Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 as an important cause of severe lower respiratory tract infection in humans, and in vitro models of the lung are needed to elucidate cellular targets and the consequences of viral infection. The SARS-CoV receptor, human angiotensin 1-converting enzyme 2 (hACE2), was detected in ciliated airway epithelial cells of human airway tissues derived from nasal or tracheobronchial regions, suggesting that SARS-CoV may infect the proximal airways. To assess infectivity in an in vitro model of human ciliated airway epithelia (HAE) derived from nasal and tracheobronchial airway regions, we generated recombinant SARS-CoV by deletion of open reading frame 7a/7b (ORF7a/7b) and insertion of the green fluorescent protein (GFP), resulting in SARS-CoV GFP. SARS-CoV GFP replicated to titers similar to those of wild-type viruses in cell lines. SARS-CoV specifically infected HAE via the apical surface and replicated to titers of 107 PFU/ml by 48 h postinfection. Polyclonal antisera directed against hACE2 blocked virus infection and replication, suggesting that hACE2 is the primary receptor for SARS-CoV infection of HAE. SARS-CoV structural proteins and virions localized to ciliated epithelial cells. Infection was highly cytolytic, as infected ciliated cells were necrotic and shed over time onto the luminal surface of the epithelium. SARS-CoV GFP also replicated to a lesser extent in ciliated cell cultures derived from hamster or rhesus monkey airways. Efficient SARS-CoV infection of ciliated cells in HAE provides a useful in vitro model of human lung origin to study characteristics of SARS-CoV replication and pathogenesis.
Infection of human respiratory airways by classic human coronaviruses (HCoV) (e.g., HCoV-229E and HCoV-OC43) typically produces mild common cold symptoms, although more serious disease has been reported to occur in infants and individuals with underlying comorbidities (42). However, infection by a recently emerged HCoV can lead to a fatal pneumonia characterized as severe acute respiratory syndrome (SARS), and the newly identified HCoV is designated SARS coronavirus (SARS-CoV) (11, 25). SARS-CoV has caused about 8,000 cases and ∼800 deaths worldwide, with an ∼10% overall mortality rate prior to successful containment of the epidemic. SARS-CoV has been isolated from humans, civet cats, raccoon dogs, bats, and swine, suggesting that several animal species may function as natural reservoirs for future outbreaks (17, 28). In some cases, humans may display mild disease, raising the possibility for asymptomatic carriage and maintenance of virus in human populations (22, 52). The SARS-CoV discovery also led to the identification of two new HCoV, HCoV-NL63 and HCoV-HKU1, both of which are associated with more-serious lower respiratory tract infections in humans (62, 67). Given the growing importance of HCoV as pathogens that produce severe human respiratory diseases, relevant model systems are needed to elucidate the underlying molecular mechanisms governing coronavirus pathogenesis and virulence in the human lung. SARS-CoV infection is an attractive model for HCoV infection, as it produces severe disease in the human lung, it replicates efficiently in vitro, a molecular clone is available to identify the genetic determinants governing pathogenesis and virulence, and a variety of animal models are under development (16, 34, 41, 44, 45, 70).
The SARS-CoV virion contains a single-stranded, positive-polarity, 29,700-nucleotide RNA genome bound by the nucleocapsid protein (N). The capsid is packaged by a lipid bilayer containing at least three structural proteins, including a 180-kDa spike glycoprotein (S) that interacts with human angiotensin 1-converting enzyme 2 (hACE2) to mediate viral entry into cells (30). In addition to the 23-kDa membrane glycoprotein (M), the envelope (E) protein is likely essential for efficient virion maturation and release. More recently, open reading frame 3a (ORF3a) was shown to encode a fourth virion protein, although its functions in maturation and release are unknown (72). The SARS-CoV genome contains nine ORFs, the first of which encodes the viral replicase proteins required for subgenomic- and genome-length RNA synthesis and virus replication (see Fig. 2A) (32, 46, 48). Based on studies with other coronaviruses, it is likely that SARS-CoV uses a transcription attenuation model to synthesize both full-length and subgenomic-length negative-strand RNAs containing antileader RNAs, which then function as templates for the synthesis of like-sized mRNAs (2, 49, 50). ORF2 to ORF8 are located on eight subgenomic mRNAs synthesized as a nested set of 3′ coterminal RNA species in which the leader RNA sequences on the 5′ end of the genome are joined to body sequences at distinct transcription regulatory sequences containing a highly conserved consensus sequence (53, 59, 70). The SARS-CoV consensus sequence is ACGAAC (32, 46, 53, 59, 70). The virion structural gene products S, the ORF3a protein, M, E, and N are encoded in mRNA transcripts 2, 3, 4, 5, and 9a, respectively, while the group-specific ORFs are located on mRNAs 3, 6, 7, 8 and 9. Interspaced among the SARS-CoV structural genes are the group-specific genes (ORF3b, ORF6, ORF7a/7b, ORF8a/8b, and ORF9b), which are not conserved in other coronaviruses and whose functions in replication and pathogenesis are generally unknown (32, 46, 53). Group-specific ORFs from other coronaviruses, such as mouse hepatitis virus, feline infectious peritonitis virus, and transmissible gastroenteritis virus, usually encode luxury functions for replication in vitro (9, 10, 66). In the case of these viruses, group-specific genes can be deleted, oftentimes attenuating pathogenesis in vivo (9, 10, 18). The development of a molecular clone for SARS-CoV provides a useful tool to study replication and pathogenesis by allowing direct manipulation of the viral genome (70).
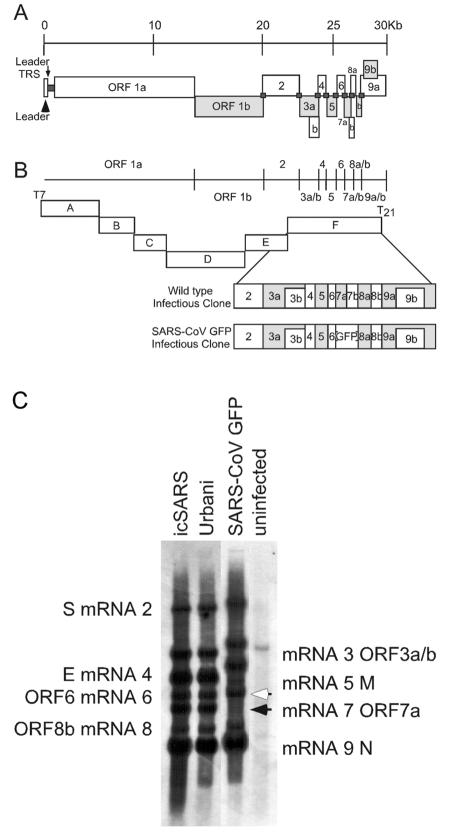
FIG. 2.
Schematics of SARS-CoV GFP construct and SARS-CoV infectious cloning strategy and subgenomic RNA transcription levels. (A) Schematic representation of the SARS-CoV genome, with all defined ORFs indicated by rectangles. Viral leader sequences are indicated by dark gray squares at the 5′ end of the genome (arrowhead) and between each ORF. The arrow indicates the 5′ viral leader transcription regulatory sequence (TRS). (B) Schematic representation of the SARS-CoV infectious cloning strategy and the mutations to engineer the GFP into ORFs7a/7b. The fragments of the genome are indicated by rectangles with the ORFs in each, as shown in panel A. The ORFs of the F clone have been expanded to indicate the location of GFP within the SARS-CoV GFP infectious-clone construct. (C) To determine levels of viral RNA synthesis from wild-type and SARS-CoV GFP viruses, total RNA was isolated from infected cells at 12 h postinfection and probed with a nucleocapsid-specific leader-containing probe. mRNAs 2 through 9 were detected for all samples, demonstrating that deletion of ORF7a/7b allowed the synthesis of all subgenomic RNA species. The filled arrowhead indicates wild-type mRNA 7, and the open arrowhead indicates the increased size of mRNA 7 associated with ORF7a/b excision and replacement with GFP. Samples are indicated across the top of the gel, and the individual RNA species are specified to the left and right of the image.
The predominant pathological features of SARS-CoV infection of the human lung include diffuse alveolar damage, atypical pneumonia with dry cough, persistent fever, progressive dyspnea, and in some cases acute exacerbation of lung function. Major pathological lesions include inflammatory exudation in the alveoli and interstitial tissue, with hyperplasia of fibrous tissue and fibrosis (5, 25, 27). In tissues isolated from fatal cases of SARS-CoV, virus was localized by fluorescence in situ hybridization within alveolar pneumocytes (primarily type II) and within alveolar spaces. Pathological assessment of histological samples of airway regions other than the alveolus have been less rigorously studied, as the alveolar regions have consistently demonstrated the most significant disease in late-stage fatal cases (7, 60). However, there are now several reports that examined early disease associated with SARS-CoV infection which noted marked bronchiolar disease with respiratory epithelial cell necrosis, loss of cilia, squamous cell metaplasia, and intrabronchiolar fibrin deposits. In fact, it has been suggested that early diffuse alveolar damage as a result of SARS-CoV infection may actually initiate at the level of the respiratory bronchioles (13, 39).
We have previously described the use of an in vitro model of human airway epithelium that recapitulates the morphological and physiological features of the human airway epithelium in vivo (43, 74). By use of this model, it has been shown that common human respiratory viruses, e.g., human respiratory syncytial virus (RSV) and human parainfluenza virus type 3 (PIV3), exclusively infect ciliated cells after intraluminal inoculation, raising the possibility that these cells play a major role in pathogenesis of respiratory virus infection. Here, we test the ability of SARS-CoV to infect human ciliated airway epithelia (HAE) in vitro to determine whether infection and spread of SARS-CoV throughout the ciliated conducting airway epithelium may provide a valid model for understanding the pathogenesis of SARS-CoV lung disease. To aid assessment of SARS-CoV infection of HAE, we have generated a recombinant clone of the Urbani strain of SARS-CoV that expresses green fluorescent protein (GFP), SARS-CoV GFP, to allow infection and spread to be monitored over time. We demonstrate that SARS-CoV infects ciliated airway epithelial cells via an interaction with hACE2 on the apical surface of ciliated cells. Although progeny virus was initially shed into the luminal compartment of the epithelium, at later times postinfection, virus was also shed into basolateral compartments. Since ciliated airway epithelial cells possess unique physiological and innate defense functions in the human lung (e.g., mucociliary clearance), it is important to identify the ciliated cell tropism of SARS-CoV and the pathological consequences of infection of these cells.
MATERIALS AND METHODS
Construction of SARS-CoV GFP recombinant virus.
To replace ORF7a/7b with GFP, primer sets (i) no. 47 (5′-GTGCTTGCTGTTGTCTACAG-3′) and SARS Mu6 (5′-ATCGATCACCTGCCATGTTCGTTTTATGGATAATCTAACTCCATAGGTTC-3′) and (ii) SARS Mu7 (5′-ATCGATTTAATTAAGAGCTCACTTTAATTGACTTCTATTTGTG-3′) and 3′-Ngl(−) (5′-CTTTGCTCTCAAGCTGGTTC-3′) were used to generate amplicons flanking nucleotides 27,304 to 27,671 in the SARS-CoV genome. The amplicons were digested with ClaI to create a cassette in which ORF7a/7b was removed, making available AarI, ClaI, and PacI restriction sites. An AvrII cut amplicon was purified and ligated back into the SARS-CoV F cDNA clone (70), systematically deleting nucleotides 27,276 to 27,643 and inserting an AarI, ClaI, and PacI multiple cloning site in the SARS-CoV genome. The sequence-confirmed F fragment was used as the base plasmid for GFP insertion. A GFP amplicon was generated using primers 5′GFP(+) (5′-CACCTGCTAAACGAACAAATTAAAATGGTGAGCAAGGGCGAGGAG-3′) and 3′GFP(−) (5′-TTAATTAATTACTTGTACAGCTCGTCCATGC-3′). The GFP amplicon was then subcloned into the F fragment missing the ORF7a/7b sequence by use of the AarI and PacI restriction sites and sequenced, and this mutant F fragment was used to build the SARS-CoV GFP.
To assemble full-length SARS-CoV GFP cDNA, the SARS A through F inserts were restricted, separated through 0.8% agarose gels, visualized with a Dark Reader light box (Claire Chemical), excised, and purified using a Qiaex II DNA purification kit. The A through F fragments were ligated overnight, phenol-chloroform extracted, and precipitated with isopropyl alcohol. Full-length transcripts were generated in vitro as described by the manufacturer (mMessage mMachine; Ambion) with certain modifications. To produce full-length capped SARS-CoV N gene mRNA transcripts, 1 μg of plasmid DNA encoding the N gene forward primer (5′-NNGGCCTCGATGGCCATTTAGGTGACACTATAGATGTCTGATAATGGACCCCAATC-3′) and reverse primer (5′-NNNTTTTTTTTTTTTTTTTTTTTTTTTTTATGCCTGAGTTGAATCAGCAG-3′) was transcribed by SP6 RNA polymerase with a 2:1 ratio of cap analog to GTP.
Full-length RNA transcripts were transfected into 800 μl of Vero E6 cells (8.0 × 106) in an electroporation cuvette, and four electrical pulses of 450 V at 50 μF were given with a Gene Pulser II electroporator (Bio-Rad), similarly to approaches previously described by our laboratory (70, 71). The transfected Vero E6 cells were seeded in a 75-cm2 flask, incubated at 37°C, and monitored for GFP fluorescence. Clonal virus progeny were then isolated by plaque assay and monitored for GFP expression by fluorescence with an Olympus IX51 Research inverted microscope.
Growth curve analysis in cell lines.
To determine viral titers, samples were taken at 2, 8, 12, 16, 20, and 32 h postinfection and assessed by plaque assay. Briefly, supernatants from infected plates of Vero E6, MA104, and Caco-2 cells were serially diluted, inoculated onto Vero E6 cell monolayers in 60-mm dishes, and overlaid in complete media plus 0.8% agarose, and plaques were visualized at 48 h via neutral red staining.
Northern blot analysis.
Cultures of Vero E6 cells were inoculated with wild-type Urbani, infectious-clone SARS-CoV (icSARS-CoV), and SARS-CoV GFP at a multiplicity of infection of 1 for 1 h at 37°C. At 12 h postinfection, intracellular RNA was isolated using RiboPure reagents as directed by the manufacturer (Ambion, Austin, TX). mRNA was isolated using QIAGEN′s Oligotex mRNA spin column reagents according to the manufacturer's instructions (QIAGEN, Valencia, CA). The mRNA was treated with glyoxal and separated on agarose gels by use of NorthernMax-Gly according to the manufacturer's instructions (Ambion, Austin, TX). The RNA was transferred to a BrightStar-Plus membrane (Ambion) for 4 to 5 h, and the RNA was cross-linked to the membrane by UV light. The blot was prehybridized and probed with an N-gene-specific oligodeoxynucleotide probe (5′-CTTGACTGCCGCCTCTGCTbTbCCCTbCTbGCb-3′), where biotinylated nucleotides are designated with a superscript b. Blots were hybridized overnight and washed with low- and high-stringency buffers as recommended by the manufacturer. Filters were incubated with streptavidin-AP, washed, and then incubated with chemiluminescent substrate CDP-STAR (New England BioLabs). Filters were overlaid with film and developed.
Cell cultures.
Vero E6 and MA104 cells were maintained in Eagle's minimal essential medium containing 10% fetal calf serum, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). Caco-2 cells were maintained in Dulbecco's high-glucose minimal essential medium containing 20% fetal calf serum, 0.1 M nonessential amino acids, kanamycin (0.25 μg/ml), and gentamicin (0.05 μg/ml). Urbani and icSARS-CoV wild-type viruses and SARS-CoV GFP were propagated and plaque isolated on Vero E6 cells. All virus work was performed in a biological safety cabinet in a biosafety level 3 laboratory containing redundant exhaust fans; personnel were dressed in Tyvek suits, full hoods, and powered air-purifying respirators with high-efficiency particulate air (HEPA) and organic vapor filters.
Human nasal and tracheobronchial epithelial cells were obtained from airway specimens resected from patients undergoing elective surgery under UNC Institutional Review Board-approved protocols by the UNC Cystic Fibrosis Center Tissue Culture Core. Briefly, primary cells were expanded on plastic to generate passage 1 cells and plated at a density of 250,000 cells per well on permeable Transwell-Col (12-mm-diameter) supports (14, 43). HAE cultures were generated by provision of an air-liquid interface for 4 to 6 weeks to form well-differentiated, polarized cultures that resemble in vivo pseudostratified mucociliary epithelium (43). Cultures of airway epithelial cells derived from alveolar regions of the human lung were obtained by plating primary human alveolar cells (ScienCell Research Labs, CA) or A549 cells on Transwell-Col supports and maintained for 5 days before viral inoculation in manufacturer-supplied medium or Dulbecco's high-glucose minimal essential medium, respectively.
Apical or basolateral virus inoculations were performed with 200 μl of virus stocks applied to the apical or basolateral surfaces of HAE, respectively. Prior to apical inoculation, the apical surfaces of HAE were rinsed three times over 30 min with phosphate-buffered saline (PBS) at 37°C. Basolateral inoculations were performed by inverting the Transwell-Col insert prior to addition of virus. Following a 2-h viral inoculation at 37°C, all inocula were removed and HAE were maintained with an air-liquid interface for the remainder of the experiment. To generate growth curves at specific times after viral inoculation, 120 μl of tissue culture medium was applied to the apical surface of HAE and collected after a 10-min incubation at 37°C. Basolateral samples were collected by removing 120 μl of basolateral medium at each time point. All samples were stored at −80°C until assayed for plaque formation on Vero E6 cells as described above.
Mouse, hamster, and rhesus monkey airway epithelial cell cultures.
Briefly, mouse and hamster tracheas were excised between the larynx and bronchial branches by use of a sterile technique. Epithelial cells were isolated following overnight incubation with pronase and subsequent incubation with pancreatic DNase I. Following a short incubation to allow for fibroblast adherence, mouse epithelial cells were plated in TEC Plus (TEC basic medium plus 10 μg/ml insulin, 5 μg/ml transferrin, 0.1 μg/ml cholera toxin, 25 ng/ml epidermal growth factor [Becton Dickinson, Bedford, MA], 30 μg/ml bovine pituitary extract, 5% fetal bovine serum, and freshly added 0.01 μM retinoic acid) on rat-tail-collagen-coated Transwell-Clear membranes at various cell densities ranging from 100,000 to 250,000 cells per well. Following the fibroblast adherence step, hamster cells were plated on Transwell-Col membranes at 100,000 to 200,000 cells per well. Once a confluent monolayer was established, apical medium was removed and cells were maintained at the air-liquid interface. From this point on, cells were grown in TEC/NS (NuSerum; Becton Dickinson) (47). Basolateral medium was refreshed every 2 days, and cultures were washed with PBS on the apical surface once per week. Rhesus monkey tracheobronchial specimens were a gift from Rebecca Grant (Nonhuman Primate Research Program, UPENN), and ciliated airway cultures were established exactly as described above for the human airway cultures.
Microscopy.
For detection of viral structural antigens, antisera against SARS-CoV structural S and N proteins were generated by inoculation of mice with Venezuelan equine encephalitis virus (VEE) replicon particles (VRPs) expressing the individual consensus sequences of S and N. Briefly, S and N were cloned into the pVR21 VEE replicon vector by overlapping-extension PCR with SARS-CoV- and VEE-specific primer pairs and PCR conditions previously described for the insertion of norovirus capsid genes (3). VRPs carrying the SARS-CoV S and N ORFs were produced as previously described (20, 21). The VRPs were inoculated into the footpads of 4-week-old mice and boosted at 30 days postinfection, prior to the harvesting of antiserum (20, 21). Mouse anti-S and -N titers exceeded 1:10,000.
For detection of SARS-CoV antigens in HAE, cultures inoculated with Urbani, icSARS-CoV, and SARS-CoV GFP (5 × 106 PFU/ml) were fixed in 4% paraformaldehyde (PFA) for 24 h, transferred to 70% ethanol, and prepared as paraffin-embedded histological sections by the UNC Cystic Fibrosis Center Morphology and Morphometrics Core. After deparaffinization, histological sections were incubated for 1 h in PBS containing 3% bovine serum albumin (BSA). Primary antibodies were applied at a 1:100 dilution in PBS with 1% BSA overnight and detected with anti-mouse antibody conjugated to Texas Red (Jackson ImmunoResearch). For detection of hACE2 in human airway samples and HAE, frozen sections of fresh nasal and tracheobronchial airways or HAE were fixed in ice-cold methanol, blocked in 3% BSA-PBS, and then probed overnight with goat polyclonal anti-hACE2 (AB933; R&D Systems) or an irrelevant antibody control (goat anti-biotin) at a 1:100 dilution, followed by a donkey anti-goat immunoglobulin G conjugated to Texas Red. Immunofluorescence was visualized with a Leica Leitz DMIRB inverted fluorescence microscope equipped with a cooled color charge-coupled digital camera (MicroPublisher; QImaging). A tricolor filter cube set (GFP/Texas Red/DAPI [4′,6′-diamidino-2-phenylindole]) was used to show the morphology of the tissue section (by combining low-level autofluorescence levels across the three filters), thus aiding determination of fluorescent antibody localization to specific regions of the cells. Visualization of virus with transmission scanning electron microscopy was performed by using standard techniques with HAE inoculated with Urbani, icSARS-CoV, or SARS-CoV GFP fixed at 48 h postinfection in 2% PFA-2% glutaraldehyde. Representative images infected with Urbani are shown.
Antibody blockade.
To determine whether SARS-CoV infection could be blocked with anti-hACE2, the apical surfaces of HAE were rinsed and 300 μl of a 1:10 dilution of monoclonal anti-hACE2 (MAB933; R&D Systems), polyclonal anti-hACE2 (AB933; R&D Systems), or control antibody (anti-MUC1, clone b27.29, a gift from Fujirebio Diagnostics, Inc.) (8) was applied to the apical surface of HAE; 2 h later, antisera were removed from the apical surface. SARS-CoV GFP (200 μl; 5 × 106 PFU/ml) or PIV3 expressing GFP (107 PFU/ml) was then inoculated onto the apical surface and incubated for an additional 2 h at 37°C. Viral inocula were then removed, and 30 μl of the original diluted antiserum was returned to the apical surface of HAE. As described above for SARS-CoV GFP, apical washes were collected at 2, 6, 12, 22, and 36 h postinfection to assess viral growth titers by plaque assay in Vero E6 cells. At 36 h postinfection, HAE were fixed (4% PFA) prior to quantitative assessment of GFP expression performed with image-processing tool kit plug-ins for Photoshop (ISBN 1-928808-00-X; John Russ).
RESULTS
Localization of hACE2 in human airway epithelia ex vivo and in vitro.
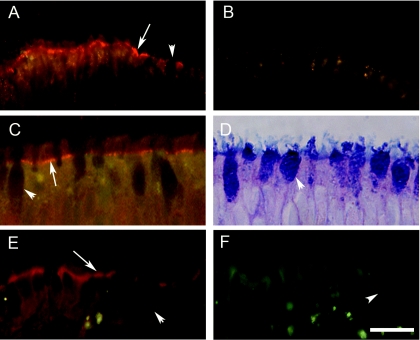
To determine if upper (nasal) and lower (tracheobronchial) regions of human conducting airway epithelia expressed the SARS-CoV receptor, hACE2, histological sections of freshly excised human airway tissue isolated from the nasal and tracheobronchial regions were probed with polyclonal antisera specific for hACE2 and immunoreactivity was assessed by fluorescent secondary antibody. In tissue sections of nasal (Fig. 1A and B) and tracheobronchial (Fig. 1C and D) regions, hACE2 immunoreactivity was detected on the luminal surface of the airway epithelia and was specifically localized to the apical membranes of ciliated airway epithelial cells. Although hACE2 was present at the apical surface of ciliated cells, it was not detected in association with the cilial shafts per se but rather was localized to regions corresponding to the apical membranes of these cells (Fig. 1A and C). To determine whether human ciliated airway epithelium cultures showed a similar distribution of hACE2 in vitro, we also probed histological sections of HAE with hACE2 antibodies. hACE2 was detected on the luminal surface of HAE and localized specifically to the apical surface of ciliated cells (Fig. 1E and F), thus recapitulating the findings for hACE2 localization in ex vivo tissue. Given the previously reported data that hACE2 serves as a receptor for SARS-CoV (30), the localization of hACE2 to the luminal membrane of ciliated cells suggests that SARS-CoV that enters the lumen of the human airway may utilize ciliated cells as a primary cell target for infection. Since hACE2 was localized to ciliated cells in vitro and ex vivo and we have previously reported that ciliated HAE are infected by several common human respiratory viruses, we predicted that the HAE model would serve as an excellent system to study characteristics of SARS-CoV infection, replication, spread, and pathogenesis (73, 74).
FIG. 1.
Localization of hACE2 on the apical surface of human airway epithelium ex vivo and in vitro. Representative histological frozen sections of freshly excised human nasal (A and B) or human tracheobronchial (C and D) airway tissues or HAE (E and F) were probed with either goat polyclonal anti-hACE (A, C, and E) or goat polyclonal anti-biotin (B and F) as a species-specific negative-control antibody. Bound primary antibody was visualized using donkey anti-goat secondary antibody conjugated to Texas Red. Immunofluorescence indicative of hACE (red) was detected in nasal and tracheobronchial tissue as well as HAE and was localized specifically to the apical membrane of ciliated cells (arrows). Nonciliated cell types present in the airway tissue or HAE were negative for hACE immunolocalization (arrowheads). Panel D shows Alcian blue (pH 2.5)- periodic acid-Schiff staining of human tracheobronchial airway tissue to highlight nonciliated cells (mucin-containing cells) present in the epithelium. Representative images are from specimens obtained from three different patients. Bar, 30 μm.
Construction and characterization of the SARS-CoV GFP virus construct.
To directly observe the extent and kinetics of SARS-CoV infection of HAE in real time, we constructed recombinant, GFP-expressing SARS-CoV (SARS-CoV GFP) to monitor infection. A schematic of the design of SARS-CoV GFP and the SARS-CoV cloning strategy are shown in Fig. 2A and B, respectively. To generate recombinant SARS-CoV GFP, the F plasmid was mutated to replace ORF7a/7b with the GFP cDNA, using the type IIS restriction enzyme approach described previously (71). The mutated F fragment was confirmed by sequence analysis and amplified in Escherichia coli with wild-type fragments A through E. Fragments were then isolated by digestion with the appropriate restriction enzyme, purified, and ligated into a genomic-length cDNA. The full-length cDNA was transcribed, and genomic-length RNA was coelectroporated with SARS-CoV N transcripts into Vero E6 cells. GFP-positive cells were detected within 24 h of transfection, and clarified supernatant from transfected cells when passed to fresh Vero E6 monolayers resulted in viral cytopathic effect and green fluorescent cells. The deletion of ORF7a/7b did not obviously affect efficient SARS-CoV replication in tissue culture, which is similar to observations with transmissible gastroenteritis virus and mouse hepatitis virus when GFP was inserted into accessory ORFs of these viruses (1, 9, 12, 54). Prior to further evaluation, five individual plaques of SARS-CoV GFP were isolated, amplified, and monitored for GFP fluorescence in addition to sequence confirmation of the mutation. All plaques contained GFP and had appropriate sequence mutations.
To determine if levels of replicated subgenomic RNA species from the SARS-CoV GFP construct were similar to those of wild-type SARS-CoV, total RNA was isolated from Urbani-, icSARS-CoV-, and SARS-CoV GFP-infected cells. Oligotex mRNA columns were used to enrich for poly(A)-containing mRNAs which were run on agarose gels and assayed by a leader-containing N-gene-specific probe (Fig. 2C). The total numbers of RNA species and relative levels of transcription were similar for Urbani, icSARS-CoV, and SARS-CoV GFP, suggesting that there was no transcriptional defect associated with deletion of ORF7a/7b. While the relative levels of subgenomic RNA were similar for all three viruses, the replacement of ORF7a/7b caused the expected shifts in size of subgenomic RNAs 2 through 7, demonstrating that the removal of ORF7a/7b was not detrimental to virus replication in vitro and that the GFP insert was stable. RNA isolated from mock-infected samples had no detectable leader-containing mRNA species.
To evaluate the growth kinetics of SARS-CoV GFP, growth curves were generated for three different mammalian epithelial cell lines. For Vero E6 cells, Urbani and SARS-CoV GFP grew to titers of ∼2 × 107 PFU/ml, while icSARS-CoV peaked at ∼1.0 × 107 PFU/ml (data not shown). For MA104 cells, all three viruses grew to titers of 3 × 107 to 5 × 107 PFU/ml by 32 h postinfection, whereas for Caco-2 cells, titers of all three viruses were reduced by approximately 1 log compared to titers of viruses with MA104 cells. Efficient replication of SARS-CoV GFP and wild-type virus was noted at both high and low multiplicities of infection (data not shown). Overall, replacing ORF7a/7b with GFP in SARS-CoV GFP was not detrimental to virus replication in any of the three cell lines evaluated, thus providing a fluorescent marker of virus infection with replication at wild-type virus levels.
SARS-CoV infection of human airway epithelia.
To determine whether SARS-CoV GFP could infect human airway epithelial cells derived from different regions of the lung, we prepared cultures of nasal and tracheobronchial ciliated epithelium as models of proximal airway epithelium and cultures of alveolar epithelium as models of distal lung regions. As a model of virus entry into the lumen of the airway, we inoculated the apical surfaces of these cultures with SARS-CoV GFP (5 × 106 PFU/ml) and assessed GFP fluorescence 48 h later. As shown in Fig. 3, HAE derived from nasal and tracheobronchial regions were efficiently infected by SARS-CoV GFP, with a large proportion of the cells expressing the marker transgene (Fig. 3A and B). In contrast, primary epithelial cells derived from alveolar regions of the human lung were poorly infected, if at all, by SARS-CoV GFP, with little evidence for GFP expression (Fig. 3C). Use of antisera against S also failed to detect SARS-CoV GFP or icSARS-CoV infection of these cells (data not shown). To determine if these cells were susceptible to infection with other common respiratory viruses in parallel, we also inoculated alveolar cultures with recombinant PIV3 or RSV expressing GFP (106 PFU). Both PIV3 (Fig. 3D) and RSV (not shown) efficiently infected primary cultures of alveolar cells. Similar data were obtained for SARS-CoV GFP, PIV3, and RSV inoculations of the alveolar cell line A549 (not shown). These data demonstrate that the human airway epithelium that lines the conducting airways of the lung was susceptible to infection by SARS-CoV GFP and that, in our hands, cells derived from the distal alveolar regions were poorly infected by SARS-CoV GFP.
FIG. 3.
SARS-CoV GFP infects human airway epithelial cells derived from nasal and tracheobronchial epithelia but not alveolar epithelium. The apical surfaces of HAE derived from nasal (A) or tracheobronchial (B) airway tissues or alveolar regions of the human lung (C and D) were inoculated with SARS-CoV GFP (A, B, and C) or human PIV3 expressing GFP (D), and 48 h later, GFP-positive cells were assessed with fluorescent microscopy. Although nasal and tracheobronchial HAE were efficiently infected by SARS-CoV GFP, alveolar-derived cells were poorly infected by SARS-CoV (C) but efficiently infected by PIV3 (D). Similar data were obtained for alveolar cultures derived from A549 cells.
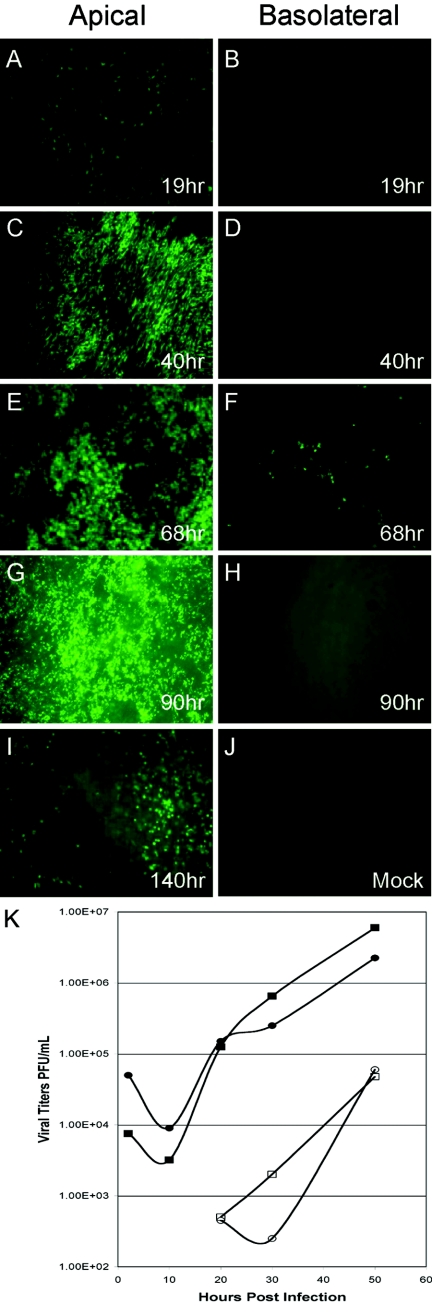
To determine whether SARS-CoV could spread throughout cells of HAE, SARS-CoV GFP was inoculated onto the apical or basolateral surface of cultures for 2 h at 37°C (5 × 106 PFU/ml) and levels of GFP fluorescence were monitored from 19 to 140 h postinfection. Inoculation of SARS-CoV GFP onto the apical surface of the HAE resulted in detectable levels of GFP as early as 19 h postinfection (Fig. 4A), with peak viral replication occurring between 68 (Fig. 4E) and 90 (Fig. 4G) h postinfection and GFP still detected at 140 h postinfection (Fig. 4I). In contrast, inoculation of the basolateral membrane of HAE with SARS-CoV GFP resulted in only a low number of fluorescent cells at 68 h postinfection, indicating that this was not the most productive route for SARS-CoV infection of HAE (Fig. 4B, D, F, and H). These data show that HAE are efficiently infected by SARS-CoV via the apical surface whereas basolateral surface inoculation is comparatively resistant to infection. Interestingly, by 140 h significant loss of GFP fluorescence was observed, an effect most likely related to the loss of SARS-CoV-infected cells (see below).
FIG.4.
Infection and spread of SARS-CoV GFP infection in HAE over time after apical or basolateral inoculation. HAE were inoculated via the apical (A, C, E, G, and I) or basolateral (B, D, F, and H) compartments with SARS-CoV GFP and GFP-positive cells and assessed over time (1 to 5 days postinfection). HAE inoculated with vehicle alone showed no GFP-positive cells (J). Apical inoculation resulted in significant numbers of GFP-positive cells at 40 h postinfection (C), with efficient spread of infection by 90 h postinfection (G). In contrast, basolateral inoculation resulted in a low proportion of cells positive for GFP only at 68 h postinfection (F). These images are representative of duplicate cultures from at least three different patient sets. Original magnification, ×10. (K) Apical inoculation of HAE with Urbani or icSARS-CoV was performed, and apical washes and basolateral media were harvested at the indicated time points postinfection. Collected samples were serially diluted, and titers were determined by plaque assay with Vero E6 cells. Titers are expressed as PFU/ml. Both Urbani and icSARS-CoV replicated to high titers in the apical compartment of HAE within 24 h, whereas progeny virions were detected in the basolateral compartment at later time points and to lower levels. All infections were performed in duplicate. Filled circles, Urbani apical; open circles, Urbani basolateral; filled squares, icSARS-CoV apical; open squares, icSARS-CoV basolateral.
To determine whether shedding of progeny SARS-CoV from HAE was polarized, apical washes and basolateral media were sampled at 2, 9, 21, 31, and 48 h postinfection with Urbani or icSARS-CoV and viral titers were assessed by plaque assay with Vero E6 cells (Fig. 4K). The peak titers of Urbani and icSARS-CoV shed from the apical surface approximated 107 PFU/ml, demonstrating a high level of replication similar to that observed with Vero E6 cell monolayers (Fig. 4K). In contrast, viral titers in the basolateral compartments were low for both Urbani and icSARS-CoV, with peak titers of 104 PFU/ml (Fig. 4K). Following apical inoculation, no infectious virus was detected until after 20 h postinfection in basolateral media. Since viral titers from the apical compartment of HAE were 2 to 3 logs higher than those detected from basolateral compartments for both viruses and the numbers of GFP-positive cells after apical and basolateral inoculation with SARS-CoV GFP also correlated with the different magnitudes of viral titers from the two compartments, these data suggest that within the first 48 h of infection SARS-CoV predominately sheds from the apical surface of HAE. An apical route of infection, shedding, and reinfection with progeny virus correlates with the apical localization of hACE2 on airway tissues (Fig. 1A, C, and E). Since SARS-CoV replicates to titers similar to those in permissive cell lines, these data indicate that SARS-CoV replicates well in HAE, providing a new model of the human lung epithelium for the study of SARS-CoV replication and pathogenesis.
Identification of SARS-CoV-infected cells in HAE.
Having demonstrated that SARS-CoV can productively infect HAE after apical inoculation, we next determined whether SARS-CoV targeted specific airway epithelial cell types. Since hACE2 was detected exclusively on the apical surfaces of ciliated cells in ex vivo human airway tissue and HAE in vitro, we predicted that ciliated cells would be infected by SARS-CoV. Histological sections from HAE infected with Urbani, icSARS-CoV, or SARS-CoV GFP for 48 h were probed with mouse polyclonal antisera directed against SARS-CoV structural proteins S or N, and localization was assessed by standard immunofluorescence techniques. For all viruses, S (Fig. 5A and B) and N (Fig. 5C) immunoreactivity was detected in ciliated cells, correlating with the localization of hACE2 to ciliated cell types (Fig. 1E). In experiments performed with HAE derived from four different patients, evidence of S immunoreactivity was detected only in cells that morphologically resembled ciliated cells. In all cases, no S or N immunoreactivity was detected in nonciliated cells. Interestingly, immunoreactivity against S was most abundant at the apical surface of ciliated cells in regions corresponding to the microvillus structures but low levels of S immunoreactivity were also detected within the cytoplasm of ciliated cells (Fig. 5B). In contrast, immunoreactivity to N protein was detected throughout the cytoplasm of SARS-CoV-infected ciliated cells, a localization indicative of the association of N with sites of CoV replication and assembly (Fig. 5C). Primary cultures of HAE that were confluent and polarized but not differentiated (i.e., cultures maintained at air-liquid interface for <5 days) were not infected by SARS-CoV as determined by GFP or S immunolocalization (data not shown).
FIG. 5.
SARS-CoV infects ciliated cells after apical inoculation of HAE. Representative histological sections of HAE 48 h postinfection with icSARS-CoV (A, B, and C) probed with mouse anti-S (A and B), mouse anti-N (C), or a mouse irrelevant anti-hemagglutinin (D) and visualized with anti-mouse secondary antibodies conjugated to Texas Red (red). Detection of S or N immunoreactivity was localized specifically to the ciliated cells of HAE (arrows), indicating that SARS-CoV infects this cell type after apical inoculation. No immunoreactivity was observed for nonciliated cell types (arrowheads). Images were obtained with a tricolor fluorescent filter to define the morphology of the tissue (gray/blue), with original magnifications of ×40 (A) and ×100 (B, C, and D).
To confirm that ciliated cells could be infected by SARS-CoV, we performed transmission electron microscopy on fixed HAE 48 h postinfection with Urbani, icSARS-CoV, or SARS-CoV GFP. HAE inoculated with SARS-CoV (representative pictures are of Urbani infection) showed ciliated cells containing classic coronavirus cytoplasmic vesicles filled with viral particles (Fig. 6B, G, and H). In addition, large numbers of viral particles were seen within the spaces between the microvilli/cilia shafts as well as in the airway surface microenvironment that surrounds the apical surface of ciliated cells, suggesting mechanisms for the release of large quantities of SARS-CoV into the lumen of the conducting airway during viral replication (Fig. 6C, D, E, and F). Immuno-electron microscopy (EM) detecting S confirmed that particles detected in the airway surface microenvironment were indeed SARS-CoV virions (Fig. 6F). Therefore, SARS-CoV entry, replication, and release occur in hACE2-positive ciliated cells of HAE.
FIG. 6.
Ultrastructural localization of SARS-CoV in HAE. Representative transmission electron microscopic photomicrographs of HAE infected with Urbani SARS-CoV. (A) HAE inoculated with vehicle alone, demonstrating the typical morphological features of the apical surfaces of ciliated cells with prominent cilia and microvilli. (B to E) HAE inoculated with Urbani SARS-CoV 48 h before fixation and showing the presence of large numbers of virus particles in vesicles inside ciliated cells (B and E) and on the surface of ciliated cells (B, D, and E) or shed into pericilial regions (C). Large quantities of virions were noted on the surface of ciliated cells, where ciliated cells were identified by cilial basal bodies (E). (F to H) To confirm that the observed virions were SARS-CoV, immuno-EM was performed using polyclonal mouse antisera against S with secondary antibodies conjugated to 12-nm colloidal gold (F). SARS-CoV infection resulted in extrusion and shedding of infected ciliated cells into the airway surface microenvironment (G and H). Similar observations were seen with HAE infected with icSARS-CoV and SARS-CoV GFP. Scale bars are shown for each panel. Filled arrowheads, cilia; filled arrows, microvilli; open arrowheads, virions; thin-tailed arrow in panel F, immuno-EM colloidal gold.
SARS-CoV infection of ciliated epithelial cells is hACE2 dependent.
To determine if SARS-CoV infects ciliated cells via an interaction with hACE2, we utilized antibody blockade experiments with antisera directed against hACE2, a method that has previously been shown to block the interaction of SARS-CoV with hACE2 in Vero E6 cells (30). HAE were preincubated with polyclonal or monoclonal antisera directed against hACE2 (R&D Systems) or a control antibody that binds to the apical surface of HAE (anti-tethered mucin MUC1, clone b27.29) for 2 h prior to inoculation with SARS-CoV GFP (5 × 106 PFU/ml). Quantitative analyses of the number of GFP-positive cells 36 h postinfection revealed that HAE pretreated with polyclonal hACE2 antibody alone ablated the levels of GFP-positive cells compared to controls (Fig. 7A). However, no inhibition of SARS-CoV GFP infection was observed with the monoclonal hACE2, indicating that the epitope recognized by the monoclonal hACE2 does not sterically inhibit the interaction of this molecule with SARS-CoV. No inhibition of infection was observed with a control antibody that binds to a highly abundant epitope on the apical surface of HAE (MUC1) (55). None of these antibodies significantly affected infection of ciliated cells by a recombinant PIV3 expressing GFP, indicating that the steric hindrance of hACE2 was specific to SARS-CoV infection. To confirm that the addition of polyclonal hACE2 antisera reduced not only the number of GFP-positive cells but also the replication of viral progeny, we assessed the growth kinetics of SARS-CoV GFP generated from the apical surface of infected HAE in the absence and presence of the antibodies described above. Apical-surface sampling was performed from 2 to 36 h postinfection, and titers of virus were determined by plaque assay with Vero E6 cells. In the absence of antisera or in the presence of control antisera, SARS-CoV GFP replicated to titers of 107 PFU/ml (Fig. 7B), similar to titers detected with the wild-type Urbani and icSARS-CoV strains (Fig. 4K). In contrast, in the presence of hACE2 polyclonal antisera, viral titers were reduced by at least 2 logs. Monoclonal antisera against hACE2 again failed to effect viral growth, confirming that this antibody was not sufficient to block SARS-CoV entry into ciliated cells. Therefore, both assessment of SARS-CoV infection by analyses of GFP-positive cells and viral growth kinetics suggest that hACE2 is the predominant receptor used by SARS-CoV for infection of ciliated cells in HAE.
FIG. 7.
hACE2 is the primary receptor for SARS-CoV entry into HAE. (A) HAE were pretreated with polyclonal or monoclonal antisera directed against hACE2 (pACE2 and mACE2, respectively) or an anti-MUC1 negative-control antibody prior to inoculation with SARS-CoV GFP (black bar) or PIV3 expressing GFP (gray bar). Thirty-six hours postinfection, the numbers of GFP-positive cells for each antiserum treatment were assessed, demonstrating that pACE2 could ablate SARS-CoV infection of HAE but that mACE2 or MUC1 antisera had no effect on SARS-CoV infection. None of the antiserum treatments significantly affected PIV3 infection of HAE. (B) To assess effects of pretreatment of HAE with receptor-specific antisera on the growth kinetics of SARS-CoV infection, apical washes at the indicated time points were collected and results were determined by plaque assay with VeroE6 cells. Results represent data obtained from HAE derived from two different patients. Titers are expressed as PFU/ml. Filled squares, polyclonal hACE2 only; open squares, polyclonal and monoclonal hACE2; filled circles, monoclonal hACE2 only; filled triangles, MUC1; filled diamonds, no antiserum.
Consequences of SARS-CoV infection on ciliated epithelial cell morphology.
To determine the consequences of ciliated cell infection by SARS-CoV over time, histological sections derived from HAE infected with SARS-CoV GFP for 20 to 140 h were probed with anti-S antisera. GFP was used to assess infection in real time before HAE were fixed and sectioned. Immunoreactivity against S in ciliated cells could be detected as early as 24 h postinfection (data not shown) and by 48 h postinfection was robustly located at the apical surface of ciliated cells (Fig. 8A). By 72 h (Fig. 8B), S immunoreactivity remained at the apical surfaces of ciliated cells but was also present on the basolateral surfaces of the infected cells. Late in infection (96 and 120 h), S-positive ciliated cells appeared to extrude from the epithelium onto the luminal surface of HAE (Fig. 8C and D). Extruded ciliated cells often showed S immunoreactivity around the entire periphery of the cells (Fig. 8D). These data demonstrate that SARS-CoV infection of ciliated cells in HAE resulted in shedding of the infected ciliated cells into the airway lumen. Furthermore, the presence of SARS-CoV immunoreactivity at the basolateral surfaces at later time points postinfection suggests that SARS-CoV would also enter the basolateral compartments of HAE, as was shown in Fig. 4K.
FIG. 8.
Morphological consequences of SARS-CoV infection of ciliated cells. Representative histological sections of HAE inoculated with SARS-CoV GFP and probed with mouse anti-S conjugated to Texas Red at 48, 96, 120, and 144 h postinfection (A to D). S localization (red) was restricted to the apical surface of ciliated cells by 48 h (A), but by 96 h postinfection, in some cases S immunoreactivity was detected around the periphery of the ciliated cells (B) (arrow), suggesting basolateral shedding of SARS-CoV. By 120 h postinfection, infected ciliated cells began to extrude from the epithelium (C) (arrow), and by 144 h postinfection, infected ciliated cells or components thereof were shed into the luminal compartment of HAE (D) (arrow). Note that the shed ciliated cell was positive for S immunoreactivity around the periphery of the cell, suggesting that virus was being shed from these regions. Bar, 5 μm. Images were obtained with a tricolor fluorescent filter to define the morphology of the tissue (gray/blue), with an original magnification at ×100.
Susceptibility of animal airway epithelial cells to SARS-CoV infection.
SARS-CoV is an emerging pathogen whose natural host range has not been clearly established. However, in vivo studies have shown that SARS-CoV can replicate in the mouse, hamster, cat, civet cat, ferret, and nonhuman primate lung, and murine and hamster models for the study of SARS-CoV pathogenesis and vaccine production have been developed (34, 41, 44, 45). The civet cat likely represents an important reservoir for transmission to humans (17). Airway epithelial cell cultures similar to the human model described here but derived from susceptible species may provide an important comparative model to determine the mechanisms of pathogenesis and replication efficiency in alternative hosts. Towards this goal, we have successfully generated ciliated airway epithelium models derived from tracheobronchial airway epithelia of C57BL/6 mice (Fig. 9A), golden Syrian hamsters (HmAE) (Fig. 9B), and rhesus macaques (Fig. 9C). Inoculation of the apical surfaces of these cultures with SARS-CoV GFP indicated that SARS-CoV GFP replicated to some extent in ciliated cells of airway epithelia of rhesus macaques (Fig. 9F), to a lesser extent in HmAE (Fig. 9E), and to a much lesser extent in airway epithelia of C57BL/6 mice (Fig. 9D). Again, although fewer cells than HAE were infected, ciliated epithelial cells were the predominant cell targets for SARS-CoV GFP infection (data not shown). The growth of virus and the percentages of infected cells were greatly reduced compared with infection rates and viral titers achieved with HAE, as evidenced by titers of ∼106 PFU/ml in HmAE (e.g., SARS-CoV GFP only reached 1.4 × 106 PFU/ml after 72 h postinfection); however, a rough parallel was noted with infection of ciliated cells derived from human > nonhuman primate > hamster > mouse, roughly correlating to the severity of disease noted for each species (34, 37, 45, 56, 65). Clearly, HAE are the most robust airway model for studying SARS-CoV replication and pathogenesis in the lung.
FIG. 9.
SARS-CoV GFP infection of mouse, hamster, and rhesus monkey airway epithelial cell cultures. Ciliated airway epithelial cell cultures were derived from mice (A and D), hamsters (B and E), and rhesus monkeys (C and F) and inoculated via the apical surface with SARS-CoV GFP (106 PFU). Hematoxylin and eosin-stained histological sections are shown for each species (A, B, and C), and GFP fluorescent images were recorded 48 h postinfection (D, E, and F). Original magnifications were ×40 (A, B, and C) and ×10 (D, E, and F). Bar, 5 μm (A, B, and C).
DISCUSSION
The human conducting airway epithelia of the nasal and tracheobronchial regions consist of a pseudostratified mucociliary epithelium with predominant ciliated cells interspersed with mucus-secreting goblet cells overlaying a basal cell layer. The predominant cells of the cuboidal epithelium of the human distal bronchiolar airways are also ciliated cells. The airway epithelium is often the first tissue encountered by intraluminal pathogens, and ciliated cells are major targets for common respiratory viruses, such as RSV, PIV3, and influenza (36, 73, 74). In this study, in vitro models of human airway epithelia derived from nasal and tracheobronchial regions were susceptible to SARS-CoV infection after luminal inoculation of virus, and the cell types targeted preferentially by SARS-CoV were ciliated epithelial cells. In our hands, nonpolarized or polarized but not differentiated human airway epithelial cells derived from nasal or tracheobronchial airway specimens were not infected by SARS-CoV (data not shown). Previously, we showed that RSV and PIV3 also targeted ciliated cells and that HAE required ciliated cell differentiation for infection, since polarized but not differentiated HAE were not infected by these viruses (73, 74). Furthermore, we showed that the numbers of cells infected by RSV and PIV3 directly correlated with ciliogenesis, indicating that ciliated cell differentiation was necessary for infection (73, 74). Although we show that SARS-CoV targets ciliated cells in HAE, we cannot rule out the possibility that SARS-CoV also infects nonciliated airway epithelial cells in vivo. A variety of nonciliated cell types are present in HAE, with some cells possessing morphology and markers indicative for mucin-containing cells and others with morphology that resembles precursors of ciliated cells. Since we do not fully appreciate the range of nonciliated cells in HAE, it is possible that these cells do not represent the nonciliated cell types found in the airway epithelium in vivo. Rather, it is our intention in this paper to describe the ciliated cell tropism for SARS-CoV in the HAE model. A recent report shows that Calu-3 cells are susceptible to SARS-CoV infection (61) and that although these cells are derived from human lung epithelial cells and polarize when grown on semipermeable supports, these cells do not recapitulate the differentiated status of the airway epithelium in vivo as do the HAE reported in the current study. The fact that Calu-3 cells do not possess differentiation markers of the human airway epithelium (e.g., cilia) and are only moderately infected by SARS-CoV suggests that the HAE model may be more representative of SARS-CoV infection of the human airways.
Importantly, hACE2, a receptor for SARS-CoV, was localized to the ciliated cells of human nasal and tracheobronchial airway epithelia and to ciliated cells of HAE, reflecting the ciliated cell targeting capacity of SARS-CoV. Importantly, antibodies directed against hACE2 efficiently inhibit infection, replication, and spread of SARS-CoV in HAE (Fig. 7), similar to results reported with Vero E6 and 293 cells expressing hACE2 (30, 57), suggesting that hACE2 functions as the principle receptor for SARS-CoV infection of the conducting airway epithelium (Fig. 1 and 7). These data are in stark contrast to results obtained by Hamming et al. (19), where hACE2 expression was detected in the alveolar regions and the basal layer of the nonkeratinized squamous epithelium of the upper respiratory tract (18). Our findings of hACE2 expression and productive SARS-CoV infection in ciliated cells derived from the nasal and tracheobronchial airways dispute this claim and support the hypothesis that SARS-CoV initiates a productive infection in human ciliated airway epithelium. It is interesting to consider the biological significance of hACE2 expression on the luminal surface of ciliated cells. Recently, it has been reported that ACE2 and its role in the renin-angiotensin pathway serve a protective function against acute lung injury (24). In addition, SARS-CoV and an interaction with ACE2 have been shown to interfere with this pathway, resulting in enhanced acute lung injury in rodent models (26). The significance of hACE2 specifically on ciliated cell types remains to be explored.
Interestingly, we found that SARS-CoV poorly infected our models of airway epithelial cells derived from alveolar regions of the human lung, a finding that has also been reported by others (15). In comparison to Vero cells, A549 cells express undetectable levels of hACE2 and may account for the lack of SARS-CoV infection in these experiments. These observations may reflect that freshly isolated human alveolar cells rapidly lose their differentiation status (from type I or type II cells) during culture, resulting in cell types that do not represent cell types found in vivo. This is in contrast to HAE, for which differentiation of airway cells to a ciliated morphology requires 6 to 8 weeks of culture. Growth of primary alveolar cells for this length of time in vitro does not result in cultures that are significantly different in terms of differentiation or morphology from cultures grown for 3 to 5 days (R. J. Pickles, unpublished observations). Therefore, such studies of alveolar regions of the lung will require development of techniques that isolate and preserve the differentiation status of the epithelial cells from these regions. Alternatively, such studies may be better suited to in vivo experimentation using relevant animal models.
Another human coronavirus, HCoV-229E, which primarily causes a mild upper respiratory tract infection in humans, has been shown with other models of HAE to principally target ciliated epithelia, causing cell morphology alterations, dyskinesia, and cilium dysfunction (63). However, shedding of infected ciliated cells, noted here during SARS-CoV infection, was not reported following HCoV-229E infection. In contrast to other HCoV, SARS-CoV infection is associated with severe atypical pneumonia in adults. The major pathological findings upon autopsy suggest SARS-CoV involvement of type II pneumocytes located in the alveoli, as evidenced by detection of viral RNAs and EM detection of virus-like particles (4, 5, 75). Importantly, patients dying early following SARS-CoV exposure show marked bronchiolar disease with respiratory epithelial cell necrosis, loss of cilia, squamous metaplasia, and intrabronchiolar fibrin deposits. In fact, it has been suggested that early diffuse alveolar damage as a consequence of SARS-CoV infection may initiate in the respiratory bronchioles (13, 39). SARS-CoV has a broad host range, replicating efficiently in humans, primates, palm civets, raccoon dogs, hamsters, ferrets, cats, and mice, although most-pronounced disease is noted to occur in humans and mild persistent infections have been documented for palm civets (11, 17, 25, 34, 41, 44, 45). Significantly reduced pathology and clinical disease are noted to occur in ferrets, cats, hamsters, and mice (34, 41, 44, 45, 56). In the murine model for SARS-CoV infection, replication of virus without disease has been noted (56). Principle targets for SARS-CoV infection are the airway epithelia of the mouse, hamster, and ferret models (34, 44, 45, 56); however, specific infection of ciliated cells was not described for these species. Mechanisms of SARS-CoV cross-species transmission appear mediated in part by the orthologues of hACE2, as SARS-CoV derived from civet cats preferentially recognizes civet cat ACE2 compared to rat and human ACE2 (31, 69). In mice, the murine ACE2 orthologue functions as a receptor for SARS-CoV entry into murine cells, but inefficient usage limits virus replication and spread (29). As virus load is correlated with increased pathogenesis in humans (64), it is likely that the attenuated replication phenotypes noted for ciliated cell cultures derived from different species are closely correlated with the reduced clinical disease and pathology noted for these alternative species. Although orthologous ACE2 may in part explain the reduced replication phenotype in these species, molecular epidemiologic analysis argues that SARS-CoV host switching was associated with positive selection in the S glycoprotein gene, ORF1a replicase, and ORF3a (6). These data suggest that the ORF1a replicase proteins and the group-specific ORFs may influence replication efficiency in these alternative species. Additional studies will be needed to identify the genetic basis restricting efficient SARS-CoV replication in the mouse, hamster, and nonhuman primate in vitro and in vivo models.
Respiratory viruses induce dramatically different levels of cytopathology following infection of HAE in vitro. Influenza virus infection of HAE results in rapid and extensive cytopathology affecting ciliated and other epithelial cell types, consistent with the extensive damage in the respiratory epithelium noted with infected humans in vivo (33, 74). In contrast to the characteristic syncytium formation noted with nonpolarized epithelial cell lines, RSV and PIV3 exclusively target ciliated cells in HAE but without syncytium formation (73, 74). In addition, no shedding of progeny RSV or PIV3 from the basolateral surfaces was noted, an observation supported by the rare incidence of viremia in patients infected with these viruses. Examination of the morphology of HAE after SARS-CoV infection demonstrated the disruption of cell-cell junctions between the infected ciliated cells and the surrounding cells by 72 h postinfection (S localization to the basolateral region of the cell), and by 120 h postinfection, ciliated cells positive for S were shed onto the luminal surface of HAE (Fig. 8B, C, and D). In addition, SARS-CoV was shed albeit at a low level from the basolateral surfaces of HAE, perhaps suggesting a mechanism by which viremia can occur with this virus, explaining subsequent virus spread to the intestine and other organs (Fig. 4K) (51, 60). Mechanisms of viral spread to basolateral compartments are unknown but most likely involve disruption of tight junctions and diffusion rather than spread to the basal cell layers. By EM, large numbers of virus particles were present in ciliated cells, in budding vesicles being transported to the luminal surface, and in close proximity to the apical surface of ciliated cells (Fig. 6B to H). Abundant numbers of virus particles were also trapped in cellular debris in the luminal compartment, suggesting that cell-associated virus might contribute to SARS-CoV spread between hosts. The mechanism that specifies the predominant polarized release of SARS-CoV virions is unclear, but releasing virus particles into the lumens of airways provides a means of propagating spread via interaction with potential viral receptors also polarized to the luminal surface of ciliated cells, e.g., hACE2. Similar findings were also reported with HCoV-229E (63). As recent reports suggest that HCoV-NL63 also uses hACE2 as a receptor for entry into cells, these data indicate that many coronaviruses target this pathway while replicating in the lung epithelium (23). Our data also demonstrate that SARS-CoV assembly and release occur via budding into internal vesicles that are transported to the cell surface. Ultimately, infection results in shedding of the ciliated cell, which often contains large quantities of virus (Fig. 6D, G, and H).
EM also demonstrated extensive cytopathology and vacuolization of infected cells, suggesting that considerable cell-associated virus may be shed from infected patients (Fig. 6D, G, and H). Future studies will determine the mechanisms mediating the extrusion of infected ciliated cells from HAE and whether genetic mutants of SARS-CoV produce similar results. The immunological consequences of ciliated cells containing virus being shed into the airway lumen remain to be explored.
We have successfully deleted ORF7 from the SARS-CoV genome. SARS ORF7 is divided into two ORFs, designated ORF7a and 7b. Although no ORF7b product has been described to date, ORF7a encodes a 122-amino-acid type 1 transmembrane protein and structural studies reveal a compact seven-stranded β sandwich similar in fold and topology to members of the immunoglobulin superfamily (38). ORF7a traffics into the endoplasmic reticulum/Golgi network, the site of coronavirus budding and assembly, suggesting a possible role in maturation and release (38). Moreover, ORF7a of SARS-CoV has been shown to encode a proapoptotic activity in cells in culture (58). By use of a reverse genetic system developed for SARS-CoV, ORF7a and ORF7b were replaced with GFP, demonstrating a luxury function for these ORFs in SARS-CoV GFP replication in Vero E6, MA104, and Caco-2 cells as well as HAE. As SARS-CoV GFP replicates as efficiently as recombinant wild-type icSARS-CoV and Urbani SARS-CoV in HAE and expresses equivalent amounts of subgenomic mRNA, ORF7 does not encode a critical function for in vitro replication, although it might enhance in vivo pathogenesis, as has been described with other coronavirus accessory ORFs (9, 10, 40).
Several paradigms to explain the mechanism for spread of SARS-CoV in the human lung have been proposed. One model suggests that SARS-CoV infects lung dendritic cells that transport virus to the alveolar regions, allowing for infection of type II pneumocytes. This model is based on studies in which dendritic cells were infected by SARS-CoV via an interaction with overexpressed levels of DC-SIGN (35, 68). An alternative model would predict that SARS-CoV infection and spread could occur through some component of the respiratory epithelium. Indeed, consideration of the early pathology of SARS-CoV observed in the bronchiolar regions and the demonstration that SARS-CoV replicates efficiently in ciliated cells isolated from nasal and tracheobronchial airway regions shown here are supportive of this latter hypothesis. Moreover, the uniform gradient of hACE2 expression throughout the conducting airways of the human lung provides a suitable substrate for repeated cycles of virus amplification and spread throughout the airways, ultimately spreading to some cell component of the alveolus regions. Since progeny SARS-CoV is predominately released onto the luminal surface of HAE, we propose that infection of ciliated cells by SARS-CoV is propagated by the coordinated movement of cilium beat, as we have shown for the ciliated-cell-to-ciliated-cell spread of RSV and PIV3 in HAE (73, 74). Since specific airway epithelial cell types, especially ciliated epithelial cells, possess unique physiological and innate defense functions in the lung, it is important to determine the airway cell tropism of SARS-CoV in the human lung in order to understand the pathological consequences of infection of such cell types.
Acknowledgments
We thank the directors and team of the UNC Cystic Fibrosis Center Tissue Culture Core, the Molecular Core, and the Morphology and Morphometry Core for supplying reagents and technical expertise. In addition, we gratefully acknowledge technical assistance from the Carolina Vaccine Center in the preparation of the manuscript and the School of Public Health at UNC in remodeling a biosafety level 3 facility in support of these studies. Special thanks are extended to Ande West and Martha Collier for the production of VRPs and antisera directed against SARS-CoV structural proteins and to Alessandra Livraghi for technical support.
The work was supported by the National Institutes of Health through research grants AI059136, AI059443-01, and AI059443 to R.S.B. and by a University of North Carolina Medical Alumni Endowment Fund grant to R.J.P., and A.C.S. was supported by Infectious Disease Pathogenesis Research Training Grant 5T32AI07151-27 through NIH/NIAID.
REFERENCES
- 1.Baric, R. S., and A. C. Sims. 2005. Development of mouse hepatitis virus and SARS-CoV infectious cDNA constructs. Curr. Top. Microbiol. Immunol. 287:229-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNA function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., B. Yount, L. Lindesmith, P. R. Harrington, S. R. Greene, F.-C. Tseng, N. Davis, R. E. Johnston, D. G. Klapper, and C. L. Moe. 2002. Expression and self-assembly of Norwalk virus capsid protein from Venezuelan equine encephalitis virus replicons. J. Virol. 76:3023-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., H. T. Zhang, Y. Q. Xie, J. W. Wan, Z. H. Lu, D. T. Wang, Q. Z. Wang, X. H. Xue, W. X. Si, Y. F. Luo, and H. M. Qiu. 2003. Morphological study of severe acute respiratory syndrome (SARS). Zhonghua Bing Li Xue Za Zhi 32:516-520. (In Chinese.) [PubMed] [Google Scholar]
- 5.Cheung, O. Y., J. W. Chan, C. K. Ng, and C. K. Koo. 2004. The spectrum of pathological changes in severe acute respiratory syndrome (SARS). Histopathology 45:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinese SARS Molecular Epidemiology Consortium. 2004. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303:1666-1669. [DOI] [PubMed] [Google Scholar]
- 7.Chow, K. C., C. H. Hsiao, T. Y. Lin, C. L. Chen, and S. H. Chiou. 2004. Detection of severe acute respiratory syndrome-associated coronavirus in pneumocytes of the lung. Am. J. Clin. Pathol. 121:574-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croce, M. V., M. T. Isla-Larrain, C. E. Rua, M. E. Rabassa, S. J. Gendler, and A. Segal-Eiras. 2003. Patterns of MUC1 tissue expression defined by an anti-MUC1 cytoplasmic tail monoclonal antibody in breast cancer. J. Histochem. Cytochem. 51:781-788. [DOI] [PubMed] [Google Scholar]
- 9.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., P. S. Masters, X. Shen, S. Weiss, and P. J. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franks, T. J., P. Y. Chong, P. Chui, J. R. Galvin, R. M. Lourens, A. H. Reid, E. Selbs, C. P. McEvoy, C. D. Hayden, J. Fukuoka, J. K. Taubenberger, and W. D. Travis. 2003. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34:743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulcher, M. L., S. Gabriel, K. A. Burns, J. R. Yankaskas, and S. H. Randell. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183-206. [DOI] [PubMed] [Google Scholar]
- 15.Gillim-Ross, L., J. Taylor, D. R. Scholl, J. Ridenour, P. S. Masters, and D. E. Wentworth. 2004. Discovery of novel human and animal cells infected by the severe acute respiratory syndrome coronavirus by replication-specific multiplex reverse transcription-PCR. J. Clin. Microbiol. 42:3196-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenough, T. C., A. Carville, J. Coderre, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, and K. Mansfield. 2005. Pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am. J. Pathol. 167:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 18.Haijema, B. J., H. Volders, and P. J. Rottier. 2004. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 78:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamming, I., W. Timens, M. L. Bulthuis, A. T. Lely, G. J. Navis, and H. van Goor. 2004. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrington, P. R., L. Lindesmith, B. Yount, C. L. Moe, and R. S. Baric. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington, P. R., B. Yount, R. E. Johnston, N. Davis, C. Moe, and R. S. Baric. 2002. Systemic, mucosal, and heterotypic immune induction in mice inoculated with Venezuelan equine encephalitis replicons expressing Norwalk virus-like particles. J. Virol. 76:730-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, K. Y., K. S. Singh, A. G. Habib, B. K. Ong, T. K. Lim, E. E. Ooi, B. K. Sil, A. E. Ling, X. L. Bai, and P. A. Tambyah. 2004. Mild illness associated with severe acute respiratory syndrome coronavirus infection: lessons from a prospective seroepidemiologic study of health-care workers in a teaching hospital in Singapore. J. Infect. Dis. 189:642-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai, Y., K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi, M. A. Crackower, A. Fukamizu, C. C. Hui, L. Hein, S. Uhlig, A. S. Slutsky, C. Jiang, and J. M. Penninger. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 26.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, W., T. C. Greenough, M. J. Moore, N. Vasilieva, M. Somasundaran, J. L. Sullivan, M. Farzan, and H. Choe. 2004. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 78:11429-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 33.Martin, C. M., C. M. Kunin, L. S. Gottlieb, M. W. Barnes, C. Liu, and M. Finland. 1959. Asian influenza A in Boston, 1957-1958. I. Observations in thirty-two influenza-associated fatal cases. AMA Arch. Intern. Med. 103:515-531. [DOI] [PubMed] [Google Scholar]
- 34.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzi, A., T. Gramberg, G. Simmons, P. Moller, A. J. Rennekamp, M. Krumbiegel, M. Geier, J. Eisemann, N. Turza, B. Saunier, A. Steinkasserer, S. Becker, P. Bates, H. Hofmann, and S. Pohlmann. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 78:12090-12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matrosovich, M. N., T. Y. Matrosovich, T. Gray, N. A. Roberts, and H. D. Klenk. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA 101:4620-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAuliffe, J., L. Vogel, A. Roberts, G. Fahle, S. Fischer, W.-J. Shieh, E. Butler, S. Zaki, M. St. Claire, B. Murphy, and K. Subbarao. 2004. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelson, C. A., A. Pekosz, C. A. Lee, M. S. Diamond, and D. H. Fremont. 2005. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure (Cambridge) 13:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholls, J. M., L. L. Poon, K. C. Lee, W. F. Ng, S. T. Lai, C. Y. Leung, C. M. Chu, P. K. Hui, K. L. Mak, W. Lim, K. W. Yan, K. H. Chan, N. C. Tsang, Y. Guan, K. Y. Yuen, and J. S. Peiris. 2003. Lung pathology of fatal severe acute respiratory syndrome. Lancet 361:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osterhaus, A. D., R. A. Fouchier, and T. Kuiken. 2004. The aetiology of SARS: Koch's postulates fulfilled. Philos. Trans. R. Soc. Lond. B 359:1081-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pene, F., A. Merlat, A. Vabret, F. Rozenberg, A. Buzyn, F. Dreyfus, A. Cariou, F. Freymuth, and P. Lebon. 2003. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin. Infect. Dis. 37:929-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickles, R. J., D. McCarty, H. Matsui, P. J. Hart, S. H. Randell, and R. C. Boucher. 1998. Limited entry of adenovirus vectors into well-differentiated airway epithelium is responsible for inefficient gene transfer. J. Virol. 72:6014-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts, A., C. Paddock, L. Vogel, E. Butler, S. Zaki, and K. Subbarao. 2005. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 79:5833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, A., L. Vogel, J. Guarner, N. Hayes, B. Murphy, S. Zaki, and K. Subbarao. 2005. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 79:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 47.Rowe, R. K., S. L. Brody, and A. Pekosz. 2004. Differentiated cultures of primary hamster tracheal airway epithelial cells. In Vitro Cell. Dev. Biol. Anim. 40:303-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi, X., E. Gong, D. Gao, B. Zhang, J. Zheng, Z. Gao, Y. Zhong, W. Zou, B. Wu, W. Fang, S. Liao, S. Wang, Z. Xie, M. Lu, L. Hou, H. Zhong, H. Shao, N. Li, C. Liu, F. Pei, J. Yang, Y. Wang, Z. Han, X. Shi, Q. Zhang, J. You, X. Zhu, and J. Gu. 2005. Severe acute respiratory syndrome associated coronavirus is detected in intestinal tissues of fatal cases. Am. J. Gastroenterol. 100:169-176. [DOI] [PubMed] [Google Scholar]
- 52.Singh, K., O. E. Eong, B. Kumarsil, S. Saw, and S. Sethi. 2004. Severe acute respiratory syndrome without respiratory symptoms or abnormal chest radiograph findings. Clin. Infect. Dis. 38:585-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sola, I., S. Alonso, S. Zuniga, M. Balasch, J. Plana-Duran, and L. Enjuanes. 2003. Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 77:4357-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stonebraker, J. R., D. Wagner, R. W. Lefensty, K. Burns, S. J. Gendler, J. M. Bergelson, R. C. Boucher, W. K. O'Neal, and R. J. Pickles. 2004. Glycocalyx restricts adenoviral vector access to apical receptors expressed on respiratory epithelium in vitro and in vivo: role for tethered mucins as barriers to lumenal infection. J. Virol. 78:13755-13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W.-J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sui, J., W. Li, A. Murakami, A. Tamin, L. J. Matthews, S. K. Wong, M. J. Moore, A. S. Tallarico, M. Olurinde, H. Choe, L. J. Anderson, W. J. Bellini, M. Farzan, and W. A. Marasco. 2004. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 101:2536-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 60.To, K. F., J. H. Tong, P. K. Chan, F. W. Au, S. S. Chim, K. C. Chan, J. L. Cheung, E. Y. Liu, G. M. Tse, A. W. Lo, Y. M. Lo, and H. K. Ng. 2004. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J. Pathol. 202:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tseng, C.-T. K., J. Tseng, L. Perrone, M. Worthy, V. Popov, and C. J. Peters. 2005. Apical entry and release of severe acute respiratory syndrome-associated coronavirus in polarized Calu-3 lung epithelial cells. J. Virol. 79:9470-9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, G., C. Deering, M. Macke, J. Shao, R. Burns, D. M. Blau, K. V. Holmes, B. L. Davidson, S. Perlman, and P. B. McCray, Jr. 2000. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74:9234-9239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, W. K., S. Y. Chen, I. J. Liu, C. L. Kao, H. L. Chen, B. L. Chiang, J. T. Wang, W. H. Sheng, P. R. Hsueh, C. F. Yang, P. C. Yang, and S. C. Chang. 2004. Temporal relationship of viral load, ribavirin, interleukin (IL)-6, IL-8, and clinical progression in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 39:1071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weingartl, H., M. Czub, S. Czub, J. Neufeld, P. Marszal, J. Gren, G. Smith, S. Jones, R. Proulx, Y. Deschambault, E. Grudeski, A. Andonov, R. He, Y. Li, J. Copps, A. Grolla, D. Dick, J. Berry, S. Ganske, L. Manning, and J. Cao. 2004. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 78:12672-12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wesley, R. D., R. D. Woods, and A. K. Cheung. 1991. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J. Virol. 65:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, Z.-Y., Y. Huang, L. Ganesh, K. Leung, W.-P. Kong, O. Schwartz, K. Subbarao, and G. J. Nabel. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang, Z. Y., H. C. Werner, W. P. Kong, K. Leung, E. Traggiai, A. Lanzavecchia, and G. J. Nabel. 2005. Evasion of antibody neutralization in emerging severe acute respiratory syndrome coronaviruses. Proc. Natl. Acad. Sci. USA 102:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng, R., R. F. Yang, M. D. Shi, M. R. Jiang, Y. H. Xie, H. Q. Ruan, X. S. Jiang, L. Shi, H. Zhou, L. Zhang, X. D. Wu, Y. Lin, Y. Y. Ji, L. Xiong, Y. Jin, E. H. Dai, X. Y. Wang, B. Y. Si, J. Wang, H. X. Wang, C. E. Wang, Y. H. Gan, Y. C. Li, J. T. Cao, J. P. Zuo, S. F. Shan, E. Xie, S. H. Chen, Z. Q. Jiang, X. Zhang, Y. Wang, G. Pei, B. Sun, and J. R. Wu. 2004. Characterization of the 3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J. Mol. Biol. 341:271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, Q. L., Y. Q. Ding, J. L. Hou, L. He, Z. X. Huang, H. J. Wang, J. J. Cai, J. H. Zhang, W. L. Zhang, J. Geng, X. Li, W. Kang, L. Yang, H. Shen, Z. G. Li, H. X. Han, and Y. D. Lu. 2003. Detection of severe acute respiratory syndrome (SARS)-associated coronavirus RNA in autopsy tissues with in situ hybridization. Di Yi Jun Yi Da Xue Xue Bao 23:1125-1127. (In Chinese.) [PubMed] [Google Scholar]