Abstract
TRIM-CypA is an owl monkey-specific variant of the retrovirus restriction factor TRIM5α. Here, we exploit its modular domain organization and cyclosporine sensitivity to probe the kinetics and mechanism of TRIM5-mediated restriction. Time of addition/withdrawal experiments reveal that inhibition of incoming human immunodeficiency virus type 1 capsids by TRIM-CypA occurs within minutes of their delivery to the target cell cytoplasm. However, while TRIM-CypA restriction is partly dependent on a RING domain, restriction occurs independently of the ubiquitin/proteasome system. Moreover, tagged TRIM-CypA proteins can be fully active as restriction factors without forming cytoplasmic bodies.
Many mammalian cells express factors that inhibit retroviral replication (4, 8). One such factor is encoded by the tripartite motif 5 gene (TRIM5α) (25) and can restrict infection by targeting the incoming capsids of an array of retroviruses (9, 11, 18, 24, 25, 31). The TRIM5α protein consists of an N-terminal tripartite motif (RING, B-box, and coiled coil) which mediates restriction and is linked to a C-terminal B30.2/SPRY domain, which determines restriction specificity and is likely responsible for capsid recognition (17, 20, 23, 26, 32). A special case exists in owl monkeys, where a retrotransposition event has placed a cyclophilin A (CypA)-pseudogene into the TRIM5 locus, resulting in the expression of a protein in which CypA replaces the B30.2/SPRY domain (16, 21). Remarkably, TRIM-CypA exhibits the human immunodeficiency virus type 1 (HIV-1) CA-specific binding property of CypA (13) but retains the infection-inhibiting properties of TRIM5α.
Kinetics of TRIM-CypA-mediated restriction.
Both TRIM5α and TRIM-CypA prevent the accumulation of viral DNA (2, 3, 6, 11, 15, 21, 25, 28). However, whether restriction occurs prior to reverse transcription or causes the destruction of nascent reverse transcripts is unknown. Restriction by endogenously or exogenously expressed TRIM-CypA is abolished by cyclosporine (CsA), an inhibitor of the HIV-1 capsid-CypA interaction (16, 21, 28), and we exploited this property to determine precisely when restriction occurs. A green fluorescent protein (GFP)-expressing HIV-1 reporter virus bearing a gp120 envelope from the HXB2 strain (33) was bound to owl monkey kidney cells (OMK) stably expressing CD4 and CXCR4, on ice. After cells were washed, virus entry was initiated by shifting to 37°C, and infection events were counted by fluorescence-activated cell sorter (FACS) analysis 48 h later (Fig. 1A). The progressive loss of viral sensitivity to potent inhibitors of entry (dextran sulfate) or reverse transcription (UC781) in the minutes or hours following the temperature shift was taken as a measurement of the time at which these events occurred. Importantly, CsA was included throughout to measure these parameters in the absence of restriction. Using these criteria, virus entry began within 7 min and was 50% complete 30 to 40 min after the temperature shift (Fig. 1B and C). Completed reverse transcripts (UC781-insensitive infection events) first appeared 4 h after the temperature shift and achieved 50% of their maximal value by approximately 6 h (Fig. 1B and C).
FIG. 1.
Kinetic analysis of HIV-1 restriction by TRIM-CypA. (A) Experimental strategy. HXB-enveloped HIV-1 particles were bound to OMK-CD4/CXCR4 cells, and infection was initiated by a temperature shift. Infection events were quantitated 48 h after the temperature shift by enumerating GFP-positive cells using fluorescence-activated cell sorter analysis. The time taken to acquire resistance to subsequently applied inhibitors of entry and reverse transcription and the opposing effects of CsA application and removal were measured. (B) Quantitation of infection events (percent GFP-positive cells at 48 h after the temperature shift) following timed addition of an entry inhibitor, dextran sulfate (Dex S) or UC781, a nonnucleoside reverse transcriptase (RT) inhibitor (dashed lines). Addition of either inhibitor at the time of the temperature shift (0 h) reduced infection to undetectable levels (>1,000-fold). Alternatively, TRIM-CypA restriction was activated or inhibited by the withdrawal or addition of CsA (solid lines). (C) A portion of the data in panel B is shown, except that only early time points are plotted on an expanded time axis and infection events are plotted on a linear scale as a percentage of their maximum uninhibited level.
The presence or absence of CsA during virus binding, i.e., prior to the temperature shift, did not affect restriction or infectivity, indicating that restriction rapidly became active after CsA removal (Fig. 1B). However, removal of CsA (i.e., TRIM-CypA activation) at timed intervals after the temperature shift revealed that escape from restriction required CsA to be present for only about 30 min after virus entry. In a reciprocal experiment, HIV-1 entry was initiated in the context of active restriction (the absence of CsA). Thereafter, CsA was applied at measured times after the temperature shift in an attempt to rescue the infectivity of restricted capsids. Infectivity could be completely rescued by application of CsA only up to about 15 min after the temperature shift. Thereafter, “restorable” infectivity decayed rapidly (Fig. 1B and C). In fact, this decay occurred with approximately the same kinetics as resistance to restriction was acquired by incoming virions in the CsA withdrawal experiment (Fig. 1C). Specifically, only about 40%, 10%, and 2% of the applied infectivity could be rescued by CsA application 1, 2, and 4 h after the temperature shift, respectively. Residual infectivity that was rescued by CsA at these later time points was largely accounted for by residual dextran sulfate-sensitive infectivity (i.e., virions that were yet to enter the target cell). Overall, therefore, TRIM-CypA restriction of HIV-1 infection occurred rapidly after virus entry (within minutes) and was apparently irreversible.
TRIM-CypA restriction does not require cytoplasmic bodies.
TRIM5α and TRIM-CypA can form large aggregated structures termed cytoplasmic bodies (1, 19), but one study has suggested that N-tropic murine leukemia virus (N-MLV) and HIV-1 can be inhibited by TRIM5α without cytoplasmic body formation (17). To address this issue in the context of TRIM-CypA, we generated Mus dunni (MDTF) cell clones stably expressing C-terminally hemagglutinin (HA)-tagged TRIM-CypA (Fig. 2A and B) or, alternatively, cyan fluorescent protein (CFP)-tagged proteins at two different levels of expression (clones 1 and 2) (Fig. 2A and C). Both TRIM-CypA-HA and TRIM-CypA-CFP were highly active, and MDTF cells expressing these proteins resisted HIV-1 infection but not infection with HIV(SIV CA), which carries an SIVMAC capsid in an otherwise HIV-1 background (9), as effectively as OMK cells (Fig. 2A). Importantly, deconvolution microscopy showed that TRIM-CypA-HA and TRIM-CypA-CFP were primarily distributed throughout the cytoplasm in a diffuse or fine punctate pattern (Fig. 2D). Visibly aggregated TRIM-CypA, or “cytoplasmic bodies,” were observed only rarely in about 5% of TRIM-CypA-HA and TRIM-CypA-CFP1 cells. Conversely, in TRIM-CypA-CFP2 cells, which express higher levels of TRIM-CypA-CFP (Fig. 2C), filamentous accumulations and cytoplasmic bodies were observed in about 30% of cells (Fig. 2D). In contrast, when cells were treated with sodium butyrate, which resulted in 4- to 10-fold-increased levels of expression (Fig. 2B and C), TRIM-Cyp-HA and TRIM-Cyp-CFP more frequently localized in intensely fluorescent cytoplasmic bodies (Fig. 2D). This effect was especially evident in highly expressing, sodium butyrate-treated TRIM-CypA-CFP2 cells, where most of the TRIM-CypA resided in cytoplasmic bodies in >90% of cells. Thus, while TRIM-CypA could adopt dramatically different localizations, depending primarily on expression level, cytoplasmic body formation was clearly not required for potent restriction (Fig. 2A).
FIG. 2.
Cytoplasmic body formation by TRIM-CypA occurs only at high expression levels and is not required for restriction. (A) OMK cells, unmodified MDTF cells (none), or cloned MDTF cells stably expressing TRIM-CypA-HA or TRIM-CypA-CFP (clones 1 and 2) using LNCX2-derived retroviral vectors were infected with VSV-G-pseudotyped HIV-1 or HIV(SIV CA) vectors, as indicated. The percentage of infected (GFP-positive) cells as determined by FACS analysis is plotted, and results of one representative of two independent experiments are shown. (B) Comparison of the expression levels of TRIM-CypA-HA in untreated and sodium butyrate-treated MDTF cells. Cell lysates were fourfold serially diluted (beginning at 105 cell equivalents per lane) and subjected to Western blot (WB) analysis using an anti-HA (α-HA) tag monoclonal antibody. (C) Same as panel B except that TRIM-CypA-CFP (clones 1 and 2) we analyzed using an anti-GFP (αGFP) antibody. Note that the anti-HA antibody permits more-sensitive detection than the anti-GFP antibody; therefore, the signals in panels B and C cannot be meaningfully compared with each other. (D) Localization of TRIM-CypA-HA and TRIMCyp-CFP in untreated cells (left) and cells treated with 10 mM sodium butyrate for 16 h prior to fixation (right). TRIM-Cyp-HA localization in fixed and permeabilized cells was determined using anti-HA primary antibodies and Alexa-594-congugated secondary antibodies. TRIM-CypA-CFP localization was determined in fixed but otherwise unmanipulated cells. A single 0.25-μm deconvolved optical section is shown in each image, and selection of three representative images is shown for each cell line/condition. IF, immunofluorescence.
TRIM-CypA restriction does not require ubiquitin-activating (E1) enzyme or proteasome activity.
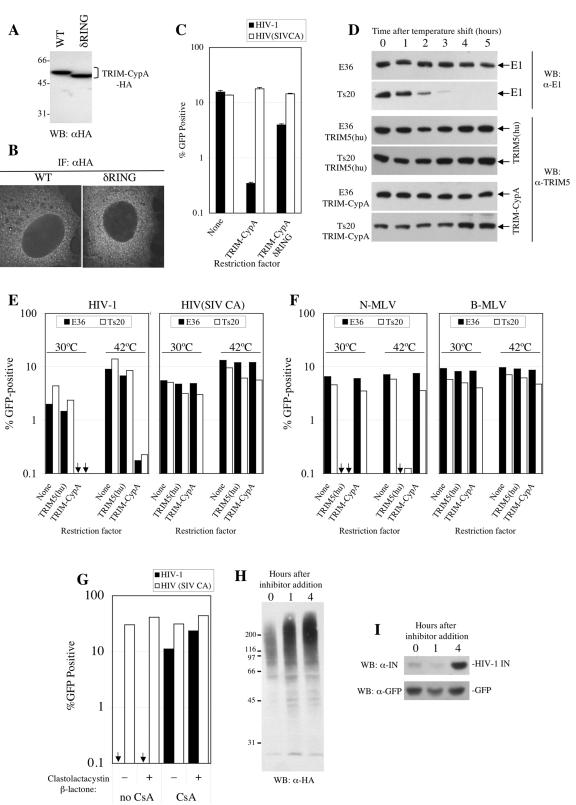
Several potential mechanisms by which restriction factors inhibit infection at a very early postentry phase could be envisaged, including ubiquitination and proteasome-mediated degradation of capsid components. Notably, a RING domain at the amino terminus of TRIM5 proteins has been shown to exhibit self-ubiquitination activity (30). Previous studies demonstrated a requirement for the RING domain for full TRIM5α activity (10, 17, 25) but could not distinguish whether it is required for capsid recognition or for secondary activities associated with restriction. Because TRIM-CypA carries an autonomous CA-binding domain (CypA), that should not, in principle, require the RING domain for capsid recognition, we tested the HIV-1-restricting activity of full-length and δRING versions of C-terminally HA-tagged TRIM-CypA. Each protein was stably expressed at equivalent levels in MDTF cells (Fig. 3A) and was similarly localized in a diffuse or fine punctate pattern in the cytoplasm (Fig. 3B). As expected, TRIM-CypA inhibited vesicular stomatitis virus G (VSV-G)-pseudotyped HIV-1 vector infection by about 40-fold but did not inhibit infection by HIV-1(SIV CA) (Fig. 3C). The δRING TRIM-CypA protein also specifically inhibited HIV-1 infection, albeit modestly (approximately fourfold). Thus, TRIM5 proteins lacking the RING domain can be weak restriction factors, but as is the case in TRIM5α restriction of HIV-1 or MLV, the RING domain is required for full activity.
FIG. 3.
Requirement for the RING domain but not ubiquitin-activating (E1) enzyme or proteasome activity in TRIM-CypA restriction. (A) Expression of TRIM-CypA and δRING TRIM-CypA (lacking residues 2 to 60). Lysates from G418-resistant pools of Mus dunni (MDTF) cells transduced with LNCX2-derived retroviral vectors expressing the indicated proteins were analyzed by Western blot (WB) analysis using an anti-HA (α-HA) monoclonal antibody. WT, wild type. (B) Localization of HA-tagged TRIM-CypA or δRING TRIM-CypA. The same MDTF cells analyzed in panel A were subjected to immunofluorescence (IF) analysis as described for Fig. 2D. (C) Restriction activity of TRIM-CypA and δRING TRIM-CypA in MDTF cells. Unmodified MDTF cells or MDTF cells stably expressing HA-tagged TRIM-CypA or δRING TRIM-CypA were infected with VSV-G-pseudotyped HIV-1 or HIV(SIV CA) vectors, as indicated. The percentage of infected (GFP-positive) cells as determined by FACS analysis 48 h later is plotted. (D) Specific E1 removal from Ts20 cells. Unmodified E36 or Ts20 cells or cells engineered to express either TRIM5α or TRIM-CypA were cultured at 30°C (time zero) and subsequently shifted to 42°C for the indicated number of hours. Cell lysates were subjected to Western blotting with antibodies specific for ubiquitin-activating (E1) enzyme or TRIM5, as indicated. (E) Lack of requirement of E1 in TRIM-CypA restriction. Unmodified Ts20 or control E36 cells (None) or cells stably expressing either TRIM-CypA or human TRIM5α [TRIM5(hu)] were incubated at the indicated temperatures for 3 h prior to and 2 h subsequent to infection with a VSV-G-pseudotyped HIV-1 or HIV(SIV CA) vector, as indicated, before being washed and returned to 30°C. The percentages of infected (GFP-positive) cells as determined by FACS analysis 48 h later are plotted. (F) The same experiment as described for panel E was performed except that the various E36- and Ts20-derived cell lines were infected with TRIM5(hu)-restricted N-MLV or unrestricted B-tropic MLV vectors, as indicated. (G) Lack of effect of proteasome inhibition on TRIM-CypA restriction in OMK cells. Cells were infected with VSV-G-pseudotyped HIV-1 or HIV(SIV CA) vectors as indicated in the presence (+) or absence (−) of 10 μM clastolactacystin-β-lactone for 1 h prior, 2 h during, and 1 h after exposure to the inoculum. Infections were done under HIV-1-restricting conditions (no CsA) or in the absence of restriction (with CsA). (H, I) Effect of clastolactacystin-β-lactone on HA-ubiquitin conjugate accumulation or HIV-1 integrase expression. OMK cells were transfected with an HA ubiquitin expression plasmid (H) or pEGFP*IRES-Ub-F-IN CTE (14), which expresses both GFP and an HIV-I integrase bearing an N-terminal Phe residue (I). Cells were treated with clastolactacystin-β-lactone for the indicated number of hours, and lysates were subjected to Western blot analysis with antibodies to the HA tag (H), HIV-1 integrase (IN), or GFP (I).
Therefore, to address whether ubiquitination and proteasome activities are required for TRIM-CypA restriction, we took two approaches. The first employed a mutant of Chinese hamster E36 cells, termed Ts20, which carries a temperature-sensitive allele of the sole ubiquitin-activating (E1) enzyme in mammals, which is fully active at 30°C but completely inactivated within 1 h at 40°C (12). While incubation of Ts20 cells above 40°C has been shown to inactivate E1, block de novo ubiquitination activity, and inhibit ubiquitin-dependent processes (12, 27), we adopted a more stringent Western blot assay to monitor E1 protein depletion. Incubation of Ts20 cells but not control wild-type E36 cells at 42°C resulted in nearly complete removal of E1 protein within 3 h (Fig. 3D). Conversely, engineered expression of TRIM-CypA or TRIM5α was only marginally affected, if at all (Fig. 3D). Incubation of Ts20 or E36 cells at 42°C for 3 h followed by challenge with a VSV-G-pseudotyped HIV-1 vector for 2 h resulted in a severalfold increase in infection, but this was independent of the presence or absence of E1 or TRIM-CypA (Fig. 3D and E) and also occurred to some extent with unrestricted HIV-1(SIV CA) (Fig. 3E). Indeed, TRIM-CypA conferred strong (>40-fold) and specific resistance to HIV-1 at either 30°C or 42°C (Fig. 3F). A similar experiment carried out using E36 and Ts20 cells expressing human TRIM5α showed that N-MLV-specific restriction by human TRIM5α is also efficient in the absence of E1 (Fig. 3D and E).
A second approach was to treat target cells with a proteasome inhibitor, lactacystin. As was previously reported (5, 22), proteasome inhibition caused modest enhancement of HIV-1 infection under nonrestricting conditions (Fig. 3G). However, treatment of OMK target cells with clastolactacystin-β-lactone for 1 h prior to, 2 h during, and 1 h after challenge with VSV-G-pseudotyped HIV-1 (4 h total treatment) failed to influence restriction in OMK cells endogenously expressing TRIM-CypA (Fig. 3G) or MDTF cells engineered to express TRIM-CypA-HA (data not shown). To verify that proteasome activity was inhibited by clastolactacystin-β-lactone treatment of OMK cells, they were transfected with plasmids expressing HA-tagged ubiquitin (29) or HIV-1 integrase, an unstable proteasome substrate (14). Subsequent clastolactacystin-β-lactone treatment, for 1 h or 4 h, resulted in the marked accumulation of high-molecular-weight HA-ubiquitin conjugates that would normally be removed as a result of proteasome activity (Fig. 3H). Similarly, expression of HIV-1 integrase (which bears an N-terminal Phe residue and, because of the N-end rule, is rapidly degraded by proteasomes [14]) was markedly stabilized by 4 h of clastolactacystin-β-lactone treatment of OMK cells (Fig. 3I). Conversely, expression of a relatively stable protein (GFP) was unaffected (Fig. 3I). Thus, we conclude that while full TRIM-CypA restriction activity requires the RING domain, neither an E1 enzyme nor active proteasomes appear essential. However, we cannot definitively exclude the possibility that long-lived ubiquitin-E2 conjugates and ubiquitin-dependent but proteasome-independent mechanisms might have a role in restriction.
Conclusions.
The rapidity with which TRIM-CypA restriction occurs suggests that the subviral structure that is targeted either exists only transiently after virus entry or moves quickly to locations within the cell where restriction cannot occur. Although it is possible that the TRIM-CypA-capsid interaction is not reversible by CsA because of stable polyvalent binding (7), the apparently irreversible nature of TRIM-CypA restriction implies destruction or some other form of inactivation of the incoming capsid, perhaps the promotion or inhibition of capsid disassembly. While it seems likely that the restriction mechanisms are largely shared by TRIM-CypA and TRIM5α, the different modes with which incoming capsids are recognized could impact restriction kinetics and reversibility. However, two suspected activities of TRIM-CypA, shared by TRIM5α, namely, cytoplasmic body formation and ubiquitin ligation, appear not to be required for restriction.
Acknowledgments
We thank Annie Yang for technical assistance, Peter Lopez for assistance with microscopy, and D. Bohmann, Ben Mulder, and Bryan Cullen for reagents.
This work was supported by a grant from the NIH (RO1AI6400 to P.D.B.) and a postdoctoral fellowship from amFAR (to T.H.) P.D.B is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation.
REFERENCES
- 1.Berthoux, L., S. Sebastian, D. M. Sayah, and J. Luban. 2005. Disruption of human TRIM5α antiviral activity by nonhuman primate orthologues. J. Virol. 79:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2004. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J. Virol. 78:11739-11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5:1109-1115. [DOI] [PubMed] [Google Scholar]
- 5.Butler, S. L., E. P. Johnson, and F. D. Bushman. 2002. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J. Virol. 76:3739-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forshey, B. M., J. Shi, and C. Aiken. 2005. Structural requirements for recognition of the human immunodeficiency virus type 1 core during host restriction in owl monkey cells. J. Virol. 79:869-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16:849-859. [DOI] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 11.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 13.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 14.Mulder, L. C., and M. A. Muesing. 2000. Degradation of HIV-1 integrase by the N-end rule pathway. J. Biol. Chem. 275:29749-29753. [DOI] [PubMed] [Google Scholar]
- 15.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human TRIM5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reymond, A., G. Meroni, A. Fantozzi, G. Merla, S. Cairo, L. Luzi, D. Riganelli, E. Zanaria, S. Messali, S. Cainarca, A. Guffanti, S. Minucci, P. G. Pelicci, and A. Ballabio. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sebastian, S., and J. Luban. 2005. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology 2:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, B., H. Javanbakht, M. Perron, H. Park do, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 26.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strous, G. J., P. van Kerkhof, R. Govers, A. Ciechanover, and A. L. Schwartz. 1996. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 15:3806-3812. [PMC free article] [PubMed] [Google Scholar]
- 28.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 29.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 30.Xu, L., L. Yang, P. K. Moitra, K. Hashimoto, P. Rallabhandi, S. Kaul, G. Meroni, J. P. Jensen, A. M. Weissman, and P. D'Arpa. 2003. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Exp. Cell Res. 288:84-93. [DOI] [PubMed] [Google Scholar]
- 31.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Y. J., T. Hatziioannou, T. Zang, D. Braaten, J. Luban, S. P. Goff, and P. D. Bieniasz. 2002. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 76:6332-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]