Abstract
The largest tegument protein of herpes simplex virus 1 (HSV-1), UL36, contains a novel deubiquitinating activity embedded in it. All members of the Herpesviridae contain a homologue of HSV-1 UL36, the N-terminal segments of which show perfect conservation of those residues implicated in catalysis. For murine cytomegalovirus and Epstein-Barr virus, chosen as representatives of the beta- and gammaherpesvirus subfamilies, respectively, we here show that the homologous modules indeed display deubiquitinating activity in vitro. The conservation of this activity throughout all subfamilies is indicative of an important, if not essential, function.
Modification of proteins by ubiquitin (Ub) plays a pivotal role in a multitude of cellular processes, including proteolysis, cell cycle control, receptor internalization, and sorting within the endo/lysosomal system (7, 14, 16). Ubiquitination is achieved by an enzymatic cascade comprising a Ub-activating enzyme (E1), several Ub-carrier proteins (E2s), and hundreds of Ub ligases (E3s). Ubiquitination can be reversed by several families of enzymes collectively designated deubiquitinating enzymes (DUBs) (1, 15).
A number of viruses have evolved strategies to manipulate the ubiquitination status of host cell proteins, both through conjugation and deconjugation (2, 4, 6, 10, 13). Recently, we reported the identification of a novel viral ubiquitin-specific protease (USP), UL36USP, encoded by the herpes simplex virus 1 (HSV-1) genome (9). UL36USP is a polypeptide of approximately 420 amino acids (aa) carried within the N-terminal portion of UL36, the largest tegument protein (3,164 aa) of HSV-1. This activity was detected through the use of mechanism-based, active-site-directed probes and confirmed by expression in Escherichia coli of a corresponding fragment that cleaves ubiquitin-based substrates. UL36USP activity peaks at late stages of viral replication and appears to require proteolytic processing from full-length UL36 (9). The N-terminal UL36 fragment is well conserved in alphaherpesviruses, and a low homology to corresponding genes of the betaherpesvirus and gammaherpesvirus subfamilies was apparent in sequence alignments, but with strict conservation of the proposed catalytic residues. DUB activity may therefore be well conserved across the herpesvirus family and, if this is proven to be correct, would suggest an important function for this type of activity.
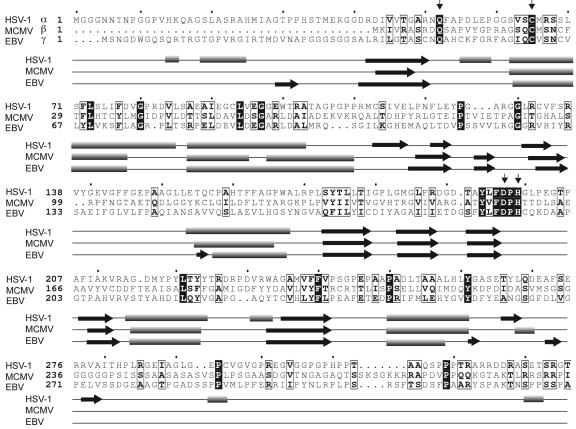
We therefore set out to investigate the possible DUB activity of two phylogenetically distant homologues of HSV-1 UL36USP, each representing a different subfamily of the Herpesviridae. We chose UL36 homologues encoded by mouse cytomegalovirus (MCMV, M48) and Epstein-Barr virus (EBV, BPLF1) as representatives of the beta- and gammaherpesvirus subfamilies, respectively. In order to assess the degree of homology between UL36 from HSV-1 and its MCMV and EBV counterparts, a sequence alignment was generated, covering the first 336 aa (the numbering refers to HSV-1) of UL36 (Fig. 1). Overall, the homology to HSV-1 is rather low, with only 10% and 15% sequence identity for MCMV and EBV, respectively. Nevertheless, the putative catalytic triad Cys-His-Asp is strictly conserved, along with a putative oxyanion hole-forming Gln residue (Fig. 1). The conserved Cys65 is the active-site cysteine in HSV-1 UL36USP (9). We therefore propose that the two homologues under investigation likewise represent cysteine protease-type DUBs. In support of this notion, a secondary structure prediction (11) shows a high degree of structural similarity despite limited sequence identity, suggesting that all members may adopt a similar tertiary structure (Fig. 1). Based on the observations that (i) the putative catalytic triad is contained within a stretch of ∼200 aa and (ii) the secondary structure prediction diverges beyond position ∼280, we speculate that a fragment of 280 aa or less should include the minimal domain necessary and sufficient for catalysis.
FIG. 1.
Key catalytic residues and the secondary structure of a ubiquitin-specific protease are conserved in three herpesvirus subfamilies. The sequence alignment of UL36 of HSV-1 (alphaherpesvirus subfamily) and its MCMV (betaherpesvirus subfamily) and EBV (gammaherpesvirus subfamily) homologues is shown. For clarity, only the N-terminal portion of UL36 is shown. Identical residues are white on a black background, and conserved residues are boxed. Putative active-site residues are marked by vertical arrows. The predicted secondary structure is depicted in alignment with the primary sequence. Symbols: cylinder, helix; horizontal arrow, strand; line, coil.
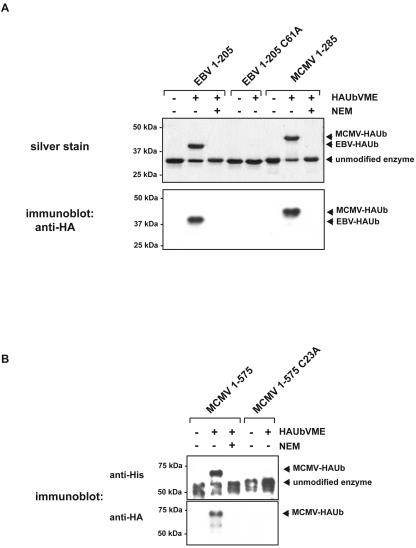
To investigate the functionality of this minimal DUB consensus domain, we cloned the relevant genomic fragments for heterologous expression and subsequent biochemical characterization. The corresponding DNA fragments, specifying aa 1 to 205 of EBV (EBV205) and aa 1 to 285 of MCMV (MCMV285), were PCR-cloned into pET28 according to standard procedures, using MCMV strain Smith and EBV B958 type 1 DNA as templates. Following expression in E. coli BL21-DE3, the fragments, equipped with an N-terminal His tag, were purified, using a Ni-NTA resin (QIAGEN), and subsequently subjected to gel filtration (S75 HiLoad; Amersham Pharmacia Biotech) in 50 mM Tris, 50 mM KCl, 50 mM NaCl, 0.5 mM EDTA, 2 mM dithiothreitol, and 10% (vol/vol) glycerol (pH 7.5) to achieve apparent homogeneity. To examine whether the purified constructs indeed display DUB activity, we used hemagglutinin (HA)-tagged Ub-vinylmethyl ester (HAUbVME), a probe that acts as a suicide inhibitor for DUBs by forming a thioether bond with the active-site cysteine (3, 12). Reactions were performed using final concentrations of 1 μM enzyme (EBV205 or MCMV285) in the absence and presence of 2 μM HAUbVME in reaction buffer (50 mM Tris, 100 mM NaCl, 1 mM dithiothreitol pH 7.5) for 30 min at 37°C. Following incubation, samples were boiled in reducing sample buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A shift in electrophoretic mobility, indicative of covalent modification by HAUb, was observed for both the MCMV and EBV constructs after silver staining (Fig. 2 A, upper panel). The identity of the covalent enzyme-HAUb adducts was confirmed by immunoblotting (Fig. 2 A, lower panel), using an HRP-conjugated anti-HA antibody (3F10; 1:4,000 dilution ratio; Roche) in conjunction with the Western Lightning chemiluminescence reagent kit (Perkin Elmer).
FIG. 2.
MCMV and EBV encode cysteine proteases that are targeted by a ubiquitin-derived probe. (A) Labeling of MCMV and EBV protease/tegument protein domains by HAUbVME. EBV205, EBV205 C61A, and MCMV285 were incubated with a twofold molar excess of HAUbVME in the absence or presence of NEM and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Silver stain was used for the upper panel. Anti-HA antibody directed against the probe was used for the immunoblot in the lower panel. Positions of the unmodified enzymes and the covalent HAUb adducts are indicated by arrowheads on the right. (B) Labeling of MCMV575 and its active-site mutant as for panel A, visualized by immunoblots using anti-His (upper panel) and anti-HA (lower panel) antibodies.
Are the proteins under investigation indeed cysteine proteases? Pretreatment of EBV205 or MCMV285 with 10 mM N-ethylmaleimide (NEM) for 10 min completely blocked labeling with the probe (Fig. 2 A). Replacement of the putative active-site cysteine by alanine (C61A) abrogated labeling with the probe for EBV205 (Fig. 2 A). In the case of MCMV, we also constructed a longer variant that terminates at position 575 (MCMV575). Although we were unable to obtain a pure preparation owing to low expression levels and susceptibility to proteolytic attack, a polypeptide of the expected molecular mass (62 kDa) was clearly detectable by immunoblotting, using a penta-His (QIAGEN) antibody (Fig. 2B). This variant likewise reacted with HAUbVME in NEM-sensitive fashion, and a nonconservative mutation of the putative active site (C23A) resulted in complete loss of labeling with the probe (Fig. 2B). Taken together with the established specificity of the electrophilic derivatives of Ub, we conclude that both the MCMV- and EBV-derived fragments are Ub-specific cysteine proteases.
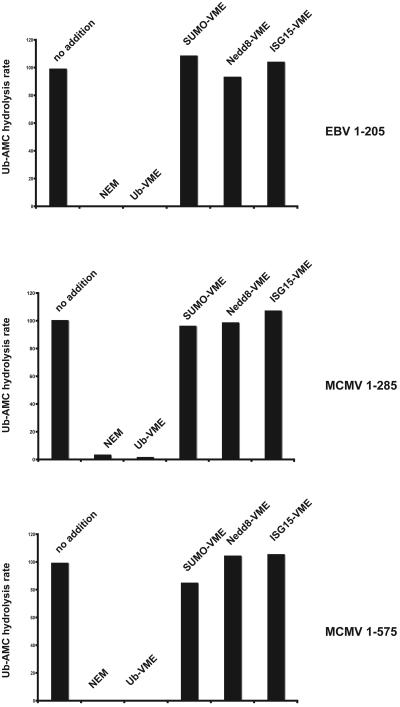
We next tested the ability of the enzymes to cleave Ub C-terminal 7-amido-4-methylcoumarin (Ub-AMC; Boston Biochem) (5). Ub-AMC hydrolysis assays were performed in reaction buffer supplemented with bovine serum albumin (50 μg/ml) by incubating the enzymes (100 pM) with a 1,000-fold excess of Ub-AMC (100 nM). EBV205, MCMV285, and MCMV575 efficiently hydrolyzed Ub-AMC, as had been shown for UL36USP (9). In accordance with HAUbVME labeling experiments, Ub-AMC hydrolysis was sensitive to NEM and was not observed for the active-site mutants, corroborating the identity of the proposed catalytic cysteine residue (Fig. 3 and data not shown). While a minimal DUB domain extending little beyond the active-site residues is active, we sought to determine whether the shorter constructs are also sufficient to confer substrate specificity. To address this issue, we tested the potential of several inhibitors to block DUB activity in Ub-AMC hydrolysis assays. Enzymes were preincubated for 1 h with a 100-fold molar excess of electrophilic derivatives of Ub, SUMO, Nedd8, and ISG15, all carrying a VME moiety as the electrophile at their C terminus (8). While UbVME completely inhibited hydrolytic activity, none of the Ub-related inhibitors impaired DUB activity (Fig. 3). We conclude that a domain consisting of little more than 200 aa, as exemplified by EBV205, mediates both catalytic activity and specificity towards Ub.
FIG. 3.
UL36 homologues of EBV and MCMV contain a protease module highly specific for ubiquitin. EBV and MCMV protease/tegument protein domains used for Ub-AMC hydrolysis are indicated on the right. Enzymes were either untreated or preincubated with a 100-fold molar excess of specific inhibitors (indicated on top) for 1 h at RT prior to the addition of Ub-AMC. The rate of Ub-AMC hydrolysis by the respective untreated construct was set to 100%.
In summary, we have demonstrated that a DUB activity previously identified in HSV-1 is conserved across all subfamilies of the Herpesviridae. Unexpectedly, a comparatively small structurally conserved module of ∼200 aa is sufficient to confer both hydrolytic activity and Ub specificity. Sequence elements adjacent to the 200-aa core domain may be required to confer additional specificity for ubiquitinated substrates in vivo or to mediate other interactions, for example, those required for subcellular localization. It will be necessary to raise antibodies against the N-terminal domain of these tegument proteins to establish their occurrence in virus-infected cells, to explore precursor-product relationships, and to relate these parameters to the deubiquitinating activity associated with them. Having demonstrated the enzymatic activity embedded within the tegument proteins of the alpha-, beta-, and gammaherpesviruses, we postulate an important and conserved function for this activity, the identification of which will require the creation of suitable mutants in the context of the intact genomes of the corresponding viruses.
Acknowledgments
This work was carried out with financial support from the National Institutes of Health (NIH) and from Astra Zeneca. C.S. is supported by a EMBO long-term fellowship (ALTF 187-2005); G.A.K. is supported by a NIH postdoctoral fellowship (GM072352-02); and L.M.K is supported by a NIH training grant (T32 AI07638).
EBV DNA was a kind gift of Eric Johannsen and Elliot Kieff.
REFERENCES
- 1.Amerik, A. Y., and M. Hochstrasser. 2004. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 1695:189-207. [DOI] [PubMed] [Google Scholar]
- 2.Banks, L., D. Pim, and M. Thomas. 2003. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 28:452-459. [DOI] [PubMed] [Google Scholar]
- 3.Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20:5187-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutell, C., and R. D. Everett. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 278:36596-36602. (First published 9 July 2003; 10.1074/jbc.M300776200.) [DOI] [PubMed] [Google Scholar]
- 5.Dang, L. C., F. D. Melandri, and R. L. Stein. 1998. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37:1868-1879. [DOI] [PubMed] [Google Scholar]
- 6.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 8.Hemelaar, J., A. Borodovsky, B. M. Kessler, D. Reverter, J. Cook, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, G. Gill, C. D. Lima, H. L. Ploegh, and H. Ovaa. 2004. Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 24:84-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 10.Li, M., C. L. Brooks, N. Kon, and W. Gu. 2004. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol. Cell 13:879-886. [DOI] [PubMed] [Google Scholar]
- 11.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 12.Misaghi, S., P. J. Galardy, W. J. Meester, H. Ovaa, H. L. Ploegh, and R. Gaudet. 2005. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J. Biol. Chem. 280:1512-1520. (First published 5 November 2004; doi: 10.1074/jbc.M410770220.) [DOI] [PubMed] [Google Scholar]
- 13.Ovaa, H., B. M. Kessler, U. Rolen, P. J. Galardy, H. L. Ploegh, and M. G. Masucci. 2004. Activity-based ubiquitin-specific protease (USP) profiling of virus-infected and malignant human cells. Proc. Natl. Acad. Sci. USA 101:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnell, J. D., and L. Hicke. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278:35857-35860. (First published 14 July 2003; 10.1074/jbc.R300018200.) [DOI] [PubMed] [Google Scholar]
- 15.Soboleva, T. A., and R. T. Baker. 2004. Deubiquitinating enzymes: their functions and substrate specificity. Curr. Protein Pept. Sci. 5:191-200. [DOI] [PubMed] [Google Scholar]
- 16.Welchman, R. L., C. Gordon, and R. J. Mayer. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6:599-609. [DOI] [PubMed] [Google Scholar]