C. At 1 h p.i., the inoculum was replaced with 1 ml DMEM-10, and incubation was continued for 16 h at 37°C. Tissue culture supernatants were harvested, titrated, and passaged five times; at each passage, cells were inoculated at an MOI of 0.01 PFU/cell and harvested after 16 h.
Plaque assays were performed by applying a 2-ml solid overlay of 1.5% Select agar (Invitrogen) to inoculated cell monolayers at 1 h p.i. Monolayers were fixed at 24 h p.i. by adding 1 ml of 9% paraformaldehyde in PBS and incubating them for 1 h at room temperature. The solid agar overlay was removed, cells were washed three times with PBS-10 mM glycine, and in situ pararosanilin staining of cells for esterase activity with α-naphthyl acetate was performed according to the method of Wagaman et al. (61). For each virus combination, mixed competition experiments were performed with at least three sets of rMHVs. To determine the percentage of acetylesterase-deficient viruses, 200 to 2,200 plaques per sample were counted.
Nucleotide sequence accession number.
The nucleotide sequence of the HE gene of MHV strain S, determined in both orientations for at least two independent clones, was deposited in the EMBL database (AY771997).
RESULTS
Construction of novel acceptor virus fMHVΔ2aHE.
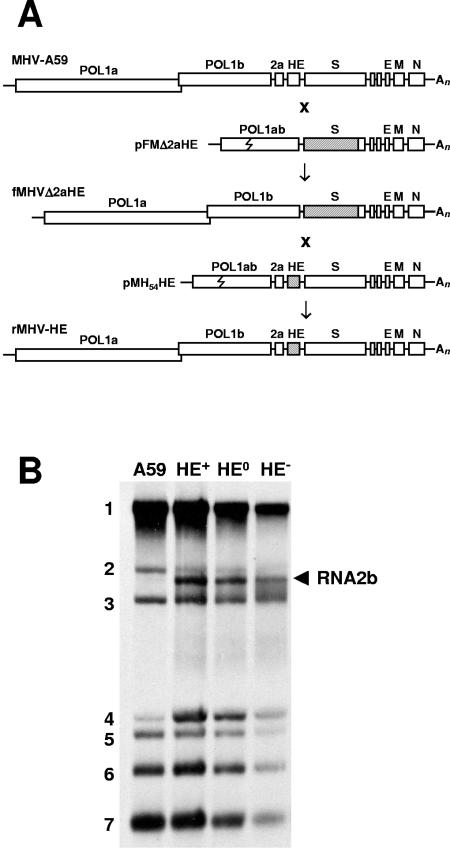
To facilitate autotopical insertion of a functional hemagglutinin esterase gene into the MHV-A59 genome via targeted RNA recombination, we employed targeted recombination to construct a novel fMHV acceptor virus variant from which the 2a and HE sequences had been deleted (Fig. 1A). The resulting recombinant virus, fMHVΔ2aHE, carried a chimeric FIPV-MHV S gene (24), and as anticipated, replicated exclusively in feline cells. Expression of the FIPV-MHV S chimera was confirmed by immunofluorescence: infected cells were stained with both MAb 23F4.5, specific for the S protein of FIPV (33), and K135, a polyclonal antibody directed against MHV (37; data not shown). Sequence analysis of RT-PCR fragments showed that in the fMHVΔ2aHE genome, ORF1b and the chimeric S gene were adjacent and the 2a and HE sequences were absent. Moreover, Northern blot analysis of intracellular fMHVΔ2aHE RNAs showed that the mRNA for 2a, RNA2, was no longer produced (data not shown).
FIG. 1.
Construction of recombinant viruses via targeted RNA recombination. (A) Schematic representation of the genome organization of MHV strain A59, the novel acceptor virus fMHVΔ2aHE, and its recombinant derivatives rMHV-HE+, -HE0, and -HE− (rMHV-HE). Also shown schematically are the structures of the synthetic transcripts, derived from transfer vectors pFMΔ2aHE and the pMH54HE series, which were used to generate fMHVΔ2aHE and the various rMHV-HE viruses, respectively. Open boxes represent the various genes, with those for the polymerase polyproteins (POL1a and POL1b) and the 2a, HE, S, E, M, and N proteins indicated. An, poly(A) tail. The jagged line in the POL1ab fusion gene in pFMΔ2aHE and pMH54HE indicates the border between ORF1a- and ORF1b-derived sequences. Shaded boxes represent S sequences derived from FIPV strain 79-1146 and the HE gene of MHV strain S. (B) mRNA profiles of MHV A59 (A59), rMHV-HE+ (HE+), rMHV-HE0 (HE0), and rMHV-HE− (HE−). Viral RNAs, [3H]uridine labeled in the presence of actinomycin D, were extracted from infected cells, separated in 0.8% agarose-MOPS-formaldehyde gels, and visualized by fluorography. Numbers indicate the MHV genome (1) and the various subgenomic mRNAs (2 through 7). The arrowhead indicates RNA2b, which is produced in cells infected with the rMHV-HE viruses but is absent from those infected with the parental virus, MHV-A59.
Construction of recombinant MHV-A59 derivatives expressing HE.
Instead of restoring the autologous A59 HE gene, we chose to use that of MHV-S, a polytropic strain naturally expressing HE. The MHV-S HE gene, as sequenced in our laboratory (GenBank accession no. AY771997), differed at six nucleotide positions from the MHV-S HE sequence published by Yokomori et al. (M64316) (66). It was 100% identical to the HE sequence of the enterotropic MHV strain DVIM reported by Morita et al. (AB008939) (31), but only 73% identical to the DVIM HE sequence submitted by Compton and Moore (AF091734).
The cloned MHV-S HE gene was used to create two mutant derivatives. In one of these, designated HE−, nucleotides 101 through 104 were deleted, causing a frameshift, as a result of which the encoded HE protein was truncated at Ser33. The other mutant gene, HE0, carried a double mutation at nucleotides 134 and 135 (AGT→ACG), resulting in the replacement of the active-site Ser45 (60) by Thr. The HE genes were used to create transfer vectors pMH54HE+, -HE0, and -HE− (Fig. 1A). To ensure wild-type levels of HE expression, the reconstruction of MHV sequences was performed such that the intergenic region between the 2a and HE genes, including the TRS, was identical to that in MHV strains S and JHM (66; A. Lissenberg and R. J. de Groot, unpublished).
rMHVs were produced by targeted RNA recombination, with fMHVΔ2aHE as the acceptor virus and with synthetic RNAs transcribed from the pMH54HE transfer vectors serving as donors (Fig. 1A). For each of the desired mutations, at least three recombinant viruses (two of which were derived from separate transfection experiments) were selected and characterized. rMHVs encoding either wild-type HE, biologically inactive HE, or truncated HE were designated rMHV-HE+, rMHV-HE0, and rMHV-HE−, respectively.
Genetic and biochemical analysis of rMHVs.
rMHVs were genetically analyzed by RT-PCR amplification of a 2.3-kb region spanning the complete 2a-HE region, using total intracellular RNA from infected cells as a template. Sequence analysis confirmed that for each of the selected rMHVs, both the 2a and HE genes had been properly inserted into the MHV-A59 genome, and that inadvertent mutations were absent (not shown).
To determine whether the recombinant viruses produced the HE-encoding mRNA, RNA2b, [3H]uridine-labeled intracellular viral RNAs were analyzed in formaldehyde-agarose gels. As shown in Fig. 1B, RNA2b was absent from A59-infected cells. However, in cells infected with the rMHVs, RNA2b was readily detected (Fig. 1B), in quantities similar to those in MHV-S-infected cells (not shown). Hence, the introduction of the native TRS restored the synthesis of RNA2b, apparently to physiologically relevant levels.
To study the expression of viral proteins, rMHV-infected LR7 cells were metabolically labeled with [35S]Met-Cys from 5 to 6 h p.i., and cell lysates were subjected to radio-immunoprecipitation (RIPA). Cells infected with strains A59 and S were used as controls. The structural proteins S, N, and M and the nonstructural protein 2a were detected for each of the tested viruses. The HE protein, however, was only found in cells infected with rMHV-HE+, rMHV-HE0, or MHV-S (Fig. 2A). Cell culture supernatants of the infected cells were tested for O-acetylesterase activity by SDS-PAGE analysis under nonreducing conditions followed by an in-gel bioassay (63). O-Acetylesterases with an apparent molecular mass of 115 kDa, the expected size for the disulfide-bonded HE dimer, were detected in the supernatants of cells infected with rMHV-HE+ and MHV-S. No acetylesterase activity was detected in cell culture supernatants of mock-infected cells or in those of cells infected with A59, rMHV-HE0, or rMHV-HE− (Fig. 2B).
FIG. 2.
Expression of HE protein in cells infected with recombinant viruses rMHV-HE+ and rMHV-HE0. (A) Analysis of intracellular viral proteins. Cells which were mock infected (m) or infected with MHV A59 (A59), rMHV-HE+ (HE+), rMHV-HE0 (HE0), rMHV-HE− (HE−), or MHV-S (S) were metabolically labeled from 5 to 6 h p.i. with 35S in vitro cell-labeling mix (Amersham). Cell lysates were subjected to RIPA with antiserum K135 (α A59), antiserum UP3 (α 2a), or MAb 4G12-2F9 (α HE). Proteins were analyzed by electrophoresis in SDS-polyacrylamide gels followed by fluorography. Only the relevant parts of the gels are shown; bands corresponding to the various proteins (S, N, M, 2a, and HE) are indicated. (B) The HE protein expressed by rMHV-HE+ is enzymatically active. Samples from tissue culture supernatants of MHV-infected cells (designations as described above) were separated in nonreducing 7.5% SDS-PAGE gels. Gels were soaked in PBS at room temperature to allow for protein refolding and then stained for acetylesterase activity, with α-naphthyl acetate as a substrate.
HE is incorporated into the envelopes of rMHV-HE+ and rMHV-HE0 virions.
Evidence for the incorporation of HE into rMHV virions was obtained by equilibrium density sucrose gradient purification. Viral infectivity, structural proteins, and acetylesterase activity all colocalized in the gradient at a density of 1.18 g/ml (Fig. 3A). Upon SDS-PAGE analysis of gradient peak fractions of rMHV-HE+, rMHV-HE0, and MHV-S, HE was detected along with M and N (Fig. 3B). In accordance with these observations, rMHV-HE+ particles studied by electron microscopy displayed a dense fringe of short surface projections which was absent in virions of rMHV-HE− (Fig. 3C).
FIG. 3.
Recombinant MHVs incorporate the HE protein into their envelopes. (A) Radiolabeled preparations of MHV-A59, rMHV-HE0, rMHV-HE+, and MHV-S were subjected to equilibrium density gradient centrifugation in 20 to 50% (wt/vol) linear sucrose gradients. The gradients were fractionated from the bottom up in 25 fractions. Fractions were analyzed for (i) infectivity by end-point dilution (solid line, open squares), (ii) the presence of the M protein by SDS-PAGE, with quantitation with a STORM PhosphorImager/Fluorimager 860 (Molecular Dynamics) and ImageQuant software (broken line, open circles), and (iii) the presence of enzymatically active HE by an in-gel O-acetylesterase assay (insets in panels HE+ and S). (B) Analysis of proteins in sucrose gradient-purified virus preparations. Samples from peak fractions 5 were separated in 15% SDS-PAGE gels. Molecular masses are indicated on the left (in kDa). The locations of the HE, M, and N proteins are indicated. The S protein could not be identified unambiguously. (C). Electron micrographs of negatively stained virions of rMHV-HE− and rMHV-HE+ (courtesy of Jean Lepault, Laboratoire de Virologie Moléculaire et Structurale, Gif-sur-Yvette, France).
We next asked whether rMHV-HE+ and rMHV-HE0 virions contain HE proteins in quantities similar to those found in MHV-S virions. Cells infected with MHV-A59, MHV-S, or each of the rMHVs were metabolically labeled from 2 to 9 h p.i. in the presence of a peptide fusion inhibitor (4). Cell culture supernatants were harvested, cell debris was removed by sequential low-speed and high-speed centrifugation, and immunopurification of virions was performed with MAbs J1.3 and J7.6 (55), specific for the M and S proteins, respectively. The rationale of this experiment was that HE present in virions would be copurified along with the other viral structural proteins. Indeed, HE was detected in preparations of rMHV-HE+ and rMHV-HE0 virions immunopurified with either the S- or M-specific MAb (Fig. 4).
FIG. 4.
Immunopurified particles of rMHV-HE+ and rMHV-HE0 contain HE. Supernatants of metabolically labeled cells which had been mock infected or infected with MHV-A59 (A59), rMHV-HE+ (HE+), rMHV-HE0 (HE0), rMHV-HE− (HE−), or MHV-S (S) were subjected to immunopurification with MAb J1.3 (α M) or MAb J1.7 (α S). Precipitates were analyzed in 15% SDS-PAGE gels. Molecular masses (in kDa) are given on the left, and bands corresponding to the structural proteins M, N, HE, and S (c, cleaved; u, uncleaved) are indicated on the right.
The conditions chosen ensured steady-state [35S]Met labeling of each of the viral proteins, and it was therefore possible to estimate their stoichiometry in the virions on the basis of incorporated radioactivity. The results suggest that rMHVs include HE in their envelopes with an efficiency equal to that of MHV-S. Immunopurified particles of rMHV-HE+, rMHV-HE0, and MHV-S contained M, N, and HE at a ratio of ∼2.6:1:∼0.4, as determined by β-scanning (Table 1). The S protein was excluded from quantitative analysis because it is present in small amounts in virus particles, occurs in both cleaved and uncleaved forms, and is easily lost during virus purification.
TABLE 1.
Stoichiometry of N, M, and HE proteins in immunopurified virionsa
| Virus | Relative amt of HE | Relative amt of N | Relative amt of M |
|---|---|---|---|
| MHV-A59 | 0 | 1 | 2.5 ± 0 |
| MHV-HE+ | 0.38 ± 0.02 | 1 | 2.5 ± 0.05 |
| MHV-HE0 | 0.35 ± 0.005 | 1 | 2.7 ± 0.2 |
| MHV-HE− | 0 | 1 | 2.7 ± 0.1 |
| MHV-S | 0.37 ± 0.06 | 1 | 2.6 ± 0.2 |
MHV-infected cells were subjected to steady-state labeling with [35S]Met. Virions were immunopurified with a MAb against M or S. Virion proteins were separated by SDS-PAGE. The amount of radioactivity present in each band was determined with a STORM PhosphorImager/Fluorimager 860 (Molecular Dynamics) and ImageQuant software. Relative ratios of HE, N, and M were calculated from the number of Met residues present per protein. The data shown are based on three independent experiments. Data are means ± standard deviations.
Expression of HE decreases viral fitness in vitro.
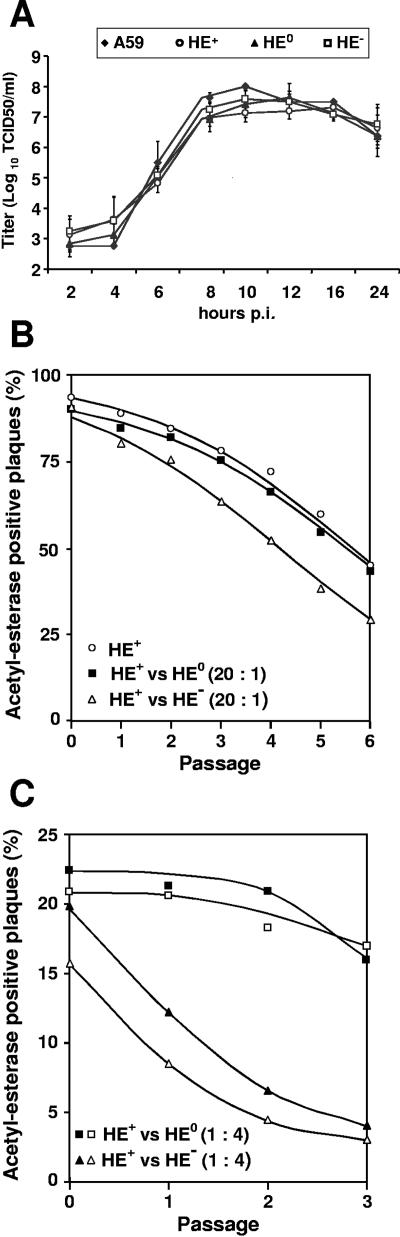
The effect of HE expression on in vitro propagation was determined by performing one-step growth experiments with LR7 cells. MHV-A59 and the rMHVs replicated with similar kinetics and to similar titers (Fig. 5A). Clearly, the expression of HE or the lack thereof did not result in gross differences in in vitro growth properties. However, during serial passaging of rMHV-HE+ viruses (with each passage involving a multistep infection of LR7 cell monolayers after inoculation at an MOI of 0.01 PFU/cell), we consistently noted the emergence and rapid accumulation of O-acetylesterase-deficient mutants (Fig. 5B). These mutants increased in the population at a relative rate of ∼1.66 per passage compared to the parental virus. To study whether this apparent difference in relative replication rate was somehow related to the enzymatic activity of HE, we performed propagation competition assays with rMHV-HE+ and rMHV-HE0 mixed at a 20:1 ratio. Accordingly, the number of O-acetylesterase-deficient plaques in the starting inoculum (passage 0) increased ∼5%. However, O-acetylesterase-deficient plaques accumulated during the subsequent five passages at a similar, and in fact, slightly reduced, rate to that seen during serial passaging of rMHV-HE+ alone (Fig. 5B). This is precisely as predicted if the increase in O-acetylesterase-deficient viruses during mixed passaging is due exclusively to the accumulation of the spontaneous HE-deficient mutants already present in the rMHV-HE+ stocks. These observations were reproducible with two additional combinations of independently isolated rMHV-HE+ and -HE0 recombinant viruses (Fig. 5C); note that in these last experiments, the initial inoculum contained HE+ and HE0 viruses mixed at ratios of approximately 1:4 to dampen the influence of the spontaneous acetylesterase-deficient mutants in the rMHV-HE+ stocks. We concluded that viruses expressing an enzymatically inactive HE replicate with equal efficiencies as syngeneic viruses with a functional O-acetylesterase.
FIG. 5.
In vitro growth properties of rMHV-HE+, -HE0, and -HE−. (A) Single-step growth kinetics of MHV recombinants. LR7 cells were infected with MHV-A59 or with each of the recombinant MHVs at an MOI of 10. Viral infectivity present in the culture medium at different times postinfection was determined by titration in LR7 cells by end-point dilution, and titers (50% tissue culture infective doses/ml [TCID50/ml]) were calculated. (B and C) Relative fitness of recombinant MHVs, as measured in mixed propagation-competition assays. rMHV-HE+ was serially passaged in LR7 cells either alone or in combination with rMHV-HE0 or rMHV-HE−, mixed at the indicated ratios. The propagation-competition experiments shown were performed with three different sets of independently isolated rMHVs (indicated by open and solid squares and triangles). Monolayers were inoculated at an MOI of 0.01. Tissue culture supernatants were harvested at 16 h p.i. and analyzed by plaque assays and in situ esterase staining. The graphs show the percentage of acetylesterase-positive plaques (y axis) in each of the passages (x axis). For graph B, at least 200 plaques were counted for each sample after every passage (average, 440 ± 170), and for graph C, at least 700 plaques were counted for each sample (average, 990 ± 340).
Conversely, in competition-propagation experiments with rMHV-HE+ and rMHV-HE−, the accumulation of O-acetylesterase-deficient viruses was accelerated (Fig. 5B), with the total population of HE-negative mutants increasing at a relative replication rate of 1.66. Again, identical results were obtained with other combinations of independently isolated rMHV-HE+ and -HE− viruses (Fig. 5C). Apparently, rMHV- HE− viruses grew to higher titers than viruses expressing intact HE and replicated with an efficiency equal to that of the spontaneous O-acetylesterase-deficient rMHV mutants endogenous to the rMHV-HE+ stocks.
To determine the defects in these spontaneous O-acetylesterase-deficient mutants, nine of them were randomly selected by plaque purification from a P8 stock of rMHV-HE+. Sequence analysis revealed that three of them (type 1) contained a one-nucleotide insertion in the HE gene; an extra cytidylate residue had been added to a stretch of five nucleotides, located at positions 1209 to 1213. This caused a frameshift at Pro404, immediately upstream of the predicted transmembrane domain. The other mutants (type 2) had a single nucleotide substitution in the HE gene, G70→T, as a result of which Gly24 was replaced by Trp. The mutant viruses expressed HE at intracellular levels equal to (type 2) or only slightly less than (type 1) that seen for rMHV-HE+ (Fig. 6, top panel), yet they failed to incorporate HE into their virions (Fig. 6, bottom panel).
FIG. 6.
Spontaneous acetylesterase-deficient mutants of rMHV-HE+ produce defective HE proteins which are not incorporated into viral particles. (Upper panel) Analysis of intracellular viral proteins. Cells were mock infected (m) or infected with rMHV-HE+ (HE+), rMHV-HE− (HE−), or rMHV-HE+ mutant type 1 or 2. The cells were metabolically labeled from 5 to 6 h p.i. with 35S in vitro cell-labeling mix (Amersham), and cell lysates were subjected to RIPA with MAb 4G12-2F9 (α HE) or antiserum K135 (α A59). Precipitates were analyzed by electrophoresis in SDS-polyacrylamide gels followed by fluorography. Bands corresponding to the structural proteins M, N, HE, and S (u, uncleaved; c, cleaved) are indicated. MAb 4G12-2F9 bound a 30-kDa host cell protein in addition to the HE protein. (Lower panel) Protein content of immunopurified virions. Supernatants of metabolically labeled cells which had been mock infected or infected with rMHV-HE+ (HE+), rMHV-HE− (HE−), or mutant type 1 or 2 were subjected to immunopurification with MAb J1.3 (α M) or with MAb J1.7 (α S). Precipitates were analyzed in 15% SDS-PAGE gels. Bands corresponding to the structural proteins M, N, HE, and S (u, uncleaved; c, cleaved) are indicated.
DISCUSSION
Coronaviruses code for a “core” set of essential proteins, comprising nonstructural proteins involved in RNA synthesis as well as the structural proteins S, E, M, and N. In addition, each coronavirus produces a variable number of accessory proteins [accessory in the sense that these proteins (i) are dispensable for replication in vitro and (ii) are not expressed by all coronaviruses]. One of these is HE, an envelope glycoprotein with sialate-O-acetylesterase activity which only occurs in group 2 coronaviruses. The expression of HE is thought to increase viral fitness during natural infection. However, in various cell culture-adapted murine coronavirus strains, including MHV-A59, the HE gene has been rendered inactive by mutations (30, 66). Here we provide an explanation for this phenomenon by demonstrating that during propagation in cell culture, MHV-A59-derived rMHVs which express a properly folded HE have a replication disadvantage compared to isogenic HE-deficient viruses.
Our data show that although the parental MHV-A59 strain has lost HE expression, it can still efficiently incorporate the HE protein into its virion, thus confirming and extending observations by Liao et al. (29). rMHVs expressing either wild-type HE or the enzymatically inactive derivative HE0 incorporated the HE protein into the viral envelope in quantities similar to those found for the naturally HE-expressing strain MHV-S. In immunopurified virus particles, the HE, N, and M proteins were determined to be present at a ratio of ∼0.4:1:∼2.6. Note that the estimated stoichiometry of N and M in these MHV preparations is in good agreement with that determined for transmissible gastroenteritis virus (1:3) (14). Low-multiplicity serial passage experiments with rMHV-HE+ variants showed, however, that HE expression was not stable. In plaque-purified virus stocks, O-acetylesterase-deficient mutants arose spontaneously and rapidly replaced the parental virus. This phenomenon appeared to be cell type independent, as a progressive loss of O-acetylesterase activity was observed not only during serial passaging of rMHV-HE+ in LR7 cells, but also during propagation in Sac(−) and 17ClI cells (data not shown). The rates at which these mutants emerged and accumulated as well as their limited genetic diversity (further discussed below) strongly argue against models in which the loss of HE is explained by a probabilistic accumulation of neutral mutations. Apparently, the expression of HE affects viral fitness in vitro, with “fitness” being defined here as the relative replication capacity of virus populations during mixed propagation (13, 18).
At first glance, our findings might be interpreted to indicate that the enzymatic activity of HE has a negative effect on viral replication. However, a direct comparison in competition-propagation experiments, which allowed a reproducible quantitative assessment of differences in the relative fitness of virus populations (18), showed that rMHV-HE0 and rMHV-HE+ viruses replicated at equal rates. In contrast, HE-deficient rMHV-HE− viruses grew more efficiently than rMHV-HE+. These combined observations lead us to believe that it is not the O-acetylesterase activity, but rather the expression of an intact HE per se, that reduces viral fitness in vitro.
Conceivably, HE synthesis might exert a negative effect on viral replication in cultured cells by drawing on the cell's economy or even by directly interfering with the production of other viral proteins, in particular the S protein, e.g., by competing for folding enzymes, chaperones, and other resources in the endoplasmic reticulum. Still, this does not seem to be the case. The spontaneous mutants that replaced rMHV-HE+ during serial passaging produced large quantities of HE proteins which were defective but nevertheless (almost) full-sized. Of the two types of mutants identified, one had a frameshift mutation immediately upstream of the transmembrane domain, and as a consequence, expressed an anchorless protein; the other had suffered a single Gly24→Trp substitution in HE. Saliently, Gly24 is the C-terminal residue of the signal sequence (16). Apparently, either mutation caused the HE protein to misfold, resulting in a loss of O-acetylesterase activity and, more importantly, exclusion from the assembly process: both mutants produced virions that lacked the HE protein. While in the case of the anchorless HE, this is no surprise, it is intriguing that a single mutation within the signal peptide of HE has the same effect. Preliminary experiments indicated that the mutation does not prevent removal of the signal sequence (Lissenberg and de Groot, unpublished data) but that cleavage most likely occurs at an alternative site.
We propose that the reduction of in vitro propagation efficiency associated with the expression of wild-type HE is related to the incorporation of this protein into virus particles. HE might exert this effect at the level of virion assembly, for example, by reducing the number of progeny viruses per cell, or at a postassembly level, for instance, by decreasing the specific infectivity of virus particles. In our serial passage experiments, viruses that lacked HE in their envelopes produced approximately 1.7 times more PFU per passage than rMHV-HE+. It is noteworthy, however, that each passage entailed a multistep infection initiated at a low multiplicity (0.01 PFU/cell). Consequently, the actual difference in replication efficiency between HE+ and HE− viruses per single round of infection will be much smaller and would go unnoticed in one-step growth experiments. Thus, our data also illustrate that even modest differences in fitness may have major consequences during prolonged viral propagation.
It seems reasonable to assume that HE expression has a similar effect on the efficiency of virus propagation in vivo. Still, in naturally occurring rodent coronavirus populations, HE expression seems to be maintained. In fact, rodent coronaviruses frequently exchange HE sequences via homologous RNA recombination, which on one occasion has even led to a shift in HE substrate specificity from Neu5,9Ac2 to Neu4,5Ac2 or vice versa (47). Presumably, this diversity among murine coronaviruses is brought about by immune pressure for antigenic variation. In any event, the observation that HE genes are substituted rather than lost strongly argues for an important role of this dispensable “luxury” protein during natural infection. Apparently, the costs of maintaining HE expression, which become obvious only during in vitro propagation (see below) and during experimentally induced persistent infection of the central nervous system (68), are outweighed by the benefits under field conditions.
Precisely how HE contributes to viral propagation in vivo remains to be established. Herrler and coworkers (23, 40, 42, 51) have proposed that the binding of virions to O-acetylated sialic acids is mediated primarily by the S protein. The HE would be a minor receptor-binding protein at best and would, in analogy to the neuraminidase in influenza A and B viruses, mainly function as a virion-associated receptor-destroying enzyme. This model, however, is based on studies of BCoV and HCoV-OC43; whether it holds true for all group 2 coronaviruses remains to be seen. In fact, recent reverse genetic experiments performed in our laboratory suggest that for MHV strain DVIM, the HE protein is the main sialic acid-binding protein (M. A. Langereis, A. L. W. van Vliet, and R. J. de Groot, unpublished). In the accompanying paper (19a), we show that HE promotes the spread of MHV in the brain and that, surprisingly, this property is not related to its enzymatic activity. The combined findings lend support to the idea that MHV HE does mediate viral attachment to sialic acid after all, and by acting as a coreceptor-binding protein, might facilitate viral dissemination.
Acknowledgments
A.L. was supported by The Netherlands Digestive Disease Foundation project WS98-41.
REFERENCES
- 1.Blau, D. M., C. Turbide, M. Tremblay, M. Olson, S. Letourneau, E. Michaliszyn, S. Jothy, K. V. Holmes, and N. Beauchemin. 2001. Targeted disruption of the Ceacam1 (MHVR) gene leads to reduced susceptibility of mice to mouse hepatitis virus infection. J. Virol. 75:8173-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breslin, J. J., I. Mork, M. K. Smith, L. K. Vogel, E. M. Hemmila, A. Bonavia, P. J. Talbot, H. Sjostrom, O. Noren, and K. V. Holmes. 2003. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at 37°C. J. Virol. 77:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brian, D. A., B. G. Hogue, and T. E. Kienzle. 1995. The coronavirus hemagglutinin esterase glycoprotein, p. 165-179. In S. G. Siddell (ed.), The coronaviridae. Plenum Press, New York, N.Y.
- 7.Bridger, J. C., E. O. Caul, and S. I. Egglestone. 1978. Replication of an enteric bovine coronavirus in intestinal organ cultures. Arch. Virol. 57:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callebaut, P. E., and M. B. Pensaert. 1980. Characterization and isolation of structural polypeptides in haemagglutinating encephalomyelitis virus. J. Gen. Virol. 48:193-204. [DOI] [PubMed] [Google Scholar]
- 9.Cannon-Carlson, S., and J. Tang. 1997. Modification of the Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis procedure to eliminate artifacts on reducing and nonreducing gels. Anal. Biochem. 246:146-148. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh, D. 1995. The coronavirus surface glycoprotein, p. 73-113. In S. G. Siddell (ed.), The coronaviridae. Plenum Press, New York, N.Y.
- 11.Cornelissen, L. A., C. M. Wierda, F. J. van der Meer, A. A. Herrewegh, M. C. Horzinek, H. F. Egberink, and R. J. de Groot. 1997. Hemagglutinin-esterase, a novel structural protein of torovirus. J. Virol. 71:5277-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo, E., C. Escarmís, L. Menéndez-Aria, and J. J. Holland. 1999. Origin and evolution of viruses. Academic Press, San Diego, Calif.
- 14.Escors, D., E. Camafeita, J. Ortego, H. Laude, and L. Enjuanes. 2001. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 75:12228-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogue, B. G., T. E. Kienzle, and D. A. Brian. 1989. Synthesis and processing of the bovine enteric coronavirus haemagglutinin protein. J. Gen. Virol. 70:345-352. [DOI] [PubMed] [Google Scholar]
- 17.Hogue, B. G., B. King, and D. A. Brian. 1984. Antigenic relationships among proteins of bovine coronavirus, human respiratory coronavirus OC43, and mouse hepatitis coronavirus A59. J. Virol. 51:384-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Kazi, L., A. Lissenberg, R. Watson, R. J. de Groot, and S. R. Weiss. 2005. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 79:15064-15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, B., and D. A. Brian. 1982. Bovine coronavirus structural proteins. J. Virol. 42:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King, B., B. J. Potts, and D. A. Brian. 1985. Bovine coronavirus hemagglutinin protein. Virus Res. 2:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klausegger, A., B. Strobl, G. Regl, A. Kaser, W. Luytjes, and R. Vlasak. 1999. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 73:3737-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel, F., and G. Herrler. 1993. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology 195:195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Monica, N., L. R. Banner, V. L. Morris, and M. M. Lai. 1991. Localization of extensive deletions in the structural genes of two neurotropic variants of murine coronavirus JHM. Virology 182:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavi, E., D. H. Gilden, M. K. Highkin, and S. R. Weiss. 1986. The organ tropism of mouse hepatitis virus A59 in mice is dependent on dose and route of inoculation. Lab. Anim. Sci. 36:130-135. [PubMed] [Google Scholar]
- 28.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao, C. L., X. Zhang, and M. M. Lai. 1995. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology 208:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luytjes, W., P. J. Bredenbeek, A. F. Noten, M. C. Horzinek, and W. J. Spaan. 1988. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita, E., H. Ebina, A. Muto, H. Himeno, K. Hatakeyama, and K. Sugiyama. 1998. Primary structures of hemagglutinin-esterase and spike glycoproteins of murine coronavirus DVIM. Virus Genes 17:123-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, T. G. 2001. The three faces of paramyxovirus attachment proteins. Trends Microbiol. 9:103-105. [DOI] [PubMed] [Google Scholar]
- 33.Olsen, C. W., W. V. Corapi, C. K. Ngichabe, J. D. Baines, and F. W. Scott. 1992. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J. Virol. 66:956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker, M. D., D. Yoo, and L. A. Babiuk. 1990. Expression and secretion of the bovine coronavirus hemagglutinin-esterase glycoprotein by insect cells infected with recombinant baculoviruses. J. Virol. 64:1625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfleiderer, M., E. Routledge, G. Herrler, and S. G. Siddell. 1991. High level transient expression of the murine coronavirus haemagglutinin-esterase. J. Gen. Virol. 72:1309-1315. [DOI] [PubMed] [Google Scholar]
- 36.Regl, G., A. Kaser, M. Iwersen, H. Schmid, G. Kohla, B. Strobl, U. Vilas, R. Schauer, and R. Vlasak. 1999. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J. Virol. 73:4721-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottier, P. J., M. C. Horzinek, and B. A. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarma, J. D., L. Fu, S. T. Hingley, and E. Lavi. 2001. Mouse hepatitis virus type-2 infection in mice: an experimental model system of acute meningitis and hepatitis. Exp. Mol. Pathol. 71:1-12. [DOI] [PubMed] [Google Scholar]
- 39.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultze, B., H. J. Gross, R. Brossmer, and G. Herrler. 1991. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 65:6232-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultze, B., H. J. Gross, R. Brossmer, H. D. Klenk, and G. Herrler. 1990. Hemagglutinating encephalomyelitis virus attaches to N-acetyl-9-O-acetylneuraminic acid-containing receptors on erythrocytes: comparison with bovine coronavirus and influenza C virus. Virus Res. 16:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultze, B., and G. Herrler. 1992. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J. Gen. Virol. 73:901-906. [DOI] [PubMed] [Google Scholar]
- 43.Schultze, B., K. Wahn, H. D. Klenk, and G. Herrler. 1991. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 180:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shieh, C. K., H. J. Lee, K. Yokomori, N. La Monica, S. Makino, and M. M. Lai. 1989. Identification of a new transcriptional initiation site and the corresponding functional gene 2b in the murine coronavirus RNA genome. J. Virol. 63:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddell, S. G. 1995. The coronaviridae: an introduction, p. 1-10. In S. G. Siddell (ed.), The coronaviridae. Plenum Press, New York, N.Y.
- 46.Siddell, S. G. 1982. Coronavirus JHM: tryptic peptide fingerprinting of virion proteins and intracellular polypeptides. J. Gen. Virol. 62:259-269. [DOI] [PubMed] [Google Scholar]
- 47.Smits, S. L., G. J. Gerwig, A. L. van Vliet, A. Lissenberg, P. Briza, J. P. Kamerling, R. Vlasak, and R. J. de Groot. 2004. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of corona- and toroviral receptor-destroying enzymes. J. Biol. Chem. 280:6933-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., J. A. den Boon, M. C. Horzinek, and W. J. Spaan. 1991. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology 180:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spaan, W. J., P. J. Rottier, M. C. Horzinek, and B. A. van der Zeijst. 1981. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59). Virology 108:424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storz, J., G. Herrler, D. R. Snodgrass, K. A. Hussain, X. M. Zhang, M. A. Clark, and R. Rott. 1991. Monoclonal antibodies differentiate between the haemagglutinating and the receptor-destroying activities of bovine coronavirus. J. Gen. Virol. 72:2817-2820. [DOI] [PubMed] [Google Scholar]
- 52.Strasser, P., U. Unger, B. Strobl, U. Vilas, and R. Vlasak. 2004. Recombinant viral sialate-O-acetylesterases. Glycoconj. J. 20:551-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugiyama, K., and Y. Amano. 1981. Morphological and biological properties of a new coronavirus associated with diarrhea in infant mice. Arch. Virol. 67:241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugiyama, K., M. Kasai, S. Kato, H. Kasai, and K. Hatakeyama. 1998. Haemagglutinin-esterase protein (HE) of murine corona virus: DVIM (diarrhea virus of infant mice). Arch. Virol. 143:1523-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taguchi, F., and J. O. Fleming. 1989. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology 169:233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlasak, R., W. Luytjes, J. Leider, W. Spaan, and P. Palese. 1988. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 62:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vlasak, R., W. Luytjes, W. Spaan, and P. Palese. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. USA 85:4526-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlasak, R., T. Muster, A. M. Lauro, J. C. Powers, and P. Palese. 1989. Influenza C virus esterase: analysis of catalytic site, inhibition, and possible function. J. Virol. 63:2056-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagaman, P. C., H. A. Spence, and R. J. O'Callaghan. 1989. Detection of influenza C virus by using an in situ esterase assay. J. Clin. Microbiol. 27:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner, R., M. Matrosovich, and H. D. Klenk. 2002. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev. Med. Virol. 12:159-166. [DOI] [PubMed] [Google Scholar]
- 63.Wurzer, W. J., K. Obojes, and R. Vlasak. 2002. The sialate-4-O-acetylesterases of coronaviruses related to mouse hepatitis virus: a proposal to reorganize group 2 Coronaviridae. J. Gen. Virol. 83:395-402. [DOI] [PubMed] [Google Scholar]
- 64.Yokomori, K., M. Asanaka, S. A. Stohlman, S. Makino, R. A. Shubin, W. Gilmore, L. P. Weiner, F. I. Wang, and M. M. Lai. 1995. Neuropathogenicity of mouse hepatitis virus JHM isolates differing in hemagglutinin-esterase protein expression. J. Neurovirol. 1:330-339. [DOI] [PubMed] [Google Scholar]
- 65.Yokomori, K., S. C. Baker, S. A. Stohlman, and M. M. Lai. 1992. Hemagglutinin-esterase-specific monoclonal antibodies alter the neuropathogenicity of mouse hepatitis virus. J. Virol. 66:2865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yokomori, K., L. R. Banner, and M. M. Lai. 1991. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology 183:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yokomori, K., N. La Monica, S. Makino, C. K. Shieh, and M. M. Lai. 1989. Biosynthesis, structure, and biological activities of envelope protein gp65 of murine coronavirus. Virology 173:683-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokomori, K., S. A. Stohlman, and M. M. Lai. 1993. The detection and characterization of multiple hemagglutinin-esterase (HE)-defective viruses in the mouse brain during subacute demyelination induced by mouse hepatitis virus. Virology 192:170-178. [DOI] [PubMed] [Google Scholar]
- 69.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37°C either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang, X., D. R. Hinton, S. Park, B. Parra, C. L. Liao, M. M. Lai, and S. A. Stohlman. 1998. Expression of hemagglutinin/esterase by a mouse hepatitis virus coronavirus defective-interfering RNA alters viral pathogenesis. Virology 242:170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zoltick, P. W., J. L. Leibowitz, E. L. Oleszak, and S. R. Weiss. 1990. Mouse hepatitis virus ORF 2a is expressed in the cytosol of infected mouse fibroblasts. Virology 174:605-607. [DOI] [PMC free article] [PubMed] [Google Scholar]