Abstract
To perform a genetic analysis of the influenza A virus NS1 gene, a library of NS1 mutants was generated by PCR-mediated mutagenesis. A collection of mutant ribonucleic proteins containing the nonstructural genes was generated from the library that were rescued for an infectious virus mutant library by a novel RNP competition virus rescue procedure. Several temperature-sensitive (ts) mutant viruses were obtained by screening of the mutant library, and the sequences of their NS1 genes were determined. Most of the mutations identified led to amino acid exchanges and concentrated in the N-terminal region of the protein, but some of them occurred in the C-terminal region. Mutant 11C contained three mutations that led to amino acid exchanges, V18A, R44K, and S195P, all of which were required for the ts phenotype, and was characterized further. Several steps in the infection were slightly altered: (i) M1, M2, NS1, and neuraminidase (NA) accumulations were reduced and (ii) NS1 protein was retained in the nucleus in a temperature-independent manner, but these modifications could not justify the strong virus titer reduction at restrictive temperature. The most dramatic phenotype was the almost complete absence of virus particles in the culture medium, in spite of normal accumulation and nucleocytoplasmic export of virus RNPs. The function affected in the 11C mutant was required late in the infection, as documented by shift-up and shift-down experiments. The defect in virion production was not due to reduced NA expression, as virus yield could not be rescued by exogenous neuraminidase treatment. All together, the analysis of 11C mutant phenotype may indicate a role for NS1 protein in a late event in virus morphogenesis.
The influenza A virus genome encodes NS1, a small, nonstructural, and multifunctional protein important for virus-cell interactions (for reviews, see references 22, 33, and 53). It is translated from the colinear transcript of segment 8, which also encodes NS2 protein from a spliced mRNA (31, 34). NS1 accumulates in the nucleus early in the infection (4, 32) and when it is expressed from cDNA (26, 35, 56), but at late times in the infection it is also found in the cytoplasm (49), in association with polysomes (8, 16, 32).
Although NS1 protein is apparently nonessential, as a recombinant virus has been generated that lacks the gene (23), several virus mutants have been isolated or generated which contain mutations in the NS1 protein and are severely hampered for replication (12, 14, 17, 27, 28, 40, 60, 63-65). The phenotypes of these mutant viruses indicate that NS1 protein may be involved in several steps in the virus infectious cycle, including transcription and/or replication of virus RNA, late virus protein synthesis, and interference with the cellular gene expression, and that it eventually modulates the virulence of virus infections in vivo (2, 23, 68).
NS1 is an RNA-binding protein that has been shown to interact with virion RNA (vRNA) (29, 43), poly(A)-containing RNAs (58), and U6 snRNA (59). The RNA-binding domain is located within the N-terminal half of the protein, and the rest of the protein appears to contain an effector domain (43, 57). NS1 can also interact with a long series of viral and cellular factors. These include the virus RNP and/or polymerase (45), cellular proteins involved in translation such as hStaufen, PABPI, and eIF4G (1, 5, 16), and cellular factors involved in posttranscriptional processing of RNA such as CPSF (47) and NS1-BP, a potential splicing-related factor (72). These interactions may be responsible for the alterations in the control of cellular gene expression observed upon expression of NS1 cDNA (18, 19, 47, 57), as well as for the regulation of virus gene expression (8, 13). In addition, NS1 has been shown to block the signaling pathway mediated by Jun N-terminal protein kinase (41) and to down-regulate apoptosis (73).
Virus mutants have been generated by in vitro mutagenesis of NS segment cDNA and rescue into infectious virus. One of them contains point mutations in the RNA-binding domain (10) and showed reduced replication in tissue-cultured cells and attenuation in mice. Another mutant contains an altered effector domain (50) and also shows a reduced replication in tissue-cultured cells. The steps of the infection cycle affected by these mutations have not been reported, but cells infected by either virus showed an increased interferon response compared to the results seen with infections with wild-type (wt) virus. Other mutants show deletions in the NS1 protein (13, 17, 63). Some of them—dl12, NS1-81, and NS1-110—were temperature sensitive (13, 17), while others showed reduced replication but were not temperature sensitive (13, 63). In general, the most prominent phenotype in these mutants was a reduced translation of M1 and other late virus mRNAs, and the strength of the phenotype was dependent on the genetic background of the strain used for rescue. In this report we describe the generation of a library of influenza virus temperature-sensitive mutants with amino acid exchanges in the NS segment. One of these mutants, mutant 11C, was affected in the NS1 protein and showed no virus particle formation, while virus RNA replication and gene expression were only slightly altered. This is the first evidence for a possible role of NS1 protein in the morphogenesis of influenza virions.
MATERIALS AND METHODS
Biological materials.
The origins of the COS-1 cell line (25), 293T HEK cells (11), and the MDCK cell line have been described previously (17). The cells were cultivated as described previously (54). The plasmids containing the cDNAs of WSN virus RNAs under control of the polI promoter (48) were a gift of Y. Kawaoka. Similar plasmids with the cDNAs of A/Victoria/3/75 (VIC) strain, as well as the corresponding expression plasmids, have been described previously (17). Plasmid vector pHH21 (48) was kindly provided by G. Hobom. Mutant virus WSN A6 (55) was kindly provided by G. Brownlee. The antisera specific for nucleoprotein (NP) and NS1 proteins were generated by hyperimmunization of rats with purified His-NP and His-NS1 proteins, respectively. Monoclonal antibody E10, recognizing both M1 and M2 proteins, was a gift of A. García-Sastre. Monoclonal antibody H179, specific for the hemagglutinin (HA) protein of the WSN strain, was kindly provided by A. Douglas. The monoclonal antibody M234/1G10, specific for the NA of the VIC strain, has been described previously (38, 66).
In vitro mutagenesis and plasmid constructions.
To mutagenize the cDNA of NS segment, its sequence was amplified by PCR using plasmid pT7NSRT (52) as a template and the nucleotide analogs dPTP (25 μM) and 8-oxo-dGTP (166 μM). The amplification product was further amplified with normal deoxynucleoside triphosphates to remove the analogs. The primers used were 5′-AGCAAAAGCAGG-3′, complementary to the NS RNA 3′ end, and 5′-GCCTGGTACCTAATACGCCTCACTATAAGTAGAAACAAGG-3′, which contains an Asp718 site (italics), the T7 RNA polymerase promoter (underlined), and the NS segment 5′ terminus. The PCR fragments were digested with Asp718 nuclease and ligated into SmaI/Asp718-digested pUC19RT to generate a pT7NSmutRT library. The conditions for mutagenesis were adjusted to produce five to six nucleotide changes per molecule, as determined by random sequencing of plasmid clones in a pilot experiment. The mutant library was amplified on agar plates and used for virus rescue.
Virus techniques.
The rescue of the mutant library was carried out by competition of virus RNPs. First, an NS RNP library was generated by cotransfection of the pT7NSmutRT library together with pGPB1, pGPB2, pGPA, and pGNP into vaccinia T7 recombinant virus-infected COS1 cells (52). The NS RNP library was purified by glycerol density gradient centrifugation (42). An excess of these RNPs was cotransfected into COS1 cells with total virion RNPs of the VIC strain, purified in the same manner, using cationic liposomes (62). The transfected cells were then plated onto a 10-fold excess of MDCK cells and overlaid with agar-containing medium. For screening of virus ts mutants, the plaques were allowed to develop for 2 days at 32°C, marked, and transferred to 39.5°C for 12 h. The plaques that did not grow further at restrictive temperature were picked and plated again at both temperatures. Those plaque isolates that showed a difference of 2 or more log units in the relative titer at either temperature were subjected to a further cycle of plaque isolation at 32°C and used for preparation of a virus stock.
Rescue of cDNAs into virus by the plasmid-only technique was carried out on HEK293T cells as described previously (17, 48). Transfected cells were plated onto an excess of MDCK cells and cultivated as described above. The identity of rescued mutant viruses was ascertained by sequencing of DNAs derived from NS RNA segment by reverse transcription-PCR amplification.
For virion purifications, culture supernatants were centrifuged for 10 min at 10,000 rpm and 4°C. The supernatant was sedimented through a sucrose step gradient (TNE; 50% and 33% in 50 mM Tris-HCl-100 mM NaCl-5 mM EDTA, pH 7.5) for 1 h at 40,000 rpm and 4°C in a SW41 rotor. The 50 to 33% interphase was collected, diluted in TNE buffer, and pelleted through a cushion of 33% sucrose in TNE for 1 h at 40,000 rpm and 4°C in a SW41 rotor. The pellet was resuspended in Laemmli sample buffer and analyzed by Western blotting as indicated below.
To rescue NA-deficient viruses, cultures of MDCK cells were infected at a multiplicity of infection (MOI) of 10−3 PFU/cell and incubated in Dulbecco's minimal essential medium containing 2.5 μg/ml trypsin and 0.5 mU/ml of Clostridium perfringens neuraminidase. The supernatants were collected when cytopathic effect was apparent and titrated at permissive temperature.
Protein analyses.
For protein labeling in vivo, cultures were washed, incubated for 30 min in Dulbecco's minimal essential medium lacking methionine-cysteine (met-cys), and labeled for 1 h at various times postinfection with [35S]met-cys to a final concentration of 200 μCi/ml (74). Total extracts were prepared in Laemmli sample buffer and processed by polyacrylamide gel electrophoresis and autoradiography. In some experiments, the extracts were denatured by boiling for 10 min and treated with PNGase F (New England Biolabs) for 3 h at 37°C, as indicated by the manufacturer, before electrophoresis.
For immunofluorescence, infected or mock-infected MDCK cells were washed with phosphate-buffered saline (PBS), fixed for 20 min with 3% paraformaldehyde at room temperature, and permeabilized with 0.5% Triton X-100 for 5 min. The cells were blocked with PBS-3% bovine serum albumin (BSA) and incubated with anti-NP rabbit serum, with rat anti-NS1 serum, or with monoclonal antibodies specific for HA or NA, all diluted in PBS-0.1% BSA, for 1 h at room temperature. After washing with PBS, the bound antibodies were revealed with fluorescein isothiocyanate-conjugated or Texas Red-conjugated goat anti-rabbit, anti-rat, or anti-mouse antibodies, as appropriate, by incubation for 1 h at room temperature. Finally, the cells were washed with PBS and the preparations were mounted with Mowiol. Images were obtained using a Leica fluorescence microscope. For immunofluorescence of surface HA, the cultures were fixed for 20 min at room temperature with 3% paraformaldehyde and stained as indicated above with H179 monoclonal.
For Western blot analysis, cell extracts were separated by polyacrylamide-sodium dodecyl sulfate gel electrophoresis and transferred to Immobilon filters. The filters were blocked for 1 h at room temperature with PBS-3% BSA and incubated with the primary antibodies diluted in 0.1% BSA-0.05% Tween 20 in PBS, for 1 h at room temperature. After being washed four times for 15 min with 0.25% Tween 20 in PBS, the filters were incubated with a dilution of goat anti-rabbit, anti-rat, or anti-mouse immunoglobulin G conjugated to horseradish peroxidase in 0.1% BSA-0.05% Tween 20 in PBS. After being washed in PBS, the filters were developed by enhanced chemiluminiscence.
Immunoprecipitation was carried out essentially as described previously (16). In brief, protein A-Sepharose immune matrices were prepared with rabbit anti-mouse immunoglobulin G and M234/1G10 anti-NA monoclonal antibody and were incubated with soluble extracts prepared in TNE-1% NP-40 buffer from influenza virus-infected or mock-infected cells. After being washed with the same buffer, the bound material was eluted in Laemmli sample buffer and processed by polyacrylamide gel electrophoresis and autoradiography.
RNA analyses.
The accumulation of vRNAs and mRNAs in each fraction was determined by dot blot hybridization. The isolation of total RNA from infected and mock-infected cells was carried out using ULTRASPEC RNA isolation reagent (Biotecx) according to the manufacturer's instructions. Appropriate amounts of each sample were diluted in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-7.5% formaldehyde buffer and fixed onto nylon filters. Hybridizations were carried out overnight in 6 × SSC-0.5% sodium dodecyl sulfate-5× Denhardt's-40% formamide-100 μg/ml single-stranded DNA at 37°C. After a washing, the filters were exposed and quantified with a PhosphorImager. For detection of NP and M vRNAs, full gene riboprobes of positive polarity were used, and a negative polarity riboprobe was used for NP mRNA detection. Detection of NA vRNA and mRNA was carried out with 500-nucleotide-long riboprobes transcribed from pGEM-NA with T7 and SP6 RNA polymerase, respectively. For the control of RNA loading, each filter was further hybridized with a terminally labeled oligonucleotide probe specific for 18S rRNA (5′ GAGCGAAAGCATTTGCCAAG 3′).
RESULTS
Generation of influenza viruses with temperature-sensitive mutations in the NS segment.
In order to perform a wide-ranging analysis of possible functions for influenza virus NS1 protein we carried out a random mutagenesis of the NS segment and a screening for temperature-sensitive mutant viruses. The cDNA for NS segment of the VIC strain was mutagenized by PCR, and a library of mutant clones was generated in the vector plasmid pUC19RT (52). As our aim was to obtain a library of virus mutants, a high-efficiency rescue system was developed to transfer the collection of mutant cDNAs into infectious virus. In essence, the rescue system comprised two steps: first, the cDNA of the virus segment was used to prepare a recombinant virus RNP as described previously (52), and then an excess of purified recombinant RNP was cotransfected with purified total virion RNPs into cultured cells. Under these conditions, competition among virion-derived and recombinant RNPs of the same segment was expected to occur. To test the efficiency of the system, a recombinant RNP derived from the NS segment of WSN virus was generated and cotransfected in excess with wt RNPs of the VIC strain. The virus produced after transfection was used to infect a fresh MDCK culture, and the nature of the NS1 protein generated was determined by Western blot analysis. As the electrophoretic mobilities of NS1 proteins from either virus strain are distinguishable, we could determine that most of the virus in the stock contained the NS segment from the WSN strain (Fig. 1). To confirm these results, the mobility of NS1 protein from several independent virus clones from the mixture was determined. Ninety percent of them were characterized as recombinant viruses (data not shown).
FIG. 1.
Rescue of virus genes by competition of RNPs. The virus yield from a mixed transfection with WSN recombinant NS RNP and total VIC virion RNPs was used to infect MDCK cells (REC. MIX). Total cell extract was analyzed by Western blotting with anti-NS1 serum. As controls, cells were infected with WSN virus or VIC virus or were mock infected (MOCK). The mobility of VIC NS1 protein is indicated by stars and that of WSN NS1 protein is denoted by diamonds. Numbers to the left indicate the mobility of molecular mass markers (in kilodaltons).
This rescue system was applied to the mutant cDNAs of NS segment. The library of mutant plasmids was used to generate a mixture of mutant RNPs that was cotransfected with VIC virion RNPs. The rescued viruses were screened for temperature-sensitive growth at 32 versus 39.5°C as described in Materials and Methods. Among around 4,200 virus clones tested, 27 showed a ts phenotype in the first screening step. A total of 22 of them were analyzed further, and 9 were confirmed as ts mutants after further two plaque isolation steps. No temperature-sensitive virus was obtained when a similar procedure was performed with a nonmutagenized NS cDNA. For the viruses that behaved as truly ts, mutant cDNAs were prepared from virus particles by reverse transcription-PCR, and their sequences were determined. From the collection, five mutants contained amino acid replacements in the NS1 open reading frame (ORF) and two in both NS1 and NS2 ORFs, and one mutant contained mutations that did not alter the amino acid sequence. The distribution of mutations detected in the NS1 ORF is shown in Fig. 2 and reveals that the N-terminal region of the protein accumulated most of the amino acid changes, although some of these were also located in the C-terminal portion of NS1. One of the mutants was chosen for further studies, and its phenotype is reported here.
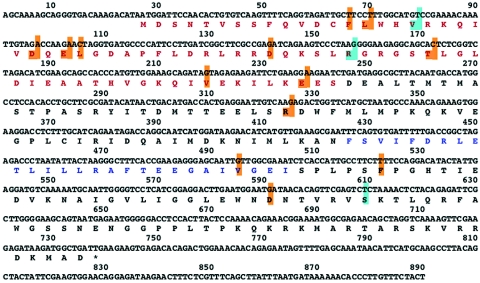
FIG. 2.
Amino acid replacements in temperature-sensitive mutants affected in NS1 protein. The NS RNA segments of influenza virus temperature-sensitive mutants isolated in this study were cloned and sequenced. The positions in which amino acid substitutions were detected in the NS1 open reading frame are boxed in light brown on the NS1 protein sequence. Those present in mutant 11C are boxed in light blue. The protein regions acting as RNA-binding domain (red) and as effector domain (blue) are indicated (37, 43, 57).
Mutations in NS1 protein are responsible for the temperature-sensitive phenotype of 11C virus.
To ensure that the observed ts phenotype was due only to the mutations in the NS1 protein, the NS cDNA derived from mutant 11C virus was transferred to vector pHH21 (48) and rescued by the plasmid-only system in a general VIC background except for the HA and M segments that were derived from the WSN strain. As a control, a wt NS cDNA was rescued in parallel in the same genetic background. The growth kinetics of both viruses were assayed at permissive and restrictive temperatures, and the results are presented in Fig. 3. Mutant 11C virus replicated at 32°C to levels indistinguishable from those of wt recombinant virus, but at 39.5°C the titers obtained were around 3 log units lower. Such a ts phenotype was also observed in other genetic backgrounds (data not shown). Thus, a plasmid-rescued 11C mutant was used thereafter for phenotype analyses. This mutant presents three amino acid changes (V18A, R44K, and S195P). The first two mutations involve conservative changes, and mutation R44K is present in the NS1 proteins from wt virus strains, while the S195P mutation has not been registered in published sequences and involves a drastic change in the predicted secondary structure of the protein (data not shown). To test whether the latter was solely responsible for the ts phenotype of mutant 11C, two additional viruses were rescued in the same genetic background, containing mutations V18A, R44K, or S195P. Contrary to predictions from the sequence comparisons, none of these mutants was temperature sensitive, and we have to conclude that all three mutations are required for a ts phenotype.
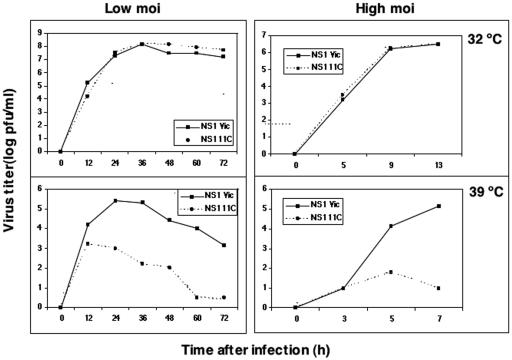
FIG. 3.
Replication of 11C virus is temperature sensitive. Cultures of MDCK cells were infected with mutant 11C or wt recombinant viruses at an MOI of 10−3 PFU/cell (left panels) or 10 PFU/cell (right panels) and incubated at either 32 or 39.5°C. At the times indicated, samples were obtained from the supernatant of each culture and assayed for virus infectivity by a plaque assay at permissive temperature.
The growth of several NS1 mutant viruses, including those with large deletions (12) and a virus lacking the NS1 gene (23), is largely affected by the cellular interferon response. To ascertain whether the ts phenotype of 11C mutant virus is related to the interferon (IFN) response we tested its ability to replicate in Vero cells, which are deficient in IFN signaling (9). Both wt and mutant recombinant viruses replicated poorly in Vero cells at 32°C (final titers of 104 to 105 PFU/ml), because they are not adapted. The wt recombinant virus grew to similar titers at 39°C, but the 11C mutant showed a reduction of around 2 log units in final titer at restrictive temperatures.
The synthesis and accumulation of NS1, M1, and M2 proteins is reduced at restrictive temperatures.
To start the characterization of mutant 11C, we analyzed the synthesis of virus proteins. Cultures of MDCK cells were infected at high multiplicity and pulse labeled with [35S]met-cys, and total cell extracts were analyzed by gel electrophoresis. The results are presented in Fig. 4 and show that M1 and NS1 proteins were labeled to a slightly lesser extent in 11C mutant-infected cells than in wt virus-infected cells, while NS2 protein was equally synthesized and NP synthesis was more efficient at restrictive temperatures in mutant than in wt virus-infected cells.
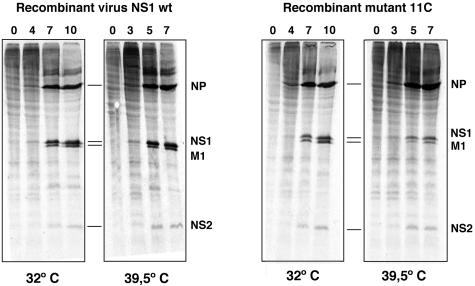
FIG. 4.
Synthesis of viral proteins in 11C mutant-infected cells. Cultures of MDCK cells were infected at an MOI of 10 PFU/cell with either wt or 11C recombinant viruses. The cultures were incubated at 32 or 39.5°C and at the times after infection indicated in the figure they were labeled with [35S]met-cys as indicated in Materials and Methods. Total extracts of each culture were analyzed by polyacrylamide gel electrophoresis and autoradiography. The position of selected viral proteins is indicated to the right of each panel.
In view of these results, the accumulation of various viral proteins in the course of mutant or wt virus infections was determined. Total cell extracts obtained at various times after high-multiplicity infection of MDCK cells with either virus were assayed by Western blot analysis. A monoclonal antibody specific for the N-terminal peptide of M1 (E10) was used that also detects M2 protein (63). In addition, the accumulation of NP and NS1 proteins was determined in parallel. The results are presented in Fig. 5. The accumulation of viral proteins tested was not affected at permissive temperature, but the accumulations of NS1, M1, and M2 were reduced for 11C mutant-infected cells at restrictive temperature.
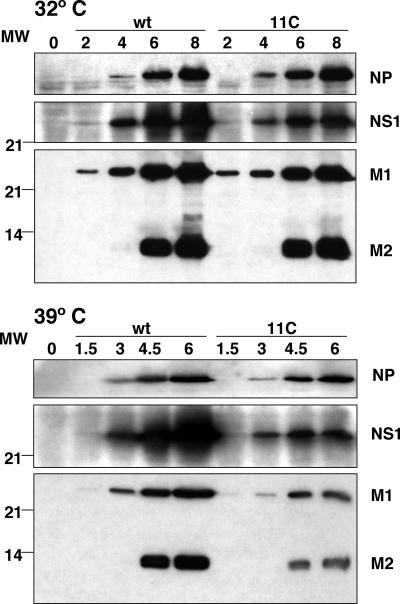
FIG. 5.
Viral protein accumulation in 11C mutant-infected cells. Cultures of MDCK cells were infected at an MOI of 10 PFU/cell with either wt virus or 11C mutant, and total cell extracts were prepared at the times (in hours) indicated in the figure. The accumulation of NP, NS1, M1, and M2 proteins was determined by Western blotting with specific antibodies, as described in Materials and Methods. The position of viral proteins is indicated to the right of the figure and the mobility of molecular mass markers (MW) is shown to the left.
Quantitative determinations indicated that, at restrictive temperature, M1 and M2 proteins accumulated to sixfold- and eightfold-lower extents in mutant-infected than in wt-infected cells, respectively. In the case of NS1 protein, accumulation in mutant-infected cells was two times lower at restrictive temperature than at permissive temperature. In contrast, twofold more NP accumulated during infection with mutant virus at restrictive temperature.
Intracellular localization of early virus proteins.
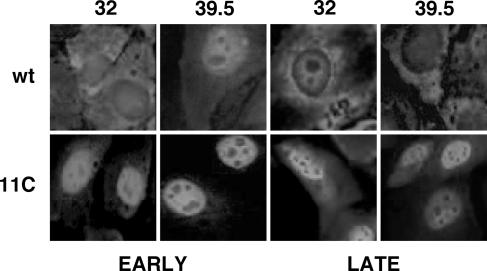
The intracellular localization of wt or mutant NS1 proteins was determined by immunofluorescence assays of infected cultures incubated at either permissive or restrictive temperature. The results are presented in Fig. 6. At both 32 and 39.5°C, mutant NS1 protein accumulated in the nucleus of the cells, independently of the time after infection used for the assay, suggesting that the mutations present in 11C virus led to a defect in nucleocytoplasmic transport of the protein, irrespective of the temperature used for virus replication. On the other hand, the nucleocytoplasmic export of virus RNPs was studied by immunofluorescence of mutant or wt virus-infected cells by use of anti-NP antibodies. No differences could be observed in the distribution of NP signal in cells infected with either virus at restrictive temperarure. At 32°C, the NP signal was detected in the cytoplasm of mutant virus-infected cells earlier than in wt-infected cells, although the significance of this observation is not clear at present (see Fig. 10B).
FIG. 6.
Intracellular localization of 11C mutant NS1 protein. Cultures of MDCK cells were infected with either wt or 11C mutant virus and incubated at permissive or restrictive temperature. At 4 and 9 hpi (32°C) or 3 and 7 hpi (39.5°C) the cells were fixed and processed for immunofluorescence to visualize NS1 protein, as indicated in Materials and Methods.
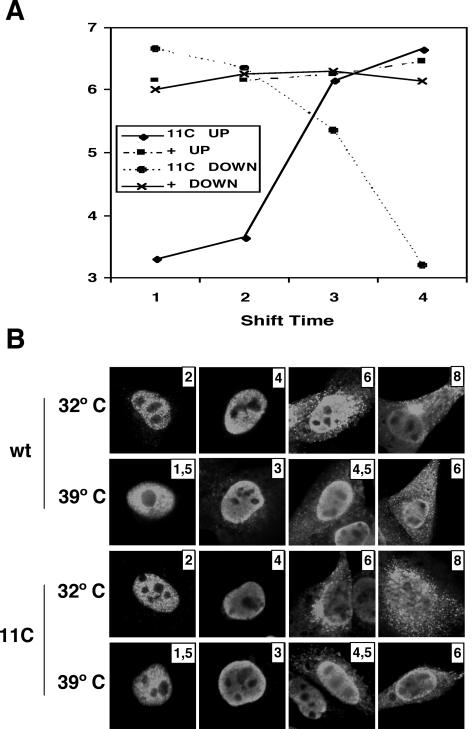
FIG. 10.
Temperature-shift experiments. (A) Cultures of MDCK cells were infected with wt or mutant recombinant viruses at an MOI of 10 PFU/cell and incubated initially at either 32 or 39.5°C. At 2, 4, and 6 hpi (32.5°C) or 1.5, 3, and 4.5 hpi (39.5°C), cultures were transferred to the alternate temperature and incubation was continued till 8 hpi (32.5°C) or 6 hpi (39.5°C). The virus titer of each culture supernatant was determined by a plaque assay at permissive temperature. (B) At each transfer time, parallel cultures were fixed and processed for immunofluorescence using anti-NP antibodies.
Expression and localization of virus membrane proteins.
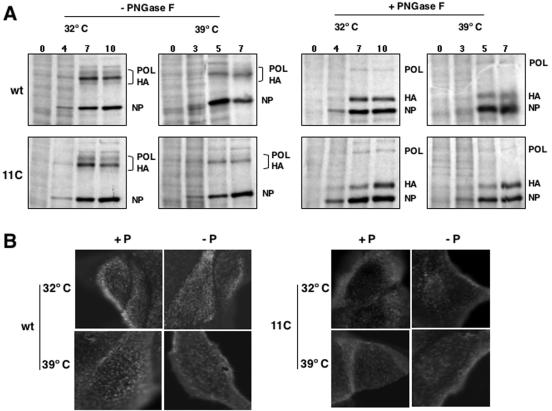
As it has been reported that expression of M2 protein can be important in some virus strains for the protection of HA during its processing and transport to the membrane (51, 67), we tested whether the low accumulation of M2 protein in 11C mutant-infected cells would affect the expression of the HA protein in the membrane. First, the synthesis of HA was determined by pulse labeling wt or mutant virus-infected cells at either permissive or restrictive temperature. To ensure the identification of the HA protein band in the gels, the cell extracts were treated with PNGase or left untreated as a control, and then they were analyzed by polyacrylamide gel electrophoresis and autoradiography. Such a treatment should eliminate the glycosidic chains, reduce the size heterogeneity, and increase the electrophoretic mobility of the protein. The results are presented in Fig. 7A and indicated that no alteration in HA synthesis could be detected in mutant virus-infected cells at either temperature. Second, wt or mutant virus-infected cells were incubated at permissive or restrictive temperature and the location of HA was determined in the cells by immunofluorescence before or after cell permeabilization. No alteration in the HA-specific signal intensity or localization was produced by the mutations present in 11C virus at either temperature (Fig. 7B).
FIG. 7.
Expression and localization of HA in wt and mutant virus-infected cells. Cultures of MDCK cells were infected with wt or mutant recombinant viruses at an MOI of 10 PFU/cell. (A) At the times postinfection indicated on the top of each lane, the cells were pulse labeled with [35S]met-cys. The cell extracts were treated with PNGase F or left untreated and were analyzed by polyacrylamide gel electrophoresis and autoradiography. The position of relevant virus protein bands is indicated to the right of the images. POL, polymerase; NP, nucleoprotein. (B) At 8 hpi (32°C) or 6 hpi (39.5°C) the cultures were fixed with paraformaldehyde. Some cultures were permeabilized with Triton X-100 (+P), and others were not (−P). The cultures were analyzed by immunofluorescence with monoclonal H179 specific for WSN HA. Mock-infected cells gave no fluorescence signal (data not shown).
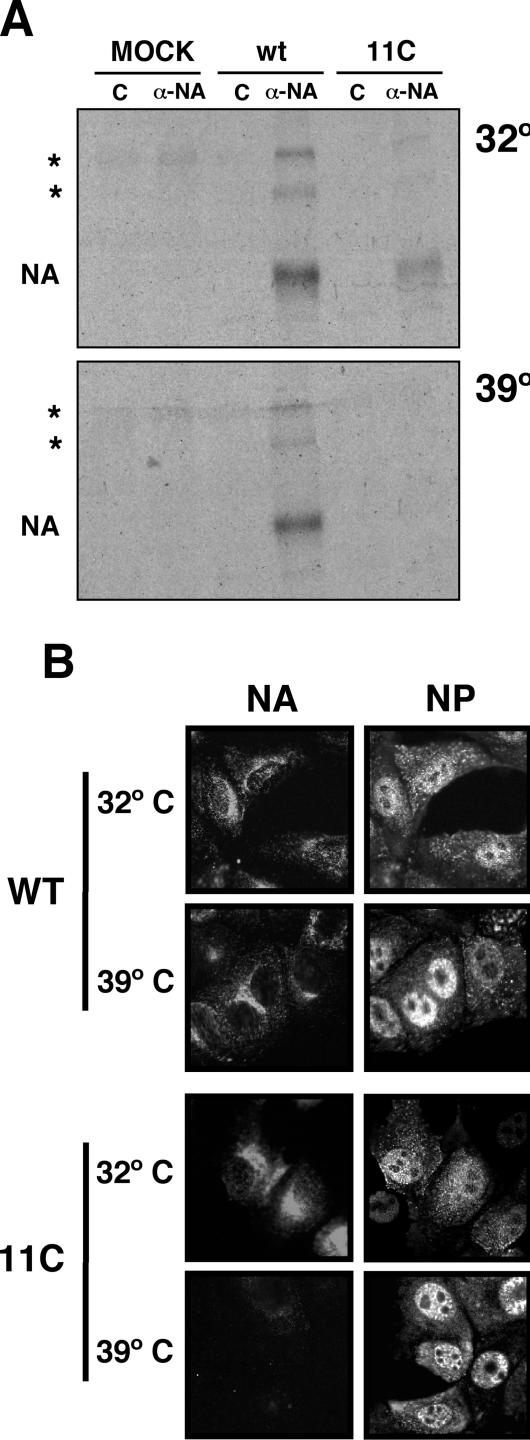
The synthesis of virus NA could not be determined with any certainty in direct labeling experiments (Fig. 7); hence, we carried out immunoprecipitation assays using specific anti-NA monoclonal antibodies. The results are presented in Fig. 8A. A strong reduction of NA expression was observed in mutant virus-infected cells at restrictive temperature compared to the results seen with a parallel infection with wt virus. This result was reinforced by immunofluorescence studies (Fig. 8B) that showed reduced NA signal in cells infected by 11C mutant virus at restrictive temperature.
FIG. 8.
Expression and localization of NA in wt and mutant virus-infected cells. Cultures of MDCK cells were infected with wt or mutant recombinant viruses at an MOI of 10 PFU/cell. (A) The cells were labeled continuously during the infection with [35S]met-cys. Cell extracts were prepared at 10 hpi (32°C) or 8 hpi (39.5°C) and immunoprecipitated with either the control or the M234/1G10 anti-NA monoclonal antibody. The protein immunoprecipitates were analyzed by polyacrylamide gel electrophoresis and autoradiography. The position of the NA band is indicated to the left of the images. The asterisks mark the positions of dimeric and tetrameric NA bands. (B) At 8 hpi (32°C) or 6 hpi (39.5°C) the cultures were fixed with paraformaldehyde. The cultures were analyzed by immunofluorescence with the M234/1G10 monoclonal, specific for NA, or a rat serum specific for NP. Mock-infected cells gave no fluorescence signal (data not shown).
RNA replication and transcription.
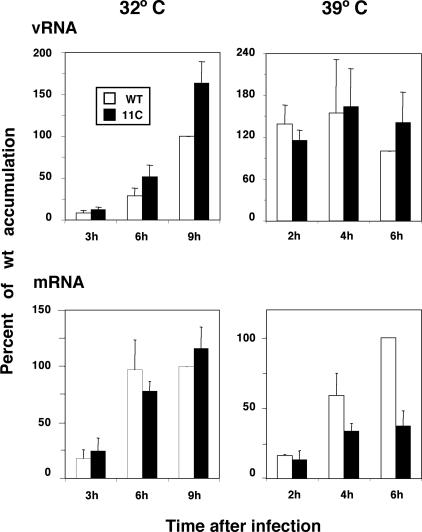
The decrease in NA expression in 11C mutant-infected cells might come as a result of defects in NA gene amplification, transcription, or translation. To ascertain which of these steps are affected by the mutations in 11C, the levels of virion and messenger RNAs were determined by dot blot hybridization in wt- and 11C mutant-infected cells at either permissive or restrictive temperature. The results obtained for the segment encoding NA are presented in Fig. 9. Limited alterations in the levels of mRNA and vRNA accumulations were observed. Thus, RNA replication of NA gene was more efficient at permissive temperature, while NA transcription was decreased two- to threefold at restrictive temperature. No alterations were observed for segment M RNA accumulations, and a slight increase in RNA replication and transcription was detected for the NP gene at both temperatures (data not shown).
FIG. 9.
Accumulation of NA-specific vRNA and mRNA. Cultures of MDCK cells were infected with wt or mutant recombinant viruses at an MOI of 10 PFU/cell and incubated at either 32 or 39.5°C. At the times indicated in the figure, total cell RNA was isolated and used for dot blot hybridization with positive- or negative-polarity riboprobes generated from NA cDNA. The RNA loading on the filters was standardized by hybridization with a 18S rRNA probe. The results presented are average values and standard deviations derived from at least three independent infection experiments. The accumulation of RNA obtained at the latest time after infection tested was defined as 100%.
Temperature shift experiments.
To determine the time point in the infection at which the virus function altered by mutations in 11C virus was required, we carried out classical temperature shift-up and shift-down experiments. Cultures of MDCK cells were infected at high multiplicity with either wt or mutant 11C viruses and incubated at 32.5 or 39.5°C. At various times after infection, cultures were transferred from one temperature to the other and incubation was continued until termination of a single infection cycle. The times for temperature shift were selected according to the length of the infection cycle at either temperature (see Fig. 3). Thus, the cultures initially incubated at 32°C were transferred at 2, 4, and 6 h postinfection (hpi), while the cultures initially incubated at 39.5°C were transferred at 1.5, 3, and 4.5 hpi. After all cultures had undergone one infection cycle (8 hpi at 32°C or 6 hpi at 39.5°C), the supernatants were collected and infectivity was determined at permissive temperature. At the same time that the cultures were transferred, parallel cultures were fixed for immunofluorescence, in an attempt to visualize the progress of the infection by the presence and localization of NP. The results are presented in Fig. 10. The final titer of wt virus-infected cells was essentially the same under all conditions of temperature shift (Fig. 10A), as expected. For mutant virus-infected cells, the decrease and increase curves corresponding to the shift-up and shift-down experiments crossed at a time after infection close to the third shift event (6 hpi at 32°C and 4.5 hpi at 39.5°C), indicating that the NS1 function affected by the mutation was required at a late stage in the infection cycle. When comparing these results with the kinetics of NP accumulation (Fig. 5) and localization (Fig. 10B), it became clear that such a function by NS1 protein is required at a time at which the NP has been expressed and a sizable fraction of it is located in the cytoplasm (see the depiction of wt virus staining at either temperature). In addition, the accumulation of NP in mutant virus-infected cells was not substantially different from the wt situation and there was no clear difference in the accumulation or localization of NP in mutant virus-infected cells at either temperature.
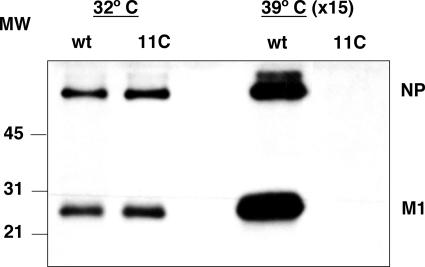
The appearance of virus particles from 11C mutant virus-infected cells is abolished at restrictive temperature.
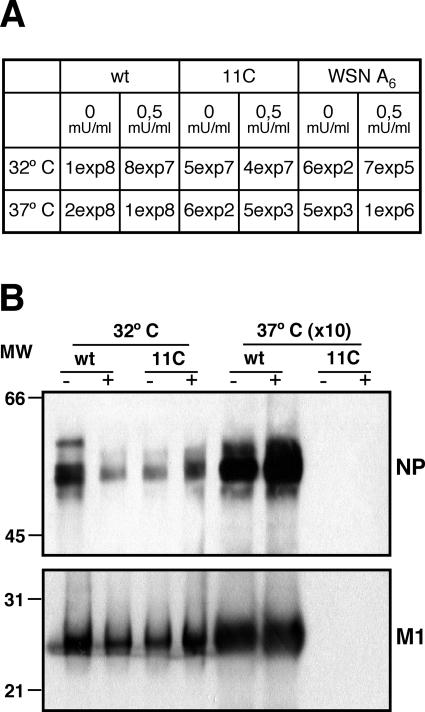
As the accumulations of NA and M2 were reduced in mutant-infected cells at restrictive temperature, we considered the possibility that the virus particles produced under these conditions would not be released from the infected cells or, alternatively, that they were defective in M2 content and hence were not able to infect new target cells. The virus particles produced in MDCK cells infected with wt and mutant viruses at either temperature were purified from the culture supernatant to estimate their relative concentration and their content in M2 protein by Western blot analysis. The results are presented in Fig. 11 and indicate that no virus particles were released from mutant virus-infected cells at restrictive temperature. Indeed, no signal for NP or M1 proteins could be detected even when 15 times more material was used for analysis than the sample obtained at permissive temperature. This result suggested a defect in virus particle production, virus release, or both. To differentiate among these possibilities, rescue experiments were performed in which MDCK cells were infected with wt virus or the 11C mutant at either temperature and bacterial NA was added to the culture supernatant. If the low expression of the NA gene were solely responsible for the ts phenotype of 11C mutant, full rescue would be expected to occur by complementation with exogenous NA. As a control, WSN mutant A6 (55) was used. It contains an oligo(A) sequence instead of the normal oligo(U) at the polyadenylation site in the NA gene and is defective for NA production. The results are presented in Fig. 12A and indicate that the ts phenotype of mutant 11C could not be rescued in this way. Thus, only a 10-fold increase in titer could be obtained by addition of exogenous NA, while the difference in virus yield at permissive versus restrictive temperatures was 5 log units. In constrast, addition of exogenous NA to an infection with WSN A6 virus led to increase in virus yield of around 3 log units. Furthermore, mutant virus particle production could not be rescued by exogenous NA treatment either, as presented in Fig. 12B.
FIG. 11.
Production of virus particles in wt and mutant virus-infected cells. Cultures of MDCK cells were infected with wt or mutant recombinant viruses at an MOI of 10 PFU/cell, and at 9 h postinfection the supernatant was collected. The virus particles were purified by centrifugation as indicated in Materials and Methods and analyzed by Western blotting with a mixture of anti-M1 and anti-NP antibodies. The position of the NP and M1 bands is indicated to the right, and the mobility of molecular weight markers in shown to the left. Material from 15 times more infected cells was loaded on the gel for samples obtained at restrictive temperature.
FIG. 12.
Rescue experiments using exogenous neuraminidase. Cultures of MDCK cells were infected with wt viruses, mutant recombinant viruses, or the WSN A6 mutant at an MOI of 10−3 PFU/cell, and the cultures were incubated at either permissive or restrictive temperature. Bacterial neuraminidase was added (+) or not (−) to the culture medium, and the supernatant was collected when cytopathic effect was apparent. (A) The virus titers were determined on MDCK cells at permissive temperature. (B) Virus particles were purified and analyzed by Western blotting as indicated in Materials and Methods and the legend to Fig. 11. The positions of the NP and M1 bands are indicated to the right, and the mobility of molecular weight markers in shown to the left. Material from 10 times more infected cells was loaded on the gel for samples obtained at restrictive temperature.
DISCUSSION
The influenza virus NS1 protein is a small, nonstructural virus factor that has been implicated in several steps during virus multiplication. Although it is not essential for virus viability (23), mutation in NS1 protein leads to defective viruses, some of which show temperature-sensitive phenotypes (12, 14, 17, 27, 28, 40, 60, 63-65). In addition, NS1 mutants are less virulent than wt viruses in experimental infections in animals (2, 15, 23, 68).
In spite of the extensive experimentation aimed at determining the possible functions of NS1, no complete picture yet exists to reveal its role during infection, possibly because of the pleiotropy of the NS1 mutations and the alteration of many aspects in the posttranscriptional processing of cell RNA caused by NS1 expression (7, 53). In this report we have undertaken a random, saturation mutagenesis approach in an attempt to reveal the possible functions of NS1 protein during an influenza virus infection. Instrumental in this experimental strategy was the development of a very efficient rescue system to introduce mutations into infectious virus. It is based on the observation that transfection of purified virion RNPs can lead to the production of infectious virus (39, 61) and the possibility of reconstituting and amplifying in vivo recombinant virus RNPs (52). When such recombinant RNPs are cotransfected in excess with total virion RNPs, around 90% of the virus progeny is a reassortant population in which the recombinant RNP has replaced the wt RNP (Fig. 1). Since the efficiency of the system is remarkable (thousands of individual recombinant virus plaques can be obtained), it was possible to rescue a preparation of mutant NS segment RNPs and produce a virus mutant library from which ts mutants could be screened. The sequencing of the mutant virus NS RNA segment indicated that 78% of the nucleotide changes led to amino acid exchanges and that, among these, 82% affected evolutionary conserved positions. The sequence data provided interesting insights about the relevance for virus infectivity of the various protein regions of NS1. Thus, most of the mutations concentrated in the N-terminal half of the protein, which is known to contain a double-stranded RNA-binding domain (37, 43) and to be sufficient to provide virus infectivity (12, 14, 17, 63). However, mutations also appeared in the C-terminal region, proposed to constitute an effector domain (57). This distribution of ts mutations lend support to the hypothesis, first proposed by Winter et al. (71), that the ancestral NS1 protein was shorter than that present in present-day viruses and that the extended C terminus might be generated by mutation of the existing termination codon. It is probable that the extended protein was evolutionary conserved, because new roles for virus replication were selected for, as supported by the isolation of ts mutants in this region (Fig. 2).
To start analyzing the role of the C-terminal portion of NS1 protein we chose mutant 11C for further study. The ts phenotype of 11C mutant could not be rescued in Vero cells, a non-IFN-responsive cell system, implying that the altered function in the mutant is not related to the NS1 interplay with the IFN cell response. However, we cannot exclude the possibility that the defect in 11C mutant NS1 is related to counteracting the cellular stress response (41), in this case the high-temperature condition.
A thorough comparison of the infection cycle of 11C mutant at permissive and restrictive temperatures indicated that several steps in the infection are slightly altered, in agreement with a multifunctional role for NS1 protein in the infection, but these alterations could not justify the dramatic reduction in virus titer at restrictive temperature. Thus, the synthesis of M1 and NS1 was slightly reduced, but this only led to a partial decrease of the accumulation of these proteins, and also of M2 protein, at restrictive temperature (Fig. 4 and 5). This is in contrast with the strong inhibition of M1 and M2 accumulation observed in other NS1 ts mutants with C-terminal deletions (17) and would not justify the ts phenotype of 11C virus. Thus, the levels of M1 protein were sufficient to allow a normal export of the virus RNPs from the nucleus at restrictive temperature (Fig. 10B), and it is known that even strong reductions in M1 protein expression induced by specific siRNAs lead to modest reductions in virus production (24, 30). On the other hand, the levels of NS1 protein itself were slightly diminished at restrictive temperature (Fig. 5); in addition, the steady-state accumulation of the mutant protein was higher in the nucleus than in the cytoplasm, at any time after infection and at either temperature, in contrast to wt NS1 results (Fig. 6).
This partial alteration in the nucleocytoplasmic distribution of mutant NS1 protein does not appear to be the only cause of the virus ts phenotype, as it is observed at both temperatures, but may contribute to diminish NS1 activity. As the S195P mutation eliminates a site with high phosphorylation potential (3), it is conceivable that such modification is important for NS1 export. Thus, an nuclear export signal sequence has been reported at positions 138 to 147 in NS1, whose activity is regulated by residues downstream in the protein (36). On the other hand, it has been reported that the nuclear export of late virus mRNA is blocked by the phosphorylation inhibitor H7 (69), but we could not observe any nuclear retention of viral mRNA (data not shown). The most important change in virus gene expression was observed for NA synthesis and accumulation (Fig. 8), but this did not correlate with similar alterations in M1 protein accumulation (Fig. 5); furthermore, HA synthesis and accumulation was not affected (Fig. 7). The reduction in NA expression could be attributed in part to a decrease in mRNA accumulation (Fig. 9). Indeed, implication of NS1 in regulating the proportion of transcription versus replication has been documented (17), although the phenotype of the NS1 deletion mutants was the opposite of that seen with 11C (17). These alterations may reflect an inappropriate interaction of NS1 with the virus RNPs or polymerase (45). However, the NA reduction cannot be fully explained by the diminished transcription, and a reduction in mRNA translation has to be assumed to account for the reduction in NA accumulation. This would be in line with the proposed role for NS1 protein in the translation of late virus mRNAs (1, 5, 8, 13, 14, 17). However, the results obtained with the 11C mutant indicate that there is a sequence-specific regulation of this NS1 function, as NA expression is affected but HA is not (Fig. 7).
The most dramatic phenotype observed with the 11C mutant was the almost absolute block in virus particle production (Fig. 11), although viral gene expression was only slightly altered. This correlated with the observation that the function affected in the 11C mutant is needed late in the infection. Thus, the synthesis and accumulation of NP and the export of RNPs are normal at restrictive temperature (Fig. 4, 5, and 9), and temperature-shift experiments documented that the function altered is required as late as 6 hpi (at 32°C) or 4.5 hpi (at 39.5°C), times at which a considerable fraction of wt progeny virus has been produced (see Fig. 2). Furthermore, these experiments showed that the ts mutation is fully reversible, an observation relevant to understanding the mechanism by which the mutation affects NS1 function.
Although a defect in virus particle release could contribute to the ts phenotype, due to the reduction of NA expression, the data presented in this report suggest that NS1 protein is involved in a late event during the morphogenesis of virus particles. Such an event would be prior to virion release but after RNP export from the nucleus and may bear a relation to the selection of the different virus RNPs for virion formation. Recent evidence indicates that the incorporation of the RNPs into virions is specific and that sequence signals located both in the segment untranslated regions and within the coding regions are important for specific RNP packaging (20, 21, 70). The phenotype of mutant 11C is compatible with a role for NS1 in the packaging process. Such a possibility is supported by the described interaction of NS1 protein with virus RNPs (45). In fact, the implication of RNA-binding proteins in the packaging of multicomponent RNA virus genomes has been previously reported for human immunodeficiency virus virions (6, 46). In this system, the association of unspliced virus RNA with Gag and human Staufen protein is important for the generation of infectious virus. In this context, it is remarkable that influenza virus NS1 protein has also been found associated with human Staufen protein (16, 44). At this point in time we can not exclude the possibility that the mutations found in the 11C mutant lead to the ts phenotype in cis, by alteration of the NS segment packaging signals, and not in trans. We deem this possibility unlikely, however, since all three mutations involve amino acid exchanges and since they lie outside of the NS RNA regions where these signals have been mapped (20).
Acknowledgments
We thank G. Brownlee, A. Douglas, A. García-Sastre, G. Hobom, A. Portela, and Y. Kawaoka for providing biological materials and A. Nieto for critically reading the manuscript. The technical assistance of Y. Fernández and J. Fernández is gratefully acknowledged.
A. M. Falcón and U. Garaigorta were fellows from Programa de Formación de Personal Investigador. This work was supported by Programa Sectorial de Promoción General del Conocimiento (grants BMC01-1223 and BFU2004-00491), Comunidad de Madrid (grant 08.2/0012.1/2001), and the European Network of Excellence ViRgil.
REFERENCES
- 1.Aragón, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Translation factor eIF4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cel. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 4.Briedis, D. J., G. Conti, E. A. Munn, and B. W. J. Mahy. 1981. Migration of influenza virus-specific polypeptides from cytoplasm to nucleus of infected cells. Virology 111:154-164. [DOI] [PubMed] [Google Scholar]
- 5.Burgui, I., T. Aragón, J. Ortín, and A. Nieto. 2003. PABP1 and eIF4GI associate to influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 6.Chatel-Chaix, L., J. F. Clément, C. Martel, V. Bériault, A. Gatignol, L. DesGroseillers, and A. J. Mouland. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 24:2637-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., and R. M. Krug. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376-383. [DOI] [PubMed] [Google Scholar]
- 8.de la Luna, S., P. Fortes, A. Beloso, and J. Ortín. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enami, K., T. A. Sato, S. Nakada, and M. Enami. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enami, M., and K. Enami. 2000. Characterization of influenza virus NS1 protein by using a novel helper-virus-free reverse genetic system. J. Virol. 74:5556-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falcón, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortín, and A. García-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817-2821. [DOI] [PubMed] [Google Scholar]
- 16.Falcón, A. M., P. Fortes, R. M. Marión, A. Beloso, and J. Ortín. 1999. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 27:2241-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falcón, A. M., R. M. Marión, T. Zürcher, P. Gómez, A. Portela, A. Nieto, and J. Ortín. 2004. Defective RNA replication and late gene expression in temperature-sensitive (A/Victoria/3/75) influenza viruses expressing deleted forms of NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortes, P., A. Beloso, and J. Ortín. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks RNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortes, P., A. I. Lamond, and J. Ortín. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 76:1001-1007. [DOI] [PubMed] [Google Scholar]
- 20.Fujii, K., Y. Fujii, T. Noda, Y. Muramoto, T. Watanabe, A. Takada, H. Goto, T. Horimoto, and Y. Kawaoka. 2005. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 79:3766-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujii, Y., H. Goto, T. Watanabe, T. Yoshida, and Y. Kawaoka. 2003. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl. Acad. Sci. USA 100:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 23.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Ge, Q., M. T. McManus, T. Nguyen, C. H. Shen, P. A. Sharp, H. N. Eisen, and J. Chen. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. USA 100:2718-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gluzman, Y. 1981. SV40 transformed simian cells support the replication or early SV40 mutants. Cell 23:175-182. [DOI] [PubMed] [Google Scholar]
- 26.Greenspan, D., P. Palese, and M. Krystal. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatada, E., M. Hasegawa, K. Shimizu, M. Hatanaka, and R. Fukuda. 1990. Analysis of influenza A virus temperature-sensitive mutants with mutations in RNA segment 8. J. Gen. Virol. 71:1283-1292. [DOI] [PubMed] [Google Scholar]
- 28.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatada, E., S. Saito, N. Okishio, and R. Fukuda. 1997. Binding of the influenza virus NS1 protein to model genome RNAs. J. Gen. Virol. 78:1059-1063. [DOI] [PubMed] [Google Scholar]
- 30.Hui, E. K., E. M. Yap, D. S. An, I. S. Chen, and D. P. Nayak. 2004. Inhibition of influenza virus matrix (M1) protein expression and virus replication by U6 promoter-driven and lentivirus-mediated delivery of siRNA. J. Gen. Virol. 85:1877-1884. [DOI] [PubMed] [Google Scholar]
- 31.Inglis, S. C., T. Barret, C. M. Brown, and J. W. Almond. 1979. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc. Natl. Acad. Sci. USA 76:3790-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krug, R. M., and P. R. Etkind. 1973. Cytoplasmic and nuclear specific proteins in influenza virus-infected MDCK cells. Virology 56:334-348. [DOI] [PubMed] [Google Scholar]
- 33.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, R. A., and P. W. Choppin. 1979. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. USA 76:4908-4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb, R. A., and C. J. Lai. 1984. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology 135:139-147. [DOI] [PubMed] [Google Scholar]
- 36.Li, Y., Y. Yamakita, and R. M. Krug. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. USA 95:4864-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 38.Lopez, J. A., M. Guillen, F. A. Sanchez, and J. A. Melero. 1986. An antigen-binding assay to determine the specificity of monoclonal antibodies against influenza virus and mapping of epitopes. J. Virol. Methods 13:255-264. [DOI] [PubMed] [Google Scholar]
- 39.López-Turiso, J. A., and J. Ortín. 1988. Transfection of MDCK cells with influenza virus ribonucleoprotein complexes. Microbiología SEM 4:167-175. [PubMed] [Google Scholar]
- 40.Ludwig, S., U. Vogel, and C. Scholtissek. 1995. Amino acid replacements leading to temperature-sensitive defects of the NS1 protein of influenza A virus. Arch. Virol. 140:945-950. [DOI] [PubMed] [Google Scholar]
- 41.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luytjes, W., M. Krystal, M. Enami, J. D. Parvin, and P. Palese. 1989. Amplification, expression and packaging of a foreign gene by influenza virus. Cell 59:1107-1113. [DOI] [PubMed] [Google Scholar]
- 43.Marión, R. M., T. Aragón, A. Beloso, A. Nieto, and J. Ortín. 1997. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 25:4271-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marión, R. M., P. Fortes, A. Beloso, C. Dotti, and J. Ortín. 1999. A human sequence homologue of staufen is an RNA-binding protein that localizes to the polysomes of the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2212-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marión, R. M., T. Zürcher, S. de la Luna, and J. Ortín. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447-2451. [DOI] [PubMed] [Google Scholar]
- 46.Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 48.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nieto, A., S. de la Luna, J. Bárcena, A. Portela, J. Valcárcel, J. A. Melero, and J. Ortín. 1992. Nuclear transport of influenza virus polymerase PA protein. Virus. Res. 24:65-75. [DOI] [PubMed] [Google Scholar]
- 50.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 51.Ohuchi, M., A. Cramer, M. Vey, R. Ohuchi, W. Garten, and H. D. Klenk. 1994. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J. Virol. 68:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ortega, J., J. Martín-Benito, T. Zürcher, J. M. Valpuesta, J. L. Carrascosa, and J. Ortín. 2000. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J. Virol. 74:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortín, J. 1998. Multiple levels of post-transcriptional regulation of influenza virus gene expression. Semin. Virol. 3:335-342. [Google Scholar]
- 54.Ortín, J., R. Nájera, C. López, M. Dávila, and E. Domingo. 1980. Genetic variability of Hong Kong (H3N2) influenza viruses: spontaneous mutations and their location in the viral genome. Gene 11:319-331. [DOI] [PubMed] [Google Scholar]
- 55.Poon, L. L., E. Fodor, and G. G. Brownlee. 2000. Polyuridylated mRNA synthesized by a recombinant influenza virus is defective in nuclear export. J. Virol. 74:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Portela, A., J. A. Melero, C. Martinez, E. Domingo, and J. Ortin. 1985. A primer vector system that allows temperature dependent gene amplification and expression in mammalian cells: regulation of the influenza virus NS1 gene expression. Nucleic Acids Res. 13:7959-7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian, X.-Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 60.Robertson, J. S., E. Robertson, I. Roditi, J. W. Almond, and S. C. Inglis. 1983. Sequence analysis of fowl plague virus mutant ts47 reveals a nonsense mutation in the NS1 gene. Virology 126:391-394. [DOI] [PubMed] [Google Scholar]
- 61.Rochovansky, O. M., and G. K. Hirst. 1976. Infectivity and marker rescue activity of influenza virus ribonucleoprotein-polymerase complexes. Virology 73:339-349. [DOI] [PubMed] [Google Scholar]
- 62.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 63.Salvatore, M., C. F. Basler, J. P. Parisien, C. M. Horvarth, S. Bourmakina, H. Zheng, T. Muster, P. Palese, and A. García-Sastre. 2002. Effects of influenza A virus NS1 protein on protein expression: the NS1 protein enhances translation and is not required for shutoff of host protein synthesis. J. Virol. 76:1206-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 65.Snyder, M. H., W. T. London, H. F. Maassab, R. M. Chanock, and B. R. Murphy. 1990. A 36 nucleotide deletion mutation in the coding region of the NS1 gene of an influenza A virus RNA segment 8 specifies a temperature-dependent host range phenotype. Virus Res. 15:69-83. [DOI] [PubMed] [Google Scholar]
- 66.Suárez, P., J. Valcárcel, and J. Ortín. 1992. Heterogeneity of the mutation rate of influenza virus: isolation of mutator mutants. J. Virol. 66:2491-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takeuchi, K., and R. A. Lamb. 1994. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 68:911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and S. A. Garcia. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogel, U., M. Kunerl, and C. Scholtissek. 1994. Influenza A virus late mRNAs are specifically retained in the nucleus in the presence of a methyltransferase or a protein kinase inhibitor. Virology 198:227-233. [DOI] [PubMed] [Google Scholar]
- 70.Watanabe, T., S. Watanabe, T. Noda, Y. Fujii, and Y. Kawaoka. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77:10575-10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Winter, G., S. Fields, M. J. Gait, and G. G. Brownlee. 1981. The use of synthetic oligodeoxynucleotide primers in cloning and sequencing segment 8 of influenza virus (A/PR/8/34). Nucleic Acids Res. 9:237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolff, T., R. E. O'Neill, and P. Palese. 1998. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 72:7170-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhirnov, O. P., T. E. Konakova, T. Wolff, and H. D. Klenk. 2002. NS1 protein of influenza A virus down-regulates apoptosis. J. Virol. 76:1617-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zürcher, T., R. M. Marión, and J. Ortín. 2000. The protein synthesis shut-off induced by influenza virus infection is independent of PKR activity. J. Virol. 74:8781-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]