Abstract
The evolution of dengue virus (DENV) is characterized by phylogenetic trees that have a strong temporal structure punctuated by dramatic changes in clade frequency. To determine the cause of these large-scale phylogenetic patterns, we examined the evolutionary history of DENV serotype 1 (DENV-1) and DENV-3 in Thailand, where gene sequence and epidemiological data are relatively abundant over a 30-year period. We found evidence for the turnover of viral clades in both serotypes, most notably in DENV-1, where a major clade replacement event took place in genotype I during the mid-1990s. Further, when this clade replacement event was placed in the context of changes in serotype prevalence in Thailand, a striking pattern emerged; an increase in DENV-1 clade diversity was associated with an increase in the abundance of this serotype and a concomitant decrease in DENV-4 prevalence, while clade replacement was associated with a decline in DENV-1 prevalence and a rise of DENV-4. We postulate that intraserotypic genetic diversification proceeds at times of relative serotype abundance and that replacement events can result from differential susceptibility to cross-reactive immune responses.
Dengue is one of the most important reemerging viral diseases, promulgated by increases in human population size and the expansion of global travel networks (10), coupled with the possibility that global climatic changes alter the distribution of the mosquito vector (11, 14). Although most cases of dengue virus (DENV) infection are subclinical or cause a febrile illness that is rarely fatal (dengue fever [DF]), increasingly large numbers of patients experience the more severe form of the illness, dengue hemorrhagic fever (DHF), associated with plasma leakage and hemorrhage. With a tendency to progress into fatal shock (dengue shock syndrome), case fatality rates of DHF/dengue shock syndrome vary from 1 to 5% (42). DENV is transmitted among humans primarily by the peridomestic mosquito species Aedes aegypti, although a transmission cycle involving sylvatic Aedes species and nonhuman primates has been described in parts of Africa and Asia (8, 21, 29). Genetically, DENV is a single-stranded, positive-sense RNA virus with a genome of approximately 11 kb that encodes 10 proteins translated as a single open reading frame and classified within the Flaviviridae. It is subdivided into four serotypes, denoted DENV-1 to DENV-4, that now cocirculate in many parts of the tropical and subtropical world. Importantly, there is growing epidemiological evidence that synergistic host interactions in response to these serotypes following sequential infection, vis-à-vis “antibody-dependent enhancement,” are instrumental in the pathophysiology of DHF (16, 34). As such, the study of epidemiological dynamics in populations in which DENV serotypes cocirculate is of special significance.
In recent years, there has been considerable interest in describing the genetic structures of DENV populations and determining their underlying evolutionary processes (13, 27). These studies have revealed that each serotype contains phylogenetically distinct clusters of viruses that have been christened “genotypes” or “subtypes” (13, 39). More-revealing observations are that (i) DENV strains from humans and nonhuman primates often fall into different genotypes, indicating that the human and sylvatic cycles are usually independent; (ii) some genotypes have widespread (or “cosmopolitan”) geographic distributions, while others are restricted to particular localities; and (iii) viral genetic diversity is greatest in Southeast Asia, suggesting that it is the “source” population for many dengue outbreaks, particularly those recently observed in the Americas (although our sampling from Africa remains sparse).
An increasingly common observation in phylogenetic studies of DENV is that of lineage turnover. This takes two forms. First, most viral lineages sampled from a particular time point fail to generate any daughter lineages, resulting in phylogenetic trees with a strong temporal (ladder-like) topology (4, 15). This most likely occurs because these lineages are defined by viruses that contain deleterious or slightly deleterious mutations that are eliminated by purifying selection (12, 15) and/or because fully functional viruses are not sampled in later years due to regular bottlenecks in viral population size, perhaps due to seasonal and spatial fluctuations in vector population size and density (30). A second, more dramatic mode of lineage turnover occurs when an entire clade of viruses that has persisted in a particular locality for a number of years is not evident on subsequent sampling, indicating that it has dropped dramatically in frequency, perhaps even experiencing extinction, and is sometimes replaced by a new clade of viruses. A number of occurrences of such dramatic clade replacement have been reported (4, 32, 41), although the evolutionary processes controlling these events remain uncertain. In particular, this large-scale phylogenetic pattern could reflect the action of either dramatic population bottlenecks or natural selection, such that clades with enhanced fitness out-compete those that are less fit.
There is growing experimental evidence that strains of DENV (that is, either entire genotypes or smaller groupings of viruses within them) differ in fitness, as required for selectively driven clade replacement. Most notably, the “American” genotype of DENV-2 appears to be less well adapted to Aedes aegypti from the Americas than are Asian genotypes of this virus (2, 3) and also does not replicate as well in certain human cell types (6). Together, these genetic differences are thought to have lowered the fitness of American genotype viruses in comparison to those viruses imported from Southeast Asia. However, such fitness differences will be manifest only at times of selective competition because the American genotype is still highly transmissible under suitable epidemiological conditions (40).
Well-characterized and archived dengue viruses serially collected within Thailand provide a unique opportunity to explore the evolutionary processes which underlie clade replacement. In particular, (i) all four DENV serotypes cocirculate with varying relative predominance from year to year, (ii) dramatic changes in clade frequency have previously been observed in DENV-2 and DENV-3, and (iii) epidemiological and clinical data are available from 1973 to the present from the metropolitan area of Bangkok (24). Herein, we undertake a detailed analysis of the temporal dynamics of DENV-1 and DENV-3 evolution over a 30-year sampling period in Thailand. We include viruses isolated from patients experiencing a variety of disease manifestations and also explore the spatial diffusion of both serotypes in geographical locations other than Bangkok. In particular, we noted a number of cases of clade replacement in these viruses and explored whether stochastic or selective forces were the cause of these major phylogenetic events. As Bangkok is considered a potential epicenter of DENV transmission throughout Thailand (7) and potentially beyond, a better understanding of the forces influencing dengue virus evolution and epidemiology within Bangkok has potential widespread implications within Southeast Asia.
MATERIALS AND METHODS
Specimen data.
DENV-1 and DENV-3 were isolated from 164 children hospitalized at the Queen Sirikit National Institute of Child Health (QSNICH) from 1974 to 2002 (see the supplemental material). Of the DENV-1 cases, 28 patients experienced DF, 43 experienced DHF, and 25 had unclassified dengue disease. Of the DENV-3 cases, the equivalent figures were 30 patients with DF and 38 with DHF. Grading of dengue disease severity associated with these diagnostic specimens was conducted by QSNICH physicians using World Health Organization (WHO) classification guidelines. Testing at the Armed Forces Research Institute of Medical Sciences (AFRIMS) characterized the specimens according to their DENV serotype by reverse transcription-PCR (RT-PCR) followed by nested PCR.
RNA extraction and RT-PCR.
Virus RNA was extracted from cell culture supernatant (samples before 2000) or serum (samples after 2000) using the QIAamp viral RNA mini kit (QIAGEN, Germany) according to the manufacturer's instructions. Oligonucleotide primers for RT-PCR amplification and sequencing were designed using the Primer3 program (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. The sequences and positions of primers used to determine the complete genomes of DENV-1 and DENV-3 are listed in the supplemental material. All specimens used for sequencing were pretested using RT-PCR and nested PCR to confirm assignment to DENV-1 and DENV-3 and to adjust virus concentration. Genomic RNA was converted to cDNA using random hexamer oligonucleotides with the SuperScript first-strand synthesis system (Invitrogen) according to the manufacturer's instructions. All DNA fragments of the envelope (E) gene and the overlapping DNA fragments which covered the entire genome for 10 strains of DENV-1 and 6 strains of DENV-3 were amplified by PCR with Proofstart (QIAGEN, Germany) and Taq DNA polymerase (Roche Ltd.). The PCR-amplified DNA fragments were purified using QIAquick PCR purification kits and the QIAquick gel extraction kit (QIAGEN, Germany) according to the manufacturer's instructions. Purified DNA fragments were used for sequencing.
Sequencing and analysis.
Cycle sequencing reactions were performed using the DYEnamic ET dye terminator sequencing kit (Amersham Pharmacia Biotech), according to the manufacturer's instructions, with sequencing primers described in the supplemental material. The sequencing products were cleaned by standard precipitation before sequencing on a MegaBACE 500 automated DNA sequencer (Amersham Pharmacia Biotech). Overlapping nucleic acid sequences were combined for analysis and edited with the aid of Sequencher software (Gene Code Corp.).
Phylogenetic analysis.
For both DENV-1 and DENV-3, a sample of 120 complete E gene sequences was used in the phylogenetic analysis. This included all the Thai DENV data newly generated here, other than four DENV-1 and two DENV-3 sequences that were identical to another sequence in the data set (ThD1-PT0214/96, ThD1-0120/00, ThD1-0049/01, ThD1-0261/01, ThD3-0007/87, and ThD3-1283/98). These Thai sequences were then combined with a sample of viruses representing the global genetic variability of DENV-1 and DENV-3. Sequence alignment was undertaken manually, which resulted in alignments of 1,485 bp and 1,479 bp for DENV-1 and DENV-3, respectively. To explore the issue of clade replacement in more detail, we also conducted a phylogenetic analysis on the complete coding regions of 65 DENV isolates representing all four serotypes. In this case, sequence alignment was undertaken using the ClustalX program (35) and then checked manually, resulting in a final data set 10,185 bp in length. All data sequences used in this study are listed in the supplemental material. Maximum likelihood (ML) phylogenetic trees were then estimated for all three data sets. In all cases, we used the most general GTR + I + Γ4 model of nucleotide substitution (where GTR is general time reversal, I is invariant sites, and Γ is rate parameters) available in the PAUP* package (33) and undertook successive rounds of branch swapping. All parameter values are available upon request. To determine the support for particular groupings on the phylogenetic trees, we performed a bootstrap resampling analysis using 1,000 replicate neighbor-joining trees estimated under the ML substitution model.
Estimating selection pressure.
We used a variety of computational methods to explore the selection pressures acting on DENV-1 and DENV-3. We also analyzed two different types of data: (i) the complete coding regions from the 10 Thai DENV-1 strains determined here and nine complete DENV-3 genomes, comprising the six Thai viruses determined here plus three other complete genomes available in GenBank, and (ii) a sample of 80 E gene sequences representative of DENV-1 and DENV-3 in Thailand. To match these selection analyses with the occurrence of clade replacement, we restricted the E gene analysis to DENV-1 strains assigned to genotype I and DENV-3 strains assigned to genotype II.
Mean ratios of nonsynonymous to synonymous substitutions per site (dN/dS) for each data set were measured using the pairwise method of Nei and Gojobori (23) as implemented in the MEGA2 package (17). To obtain codon- and lineage-specific measures of selection pressure, we employed the maximum likelihood method in the CODEML program (43). This involved the use of four models of codon evolution (44). To analyze selection pressures at individual codons, we compared the M7 and M8 models; in the former codons, we took 1 of 10 categories of dN/dS ratios, all estimated from the data but where no category had a dN/dS ratio of >1.0 so that the M7 model specified only neutral evolution. M8 allowed positive selection by adding an 11th category of codons at which dN/dS can exceed 1.0. Evidence for positive selection is obtained if M8 significantly rejects M7 in a likelihood ratio test and has at least one category of codons with a dN/dS ratio of >1. To analyze selection pressures along each lineage of the DENV-1 and DENV-3 phylogenies, we compared M0, in which each lineage has the same dN/dS ratio, with the “free-ratio” model, in which lineages are allowed to take on different values of dN/dS. For the M0 and “free-ratio” models, dN/dS is estimated from the data, and their support is again assessed using a likelihood ratio test.
We also examined the biological characteristics of the amino acid changes that were associated with the major clade replacement event in DENV-1. First, we mapped amino acid changes onto the complete genome ML phylogenetic tree for DENV-1 using the parsimony approach available in the MacClade package (19) and noted all those that occurred on the branches associated with clade replacement. To determine whether amino acid changes occurring on branches associated with clade replacement were common or rare in dengue virus evolution, we recorded the frequency of all types of amino acid changes on the ML tree of the 65 complete genomes from all four viral serotypes. This allowed us to characterize 1,909 unambiguous amino acid changes in all genes across the DENV phylogeny. We then counted the frequency of amino acid replacements from each amino acid and determined whether the types of amino acid changes that occurred on the branches associated with clade replacement were more or less common than expected, given their overall frequency in DENV evolution. For example, of the 1,909 unambiguous amino acid changes, 150 were from the amino acid Gly to another amino acid, so that the expected frequency of these types of amino acid changes is 7.9 (150 Gly amino acid changes divided by 19 possible amino acid changes). The major clade replacement event in DENV-1 is associated with a Gly-to-Ala change. As the observed frequency of this change is 9/150, we conclude that this particular amino acid change occurs at its expected frequency and, hence, is not rare in DENV evolution.
Nucleotide sequence accession numbers.
All sequences produced here have been submitted to GenBank (accession numbers AY732378 to AY732483 for DENV-1 and AY676348 to AY676421 for DENV-3).
RESULTS
Molecular epidemiology of DENV-1.
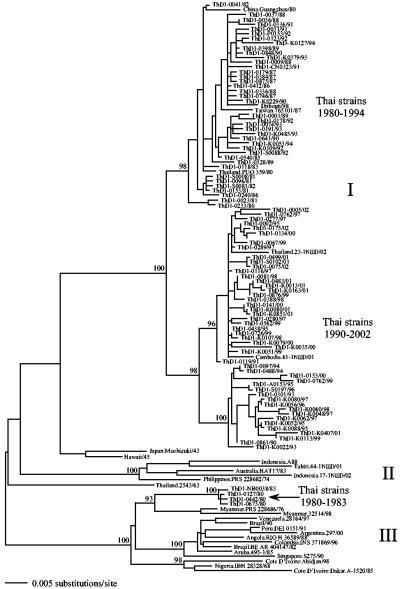
Three phylogenetically distinct groupings of viruses, which can be referred to as genotypes (1), were evident in the global phylogenetic tree of 120 DENV-1 isolates (Fig. 1). Those assigned to genotype I are principally Asian in origin, having been sampled in Cambodia, China, Taiwan, and Thailand. The only exception is a single strain from Djibouti in East Africa, which was most likely a recent importation. The genotype I viruses can be further subdivided into three well-supported groups (that is, clusters of viruses with strong bootstrap support) that are described in more detail below. A wider geographical distribution is represented by genotypes II and III. Genotype II viruses are found in diverse geographical locations in the Asia-Pacific region (Australia, Indonesia, Philippines, and Tahiti). That relatively long branches connect these viruses suggests that the viruses have experienced a history of population subdivision (1). The most geographically cosmopolitan viruses are those assigned to genotype III. These included viruses from Asia (including Thailand), Africa, and Latin America, each of which forms a strongly supported group. The phylogenetic relationships of three DENV-1 viruses—Japan.Mochizuki/43, Hawaii/45, and Thailand.2543/63—were difficult to classify into any established genotype.
FIG. 1.
ML tree of 120 strains of DENV-1, including 92 newly sampled strains from Thailand. Thai strains sampled from regions other than Bangkok are denoted as follows: A, Ayuthaya; CN, Chai Nat; K, Kamphaeng Phet; NB, Nonthaburi; P, Pichit; PT, Pathum Thani; and S, Samut Prakan. Key clades are indicated. Bootstrap support values are shown for major nodes on the tree. The tree is midpoint rooted for purposes of clarity only, and all horizontal branch lengths are drawn to scale.
The DENV-1 strains sampled from Thailand fall into two of the three genotypes. The majority clustered within genotype I. However, four strains, collected in 1980 and 1983, fell into genotype III, where they were most closely related to viruses sampled in neighboring Myanmar from 1976 to 1998. Hence, these strains form an Asian cluster within genotype III, albeit one that is rarely sampled. Equally noteworthy was that the Thai strains within genotype I fell into three distinct clades; two of them are associated with different sampling times (Fig. 1). Specifically, all those viruses sampled during the 1980s as well as a majority of those sampled from 1990 to 1994 grouped together and are herein referred to as the “1980 to 1994” clade. In contrast, a minority of the strains sampled in the early 1990s as well as all those sampled after 1994 fell into two other clusters, which form a larger phylogenetic group herein referred to as the “1990 to 2002” clade.
Molecular epidemiology of DENV-3.
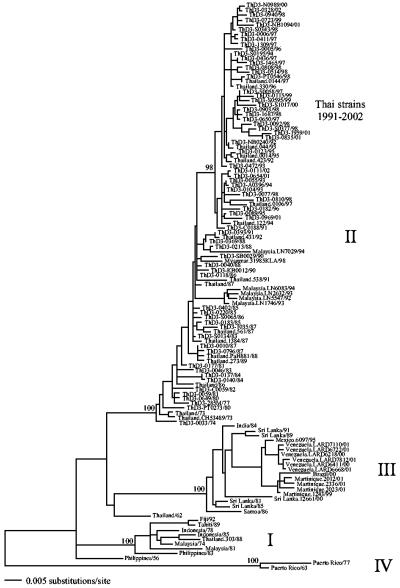
Our analysis of 120 DENV-3 isolates identified the four genotypes that have been recognized previously in this serotype (18, 38) (Fig. 2). Briefly, genotype I comprises viruses from Southeast Asia and the Pacific (although none have been sampled since 1992); the large genotype II is entirely Asian in composition, containing viruses from Malaysia, Myanmar, and Thailand; genotype III, which first invaded the Americas in the early 1990s (38), comprises viruses sampled from Asia (Sri Lanka, India), the Pacific (Samoa), and Latin America (Brazil, Martinique, Mexico, Venezuela); and genotype IV, which may have gone extinct, consists of two viruses sampled from Puerto Rico in the 1960s and 1970s. Finally, two viruses did not fall into a clear genotype: one sampled from the Philippines in 1956 and another sampled from Thailand in 1962. All the Thai DENV-3 sequences generated here fell into genotype II. Furthermore, clear phylogenetic groups associated with time of sampling were present within this genotype, with the majority of those strains sampled post-1991 (and none earlier) falling into a well-supported group. This is referred to as the “1991 to 2002” clade.
FIG. 2.
ML tree of 120 strains of DENV-3, including 66 newly sampled strains from Thailand. Thai strains sampled from regions other than Bangkok are denoted as follows: A, Ayuthaya; C, Chachoengsao; N, Nakhon Pathom; NB, Nonthaburi; KB, Kanchana Buri; PT, Pathum Thani; S, Samut Prakan; and SB, Suphan Buri. Key clades are indicated. Bootstrap support values are shown for major nodes on the tree. The tree is midpoint rooted for purposes of clarity only, and all horizontal branch lengths are drawn to scale.
Between both serotypes, there was no clear pattern of spatial separation among the Thai strains. Those sampled from Bangkok and other provinces in central and northern Thailand (ranging from 20 to 344 km from Bangkok) were interspersed throughout the tree. Furthermore, in neither serotype was there any indication that particular E gene variants were associated with serious dengue disease, as viruses associated with patients suffering from DHF fell into a variety of locations on the phylogenetic tree. This is in concordance with most previous studies of DENV (15, 31, 38).
Clade replacement in DENV-1 and DENV-3.
The most striking pattern in our phylogenetic analysis was that of clade replacement. This is most clearly seen within genotype I of DENV-1, in which the frequency of the 1980 to 1994 clade drops dramatically and may even have experienced extinction (although this will need to be confirmed with larger sample sizes), to be replaced by the 1990 to 2002 clade. Specifically, during the period from 1990 to 1994, these two clades of DENV-1 cocirculated in Thailand, but after 1994, one of them (the 1990 to 2002 clade) dominated. There is also some evidence for another replacement event in DENV-1 in Thailand during the early 1980s, in which the genotype III viruses were replaced by those assigned to genotype I. Similarly, in genotype I of DENV-3, a clade replacement took place during the early 1990s, resulting in the 1991 to 2002 clade.
To help determine whether the major clade replacement event in DENV-1 was caused by differences in viral fitness (that is, by the positive selection of one clade over the other), we examined selection pressures among all genes and lineages. We found no evidence for adaptive evolution acting on any individual gene, site, or lineage of the DENV-1 data sets (Table 1). Indeed, the overwhelming evolutionary pressure was that of purifying selection; the mean dN/dS ratios across genes were extremely low, ranging from 0.010 to 0.129, although there was a 7-fold variation in dN among genes and a 2.7-fold variation in dS. A similar picture of selective constraint across all genes was observed in DENV-3, with a 3-fold variation in dN and a 3.4-fold variation in dS (Table 2). Even more notable was the lack of compelling evidence for positive selection accompanying the lineage replacement events in DENV-1. In no gene did dN exceed dS on the branches separating the 1980 to 1994 and 1990 to 2002 clades, and in most cases, the dN/dS ratios on these branches were no greater than the means across all branches of the DENV-1 tree, further suggesting that they are not especially elevated. The exceptions to this were in the E, NS2A, and NS4A genes (Table 1), although the dN/dS values were still sufficiently low (reaching a maximum of 0.203 in NS2A) to imply that purifying selection was again the dominant evolutionary process acting on this lineage. A similar picture was seen in DENV-3. Here, the branch associated with clade replacement in 1991 had slightly elevated dN/dS ratios in the E, NS2A, NS3, and NS4A genes, although none suggested a major change in selection pressures (Table 2).
TABLE 1.
Selection pressures in DENV-1 on a gene-by-gene basisa
| Gene | Length (no. of amino acids) | dN | dS | dN/dS | dN/dS Separating genotype I clades (mean)c |
|---|---|---|---|---|---|
| Capsid | 114 | 0.012 | 0.093 | 0.129 | 0.061 (0.231) |
| Membrane | 166 | 0.006 | 0.189 | 0.032 | 0.049 (0.050) |
| Envelope | 495 | 0.008 | 0.173 | 0.046 | 0.100 (0.056) |
| Envelopeb | 495 | 0.005 | 0.086 | 0.058 | 0.078 (0.072) |
| NS1 | 352 | 0.007 | 0.214 | 0.033 | 0.029 (0.050) |
| NS2A | 218 | 0.014 | 0.235 | 0.060 | 0.203 (0.098) |
| NS2B | 130 | 0.002 | 0.201 | 0.010 | 0.0001 (0.016) |
| NS3 | 619 | 0.005 | 0.194 | 0.026 | 0.019 (0.036) |
| NS4A | 150 | 0.006 | 0.200 | 0.030 | 0.064 (0.041) |
| NS4B | 249 | 0.004 | 0.192 | 0.021 | 0.0001 (0.029) |
| NS5 | 899 | 0.006 | 0.173 | 0.035 | 0.042 (0.050) |
Estimates of dN, dS, and dN/dS are based on 10 Thai DENV-1 sequences.
Estimates are based on a sample of 80 E gene sequences.
Numbers in bold indicate cases where the dN/dS value for the genotype I clade replacement branches is greater than the mean dN/dS value for all branches of this particular gene tree.
TABLE 2.
Selection pressures in DENV-3 on a gene-by-gene basisa
| Gene | Length (no. of amino acids) | dN | dS | dN/dS | dN/dS Separating genotype II clades (mean)c |
|---|---|---|---|---|---|
| Capsid | 114 | 0.009 | 0.098 | 0.092 | NA (0.109) |
| Membrane | 166 | 0.005 | 0.116 | 0.043 | 0.0001 (0.057) |
| Envelope | 493 | 0.007 | 0.178 | 0.039 | 0.049 (0.057) |
| Envelopeb | 495 | 0.003 | 0.060 | 0.050 | 0.064 (0.056) |
| NS1 | 352 | 0.005 | 0.148 | 0.034 | 0.0001 (0.049) |
| NS2A | 218 | 0.009 | 0.205 | 0.044 | 0.116 (0.069) |
| NS2B | 130 | 0.005 | 0.150 | 0.033 | 0.0001 (0.059) |
| NS3 | 619 | 0.003 | 0.170 | 0.017 | 0.067 (0.023) |
| NS4A | 150 | 0.007 | 0.145 | 0.048 | 0.207 (0.067) |
| NS4B | 248 | 0.003 | 0.158 | 0.019 | 0.0001 (0.029) |
| NS5 | 900 | 0.004 | 0.163 | 0.025 | 0.0001 (0.043) |
Estimates of dN, dS, and dN/dS are based on six Thai DENV-3 sequences.
Estimates are based on a sample of 80 E gene sequences.
NA, not applicable (the branch in question does not exist in the capsid gene). Numbers in bold indicate cases where the dN/dS value for the genotype II clade replacement branch is greater than the mean dN/dS value for all branches of this particular gene tree.
To further explore the cause of the 1990s major clade replacement event in DENV-1, we analyzed the patterns of variation and the physicochemical properties of the amino acid changes associated with this event (with our ML tree of the 65 complete genome sequences available in the supplemental material). The branches separating the two clades of genotype I were defined by 13 amino acid changes, 5 of which were unique to the branch leading to the 1994 to 2002 clade: position 9 in the capsid (C) gene, positions 8 and 324 in the E gene, position 466 in the NS3 gene, and position 325 in the NS5 gene (Table 3). Among the 13 amino acid changes, 4 were located in the E gene, and most of these (3/4) were at positions that are invariant in the other dengue serotypes, implying that they might have major effects on fitness. In particular, changes at site 380 have previously been associated with positive selection in DENV-3 (37). However, all four amino acid changes in the E gene were conservative in nature and are frequently observed in DENV evolution, as was generally the case for the amino acid changes observed in the other genes.
TABLE 3.
Amino acid changes associated with the major clade replacement event in DENV-1
| Gene | Sitea | Change | Serotype variation(s)c | Global frequencye | Type of changef |
|---|---|---|---|---|---|
| C | 9 | Gly to Alab | DENV-2, -4 | 9 (7.9) | Conservative |
| M | 15 | Ser to Thr | DENV-2, -3, -4 | 37 (6.5) | Conservative |
| E | 8 | Asn to Serb | Noned | 29 (3.8) | Nonconservative (small polar, uncharged to small neutral, uncharged) |
| 324 | Val to Ileb | Noned | 100 (10.9) | Conservative | |
| 351 | Leu to Val | DENV-4 | 18 (8.9) | Conservative | |
| 380 | Ile to Val | Noned | 108 (10) | Conservative | |
| NS2A | 148 | Thr to Ala | None | 65 (10.4) | Conservative |
| NS3 | 466 | Gln to Hisb | Noned | 8 (2.2) | Nonconservative (relatively small polar, uncharged to large polar, positive) |
| NS4A | 76 | Lys to Arg | DENV-2 | 113 (9.2) | Conservative |
| NS5 | 108 | Pro to Ala | Noned | 9 (2.3) | Conservative |
| 325 | Arg to Lysb | None | 89 (6.5) | Conservative | |
| 367 | Arg to Gln | DENV-4 | 7 (6.5) | Nonconservative (large polar, positive to relatively small polar, uncharged) | |
| 807 | Asp to Asn | Noned | 24 (4.2) | Nonconservative (relatively small polar, negative to relatively small polar, uncharged) |
Amino acids are numbered individually for each gene.
This amino acid change is on the branch leading to the 1994 to 2002 clade of DENV-1.
Variation is assessed in terms of whether the amino acid in question varies within other serotypes. The serotypes with variation at these sites are indicated.
These amino acid sites are invariant across all four dengue serotypes.
Shown is the frequency of a specific amino acid change with respect to all types of change from that amino acid. The number in parentheses is that expected given the change's overall frequency in DENV evolution. In no case was the observed frequency of an amino acid change less frequent than expected.
Assessed in terms of weight and charge.
DISCUSSION
Although dengue virus falls into serotypes that are as phylogenetically distinct as many viral species, the analysis of increasing amounts of gene sequence data has revealed some important generalities in its evolution. In particular, our estimates of overall selection pressures (average dN/dS values) are extremely similar among both genes and serotypes and confirm that the strongest pressure shaping DENV evolution is purifying selection (12). In both DENV-1 and DENV-3, the gene with the highest average dN/dS ratio is the capsid, although in both serotypes it is the NS2A gene that has the highest rate of amino acid change, as has also been noted for DENV-4 (15). The precise cause of the increased rate of amino acid change in the NS2A gene is unknown and merits further investigation. Similarly, although the variation in synonymous rates among genes is not substantial, it is unclear why dS is relatively low in the E genes of both DENV-1 and DENV-3.
A potentially more important observation is that patterns of intraserotype genetic diversity in DENV vary greatly on a temporal scale. Most notably, through the analysis of longitudinally sampled data, it has become clear that individual lineages or entire clades of viruses frequently arise, persist for a period of time, and then disappear. Consequently, DENV phylogenetic trees often have a ladder-like structure, although it is less pronounced than that observed in the hemagglutinin gene of human influenza A virus (9), interspersed with episodes of clade replacement. In principle, clade replacement events could be produced by three contrasting evolutionary processes: (i) the clades in question differ in fitness, so the “winning” clade has a fitness advantage over the preceding clade, as suggested for influenza A virus (5); (ii) replacement events are due entirely to stochastic factors, most likely involving major fluctuations in mosquito population size and density; and (iii) clade replacement is dependent on the composite “herd” immunologic profile (antibody and T-cell priming) to each of the four serotypes. We employed a variety of computational methods in an initial attempt to tease apart the respective contributions of natural selection and genetic drift to dengue virus evolution. Strikingly, we found no evidence for positive selection in DENV-1 and DENV-3 in Thailand acting on the gene, codon, or lineage levels. Indeed, dN/dS values were usually <0.2, and frequently <0.05, indicating that the vast majority of nonsynonymous mutations that arise in DENV are slightly deleterious, fall onto the external branches (that is, the tips) of molecular phylogenetic trees, and are eventually removed from the population by purifying selection. The importance of transient deleterious mutations has been increasingly recognized in DENV evolution (12, 15, 41) and supports a “slightly deleterious” model of molecular evolution in which most mutations lower fitness and which can be fixed by genetic drift only in relatively small populations (25, 26).
Despite the preponderance of transient deleterious mutations in DENV evolution, we are unable to rule out the possibility that positive selection has acted on a small subset of amino acid changes that cannot be detected by most analytical methods, particularly where substitutions are isolated to one branch of the tree or interact epistatically. Consequently, computational methods, such as those employed here, should be used only as a way of identifying those sites of interest for future experimental analysis. This is most pertinent for those branches associated with lineage replacement. In particular, the major lineage replacement event in DENV-1, which took place in the mid-1990s, was associated with 13 amino acid changes across the viral genome. Of these, we reasoned that the changes most likely to change viral fitness were those that (i) could be classified as nonconservative in terms of their physicochemical effect; (ii) occurred at sites that rarely change in DENV evolution, suggesting that they are of great functional importance; and (iii) involved types of amino acid replacements that occur rarely in DENV evolution, again suggesting that they usually have important effects on viral protein structure. In the case of DENV-1, only one amino acid change fulfilled all these criteria—that from Gln to His at position 466 in NS3; this amino acid change is nonconservative, occurs relatively rarely in DENV evolution, and is at a site that is invariant in the other three serotypes. NS3 encodes the critical RNA helicase and has previously been shown to contain T-cell epitopes, including those that are cross-reactive for all serotypes (20, 22, 28, 45, 46), although site 466 does not fall into an epitope region defined to date. However, without further experimental verification, we are unable to determine the fitness consequences of this mutation.
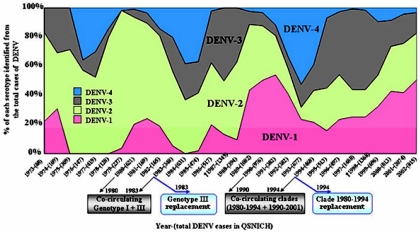
The current lack of definitive evidence for selectively driven clade replacement in dengue virus has led to the reasonable conclusion that such events are due entirely to chance processes (32). Although possible, this hypothesis is difficult to reconcile with the observation that the major clade replacement event in DENV-1 took place over a number of years in the mid-1990s rather than at a single point in time, as expected with a sudden population bottleneck. Moreover, it seems unlikely that changes in vector population size or public health measures would affect clades of DENV-1 in such a different manner. We therefore considered whether the selection pressures imposed by different serotypes cocirculating in the population, which will shape patterns of cross-immunity, also had a bearing on these events. A number of observations suggest that this is indeed the case. First, there is a striking inverse correlation between the prevalence of DENV-1 and that of DENV-4 recorded in the patients attending the QSNICH in Bangkok; DENV-1 tends to peak in prevalence when DENV-4 is at low levels and vice versa (Fig. 3). More striking is that the genetic (clade) diversity within DENV-1 also peaks at times of high prevalence of this serotype and that clade replacement is associated with periods of low prevalence. Hence, the small peak in DENV-1 prevalence during the early 1980s is associated with the cocirculation of genotype I and III viruses and low levels of DENV-4. When DENV-1 then declines in prevalence, DENV-4 increases in frequency and the genotype III viruses are not further sampled. The effect was even more dramatic during the 1990s. Again, the peak of DENV-1 prevalence is associated with very low levels of DENV-4 and the cocirculation of the two genotype I clades. The subsequent disappearance of the 1980 to 1994 clade and replacement by the 1990 to 2002 clade are associated with a trough in DENV-1 prevalence and the rise of DENV-4. From this, we propose that the DENV-1 clades that survived a decline in serotype prevalence did so because they were more antigenically distinct from DENV-4 than were the DENV-1 clades that declined in frequency, which were subject to some cross-protective immune activity. A similar proposal has recently been made to explain the replacement of genotype III by DENV-1 genotype I in neighboring Myanmar (36). Consequently, although our selection analysis suggests that the amino acid changes that distinguish the clades within DENV-1 were fixed by chance at a time of relative serotype abundance, their effect on viral fitness is not realized until the wider serologic landscape changes and selection pressures become stronger. Although this hypothesis will require rigorous experimental testing, it is notable that all clade replacement events documented to date involve viruses with amino acid differences in the immunogenic E gene, which is compatible with the notion that the viruses experience different levels of cross-protection. For example, of the four amino acid changes in the E gene that distinguish the 1980 to 1994 and 1990 to 2002 clades of DENV-1, two, at positions 8 and 324, make the latter clade more genetically different from DENV-4 than the former clade.
FIG. 3.
Relative sampling frequency of each DENV serotype over 30 years as identified from patients admitted to the QSNICH in Bangkok, Thailand, from 1973 to 2002. The predominant dengue serotype varied through time: DENV-2, 1973 to 1986; DENV-3, 1987; DENV-2, 1988 to 1989; DENV-1, 1990 to 1992; DENV-4, 1993 to 1994; DENV-3, 1995 to 1999 (24); and DENV-1, 1999 to 2002 (A. Nisalak, unpublished observations). Note the inverse relationship between the prevalence of DENV-1 and that of DENV-4, while DENV-2 and DENV-3 cocirculated relatively constantly throughout the 30 years and only DENV-3 was associated with severe dengue epidemic years. Levels of genetic diversity and clade replacement in DENV-1 are described in the context of relative DENV-1 and DENV-4 prevalence (see Discussion).
In summary, we suggest that DENV evolution is shaped by a complex interaction of selective and stochastic forces. In particular, although most mutations are deleterious and are removed by purifying selection, some are eventually fixed by stochastic processes, with their subsequent effect on viral fitness being manifest only when there are larger-scale changes in serotype prevalence. These results emphasize that the evolution of individual DENV serotypes should not be considered in isolation from broad-scale immunological patterns.
Supplementary Material
Acknowledgments
This research was supported by the U.S. Military Infectious Disease Research Program, Ft. Detrick, Maryland.
We thank the nursing staff of QSNICH and AFRIMS and the previous chiefs of the Department of Virology at AFRIMS, particularly Bruce Innis, David Vaughn, and Timothy Endy, for managing the service specimen collection. We thank two anonymous referees for useful comments.
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the U.S. government.
Footnotes
Supplemental material for this article may be found at http://www.jvi.asm.org/.
REFERENCES
- 1.A-Nuegoonpipat, A., A. Berlioz-Arthaud, V. Chow, T. Endy, K. Lowry, L. Q. Mai, T. U. Ninh, A. Pyke, M. Reid, J.-M. Reynes, S. T. S. Yun, H. M. Thu, S.-S. Wong, E. C. Holmes, and J. Aaskov. 2004. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introduction of different Asian genotypes. Virology 329:505-512. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, P. M., and R. Rico-Hesse. 2001. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 1:159-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, P. M., and R. Rico-Hesse. 2003. Efficiency of dengue serotype 2 virus strains to infect and disseminate in Aedes aegypti. Am. J. Trop. Med. Hyg. 68:539-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, S. N., E. C. Holmes, M. Chirivella, D. M. Rodriguez, M. Beltran, V. Vorndam, D. J. Gubler, and W. O. McMillan. 2003. Selection-driven evolution of emergent dengue virus. Mol. Biol. Evol. 20:1650-1658. [DOI] [PubMed] [Google Scholar]
- 5.Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox, and W. M. Fitch. 1999. Predicting the evolution of human influenza A. Science 286:1921-1925. [DOI] [PubMed] [Google Scholar]
- 6.Cologna, R., and R. Rico-Hesse. 2003. American genotype structures decrease dengue virus output from human monocytes and dendritic cells. J. Virol. 77:3929-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings, D. A. T., R. A. Irizarry, N. E. Huang, T. P. Endy, A. Nisalak, K. Ungchusak, and D. S. Burke. 2004. Travelling waves in the occurrence of dengue haemorrhagic fever in Thailand. Nature 427:344-347. [DOI] [PubMed] [Google Scholar]
- 8.Diallo, M., Y. Ba, A. A. Sall, O. M. Diop, J. A. Ndione, M. Mondo, L. Girault, and C. Mathiot. 2003. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999-2000: entomologic findings and epidemiologic considerations. Emerg. Infect. Dis. 9:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenfell, B. T., O. G. Pybus, J. R. Gog, J. L. N. Wood, J. M. Daly, J. A. Mumford, and E. C. Holmes. 2004. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303:327-332. [DOI] [PubMed] [Google Scholar]
- 10.Gubler, D. J. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100-103. [DOI] [PubMed] [Google Scholar]
- 11.Hales, S., N. de Wet, J. Maindonald, and A. Woodward. 2002. Potential effect of population and climate changes on global distribution of dengue fever: an empirical model. Lancet 360:830-834. [DOI] [PubMed] [Google Scholar]
- 12.Holmes, E. C. 2003. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77:11296-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes, E. C., and S. S. Twiddy. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect. Genet. Evol. 3:19-28. [DOI] [PubMed] [Google Scholar]
- 14.Jetten, T. H., and D. A. Focks. 1997. Potential changes in the distribution of dengue transmission under climate warming. Am. J. Trop. Med. Hyg. 57:285-297. [DOI] [PubMed] [Google Scholar]
- 15.Klungthong, C., C. Zhang, M. P. Mammen, Jr., S. Ubol, and E. C. Holmes. 2004. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 329:168-179. [DOI] [PubMed] [Google Scholar]
- 16.Kochel, T. J., D. M. Watts, S. B. Halstead, C. G. Hayes, A. Espinoza, V. Felices, R. Caceda, C. T. Bautista, Y. Montoya, S. Douglas, and K. L. Russell. 2002. Effect of dengue-1 antibodies on American dengue-2 viral infection and dengue haemorrhagic fever. Lancet 360:310-312. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Lanciotti, R. S., J. G. Lewis, D. J. Gubler, and D. W. Trent. 1994. Molecular evolution and epidemiology of dengue-3 viruses. J. Gen. Virol. 75:65-75. [DOI] [PubMed] [Google Scholar]
- 19.Maddison, D. R., and W. P. Maddison. 2000. MacClade: analysis of phylogeny and character evolution, version 4. Sinauer Associates, Sunderland, Mass.
- 20.Mathew, A., I. Kurane, S. Green, H. A. F. Stephens, D. W. Vaughn, S. Kalayanarooj, S. Suntayakorn, D. Chandanayingyong, F. A. Ennis, and A. L. Rothman. 1998. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J. Virol. 72:3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moncayo, A. C., Z. Fernandez, D. Ortiz, M. Diallo, A. Sall, S. Hartman, C. T. Davis, L. Coffey, C. C. Mathiot, R. B. Tesh, and S. C. Weaver. 2004. Dengue emergence and adaptation to peridomestic mosquitoes. Emerg. Infect. Dis. 10:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mongkolsapaya, J., W. Dejnirattisai, X.-N. Xu, S. Vasanawathana, N. Tangthawornchaikul, A. Chairunsri, S. Sawasdivorn, T. Duangchinda, T. Dong, S. Rowland-Jones, P.-T. Yenchitsomanus, A. McMichael, P. Malasit, and G. Screaton. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921-927. [DOI] [PubMed] [Google Scholar]
- 23.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 24.Nisalak, A., T. P. Endy, S. Nimmannitya, S. Kalayanarooj, U. Thisayakorn, R. M. Scott, D. S. Burke, C. H. Hoke, B. L. Innis, and D. W. Vaughn. 2003. Serotype-specific dengue virus circulation and dengue disease in Bangkok, Thailand from 1973 to 1999. Am. J. Trop. Med. Hyg. 68:191-202. [PubMed] [Google Scholar]
- 25.Ohta, T. 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23:263-286. [Google Scholar]
- 26.Ohta, T. 1998. Evolution by nearly-neutral mutations. Genetica 102/103:83-90. [PubMed] [Google Scholar]
- 27.Rico-Hesse, R. 2003. Microevolution and virulence of dengue viruses. Adv. Virus Res. 59:315-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothman, A. L., I. Kurane, and F. A. Ennis. 1996. Multiple specificities in the murine CD4+ and CD8+ T-cell response to dengue virus. J. Virol. 70:6540-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rudnick, A. 1978. Ecology of dengue virus. Asian J. Infect. Dis. 2:156-160. [Google Scholar]
- 30.Scott, T. W., A. C. Morrison, L. H. Lorenz, G. G. Clark, D. Strickman, P. Kittayapong, H. Zhou, and J. D. Edman. 2000. Longitudinal studies of Aedes aegypti in Thailand and Puerto Rico: population dynamics. J. Med. Entomol. 37:77-88. [DOI] [PubMed] [Google Scholar]
- 31.Shurtleff, A. C., D. W. C. Beasley, J. J. Y. Chen, H. Ni, M. T. Suderman, H. Wang, R. Xu, E. Wand, S. C. Weaver, D. M. Watts, K. L. Russell, and A. D. T. Barrett. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75-87. [DOI] [PubMed] [Google Scholar]
- 32.Sittisombut, N., A. Sistayanarain, M. J. Cardosa, M. Salminen, S. Damrongdachakul, S. Kalayanarooj, S. Rojanasuphot, J. Supawadee, and N. Maneekarn. 1997. Possible occurrence of a genetic bottleneck in dengue serotype 2 viruses between the 1980 and 1987 epidemic seasons in Bangkok, Thailand. Am. J. Trop. Med. Hyg. 57:100-108. [DOI] [PubMed] [Google Scholar]
- 33.Swofford, D. L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 34.Thein, S., M. M. Aung, T. N. Shwe, M. Aye, Z. Aung, K. Aye, K. M. Aye, and J. Aaskov. 1997. Risk factors in dengue shock syndrome. Am. J. Trop. Med. Hyg. 56:566-572. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thu, H. M., K. Lowry, T. T. Myint, T. N. Shwe, A. M. Han, K. K. Khin, K. Z. Thant, S. Thein, and J. Aaskov. 2004. Myanmar dengue outbreak associated with displacement of serotypes 2, 3, and 4 by dengue 1. Emerg. Infect. Dis. 10:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Twiddy, S. S., C. H. Woelk, and E. C. Holmes. 2002. Phylogenetic evidence for adaptive evolution of dengue viruses in nature. J. Gen. Virol. 83:1679-1689. [DOI] [PubMed] [Google Scholar]
- 38.Uzcategui, N. Y., G. Comach, D. Camacho, M. Salcedo, M. Cabello deq Uintana, M. Jimenez, G. Sierra, R. Cuello de Uzcategui, W. S. James, S. Turner, E. C. Holmes, and E. A. Gould. 2003. The molecular epidemiology of dengue virus type 3 in Venezuela. J. Gen. Virol. 84:1569-1575. [DOI] [PubMed] [Google Scholar]
- 39.Wang, E., H. Ni, R. Xu, A. D. T. Barrett, S. J. Watowich, D. J. Gubler, and S. C. Weaver. 2000. Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J. Virol. 74:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watts, D. M., K. R. Porter, P. Putvatana, B. Vasquez, C. Calampa, C. G. Hayes, and S. B. Halstead. 1999. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet 354:1431-1434. [DOI] [PubMed] [Google Scholar]
- 41.Wittke, V., T. E. Robb, H. M. Thu, S. Nimmannitya, S. Kalayanrooj, D. W. Vaughn, T. P. Endy, E. C. Holmes, and J. G. Aaskov. 2002. Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology 301:148-156. [DOI] [PubMed] [Google Scholar]
- 42.WHO. 2002. Dengue and dengue haemorrhagic fever. Fact sheet no. 117. [Online.] http://www.who.int/mediacentre/factsheets/fs117/en/.
- 43.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 44.Yang, Z., R. Nielsen, N. Goldman, and A. M. K. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng, L., I. Kurane, Y. Okamoto, F. A. Ennis, and M. A. Brinton. 1996. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J. Virol. 70:3108-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivna, I., S. Green, D. W. Vaughn, S. Kalayanarooj, H. A. Stephens, D. Chandanayingyong, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J. Immunol. 168:5959-5965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.