Abstract
Red sea bream iridovirus (RSIV) has been identified as the causative agent of a serious disease in red sea bream and at least 30 other marine fish species. We developed a viral DNA microarray containing 92 putative open reading frames of RSIV to monitor the viral gene transcription program over the time course of an in vitro infection and to classify RSIV transcripts into temporal kinetic expression classes. The microarray analysis showed that viral genes commenced expression as early as 3 h postinfection (p.i.) and this was followed by a rapid escalation of gene expression from 8 h p.i. onwards. Based on the expression of some enzymes associated with viral DNA replication, the DNA replication of RSIV appeared to begin at around 8 h p.i. in infected cells in vitro. Using a de novo protein synthesis inhibitor (cycloheximide) and a viral DNA replication inhibitor (phosphonoacetic acid), 87 RSIV transcripts could be classified into three temporal kinetic classes: nine immediate-early (IE), 40 early (E), and 38 late (L) transcripts. The gene expression of RSIV occurred in a temporal kinetic cascade with three stages (IE, E, and L). Although the three classes of transcripts were distributed throughout the RSIV genome, E transcripts appeared to cluster in at least six discrete regions and L transcripts appeared to originate from seven discrete regions. The microarray data were statistically confirmed by using a t test, and were also clustered into groups based on similarity in the gene expression patterns by using a cluster program.
Iridoviruses are considered infectious pathogens that are responsible for causing serious systemic diseases among aquatic animals in many parts of the world. Outbreaks of these diseases have been reported in Australia, France, Germany, Finland, Denmark, United States, Taiwan, Southeast Asia, and Japan (18). In Japan, a serious disease of red sea bream (Pagrus major) has been attributed to an iridovirus that has been named red sea bream iridovirus (RSIV). RSIV disease causes mass mortalities and huge economic losses not only in cultured red sea bream, but also in 30 other cultured marine fish species (22, 24, 25, 29, 60).
RSIV is a piscine, icosahedral, double-stranded DNA virus which is about 200 to 240 nm in diameter. RSIV can replicate in several fish cell lines at suitable temperatures from 20 to 25°C, however, its replication is inhibited by 5-iodo-2-deoxyuridine. The virus is sensitive to chloroform and ether and unstable to heat but is not affected by ultrasonic treatment or repeated freezing and thawing (22, 39). The full genome of RSIV is about 112 kbp and has been completely sequenced (31). Several open reading frames (ORFs) have been assigned putative functions based on significant matches with the potential proteins of lymphocystis disease virus 1, rock bream iridovirus (RBIV), and other better-studied viruses (14). These proteins are involved in DNA replication, DNA modification and processing, DNA transcription, and processing of viral RNA. There are also other sequences in the genome that are considered RSIV-specific genes. They include the largest submit of DNA-dependent RNA polymerase, DNA polymerase, ATPase, and putative ankyrin repeat-containing proteins. Although significant information is available on the morphology, biological and physicochemical properties, composition, and genome structure, little is known about its gene expression program.
Iridoviruses display a complex replication process that occurs in both the nucleus and the cytoplasm of infected cells and complex gene regulation strategies in which genes are expressed in three main temporal kinetic expression classes: immediate-early (IE) genes, early (E) or delayed-early (DE) genes, and late (L) genes (11, 42). IE genes are expressed immediately after primary infection and encode transcription factors associated with trans-activations. These genes are defined experimentally by their ability to produce transcripts even in the presence of protein synthesis inhibitors (33). E genes are normally expressed later and include enzymes associated with DNA replication. L genes are expressed after the onset of viral DNA replication and encode mainly structural proteins of viral particles (7, 15). Thus, inhibition of de novo protein synthesis allows expression only of IE genes. Blockage of viral DNA replication inhibits expression of L genes. Isolation of RNA at short times after infection results in abundance of IE transcripts, and isolation of RNA at longer times of viral DNA replication results in accumulation of L transcripts (15, 53). Therefore, studies of the RSIV gene expression patterns are necessary and useful for fundamental characterization of the viral replication cycle that will provide clues for a better understanding of viral replication and gene expression strategies and give additional insights on the pathogenic mechanism of the virus.
One method of examining gene expression is to use the recently developed DNA microarray technology (52). This technique allows global analysis of transcription for the complete genome and is especially useful for determining the complex transcriptional program of large DNA viruses (13). DNA microarrays have been successfully developed to study the transcriptional profiles of some viruses during an infection, such as herpes simplex virus type 1 (53), human herpesvirus 8 (HHV-8) (23, 41, 46), murine grammaherpesvirus 68 (MHV-68) (1, 15, 35), human cytomegalovirus (7), baculovirus (62), and white spot syndrome virus (30), as well as the host's transcriptional response to viral infections (2, 6, 17, 26, 27, 37, 44, 47, 54, 59).
In this study, we developed a DNA microarray for RSIV to monitor the gene temporal kinetic transcription program of the virus during an in vitro infection. Using this approach, individual RSIV ORFs were characterized at the transcriptional level and were also classified into temporal kinetic classes by their dependence on de novo protein synthesis and viral DNA replication. Such studies of RSIV infection should help to understand the gene regulation strategies and the pathogenic mechanisms of RSIV and other iridoviruses. This is the first study of gene expression for piscine viruses using DNA microarray technology.
MATERIALS AND METHODS
Cell and virus infection.
Grunt fin (GF) cells (9) were cultured at 25°C in Basal Medium Eagle (Gibco-BRL, Grand Island., NY) supplemented with 10% fetal bovine serum (JRH Biosciences, Lenexa, KS), 100 IU/ml of penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and HEPES. Japanese flounder natural embryo (HINAE) cells (28) were maintained at 20°C using Leibovitz's L-15 medium (Gibco-BRL, Grand Island, NY) containing 20% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin.
RSIV was obtained from a spleen homogenate of RSIV-infected red sea bream and was propagated in GF cells as previously described (20, 38-40). Briefly, spleens were homogenized and diluted in 10 volumes of Basal Medium Eagle supplemented with antibiotics. The suspension was centrifuged at 2,000 × g for 20 min at 4°C and then filtered using 0.45-μm low-protein-binding filter membranes (Millipore, Bedford, MA). Confluent GF cells grown in 75-cm2 flasks were inoculated with the virus suspension at 25°C. After allowing 90 min for absorption, unattached viruses were removed and infected cells were continuously cultured with fresh culture medium until cytopathic effect was complete. Subsequently, the supernatant was collected and centrifuged at 2,000 × g for 20 min. The viral suspension was filtered through 0.45 μm membrane, stored in 1 ml aliquots at −80°C and used for all experiments. The virus titer was determined using the 50% tissue culture infective dose (TCID50) method (48).
For virus infection, HINAE monolayers cultured in 75-cm2 flasks were inoculated with RSIV at a multiplicity of infection (MOI) of 5 throughout the experiments and incubated at 25°C for 90 min. Unattached virus was then removed and infected cells were continuously cultured with fresh culture medium. For mock-infected samples, HINAE cells were treated in the same manner as virus-infected cells but with fresh culture medium.
Monitoring of gene expression profiles during in vitro infection.
To examine changing RSIV gene expression levels in infected cells in vitro, RNA was isolated from both RSIV-infected and mock-infected HINAE cells at times up to 48 h postinfection (p.i.).
Inhibition of protein synthesis and polymerase activity.
To classify RSIV transcripts into temporal kinetic classes, HINAE cells were treated with drugs as previously described with some slight modifications. Briefly, HINAE monolayers were treated for 1 h prior to and throughout the viral infection with either cycloheximide (CHX) or phosphonoacetic acid (PAA), which act as de novo protein synthesis and viral DNA replication inhibitors, respectively. To obtain viral IE transcripts, CHX (50 μg/ml)-pretreated cells were mock infected or infected with RSIV (MOI of 5), and then harvested at 12 h p.i. To distinguish between viral E and L transcripts, cells infected with RSIV (MOI of 5) in the presence and absence of PAA (100 μg/ml), respectively, were harvested at 48 h p.i. for RNA extraction (8, 15, 50, 53).
IC50 assays for CHX and PAA.
To assess the effects of drugs on cell growth during assays inhibiting protein synthesis and polymerase activities, the 50% inhibitory concentrations (IC50) of CHX and PAA were established based on measurement of the cellular conversion of a tetrazolium salt into a formazan product using a commercial CellTiter 96 nonradioactive cell proliferation assay kit (Promega). Briefly, 90 μl of cell suspension (containing approximately 2 × 104 HINAE cells) was dispensed into each well of a 96-well plate and incubated at 25°C for about 24 h. The cells were then treated with twofold serial dilutions of either CHX or PAA. Living cells were detected based on formation of colored formazan product. The absorbance values of the colored formazan products were recorded at 570 nm using a dual wavelength microplate reader (Bio-Rad, Benchmark) with a reference wavelength of 630 nm. The IC50 values were calculated by locating the x axis value that represents concentrations of drug inhibitors corresponding to one-half the maximum corrected absorbance value. Duplicate experiments were performed for each drug inhibitor. Each dilution was tested in quadruplicate.
RNA preparation and labeled cDNA target construction.
Total RNA was extracted from the cells with TRIzol (Invitrogen) and subjected to DNase I treatment (Promega) according to the manufacturer's protocols. The cDNAs were synthesized from 50-μg aliquots of purified RNA with a RSIV antisense-strand-specific primer mixture and were incorporated indirectly with Cy5- or Cy3 monofunctional reaction dyes (Amersham Biosciences, England). Briefly, the cDNAs were first labeled with aminoallyl-dUTP (LabelStarArray kit, cDNA labeling module, QIAGEN) and purified with QIAquick Spin columns (QIAquick PCR purification kit, QIAGEN) following the manufacturer's recommendations. The aminoallyl-cDNAs were then coupled with Cy5 or Cy3 monofunctional dyes and purified with MinElute Spin columns (LabelStarArray kit, cDNA cleanup module, QIAGEN) according to the manufacturer's instructions.
Construction of RSIV DNA microarrays.
A DNA microarray containing 92 RSIV ORFs was constructed following a PCR product-based microarray method described elsewhere (63). Briefly, specific primer sets were designed (Table 1) to amplify approximately 300 to 1,500-bp fragments of each RSIV ORF using viral genome as a template. All PCR products showing a single band of the appropriate size by gel electrophoresis were purified, and reconstituted in TE buffer at a final concentration of about 500 μg/ml for spotting in duplicates onto the glass slides. Piscine β-actin genes from Japanese flounder, yellow tail, and red sea bream and HINAE cells were included as internal controls to normalize the microarray data. In addition, distilled water was also used as a negative control.
TABLE 1.
Primer sequences
| No. | ORF | Forward primer (5′→3′) | Reverse primer (5′→3′) | PCR product size (bp) |
|---|---|---|---|---|
| 1 | ORF016L | CGAATGCTTGCCGAACGGAT | GCGATATAGTGACAGGCTCC | 800 |
| 2 | ORF018R | ATGGACCGCGCCCTTCATAA | CTACACGGAAGGCACGTCTT | 300 |
| 3 | ORF029R | TATGCCAACGGTCGTGTGCC | TGTCGAGAGCATGGTCCTAA | 700 |
| 4 | ORF033R | TCCAGGAAGTGTATGACCTG | ACTGCATCAGCTCGTGCTGC | 1,200 |
| 5 | ORF037R | GACGCCATCGTCATGGGCAT | AGGCGTGTATATGGCAGGTC | 600 |
| 6 | ORF042R | ATGACTGGTGGCACACCAAA | CCGCAAGCTCAGGTCATTCA | 1,000 |
| 7 | ORF049R | ATGCCTACGGGATCTAGTGC | GACAGCAGGTCCTGCATTAG | 500 |
| 8 | ORF054R | CGCTATGACACGTACATTGC | ACCACATGTCCTGCCATAGC | 1,200 |
| 9 | ORF063R | CGTGTACTGCGGTGGCCATT | GCCATAGCCGTGCGTAACAT | 1,200 |
| 10 | ORF077R | GTCACAGGCCTGTCGCATGT | TTGCGCTTGAGCCCACTCAT | 1,200 |
| 11 | ORF092R | ATGACCACAGGGCACACTGA | CGCGAAATGTCAGCCTGTAC | 500 |
| 12 | ORF097R | ATGCTTGCGCTGGCCATCGT | CCATGACATCATCCGGGTGC | 450 |
| 13 | ORF101R | GCATCCAGACATACTCAGCC | CATGTCCACCAGGTTCACAT | 600 |
| 14 | ORF106R | GCCGAGCACCACTTCCAAGT | CGCACAGGGCTACGCTTTAT | 900 |
| 15 | ORF111R | ATGTCATCGTACCGGTGCCG | GTCCATGTCACGCGCAATGG | 900 |
| 16 | ORF122R | ATAAAGCACCCTCGTGCGGC | CGATGTCCGACAAATGCTCA | 300 |
| 17 | ORF128R | ACTGTCCCAGAATGGAAGAC | CATGCAATGCACAGAGCACC | 400 |
| 18 | ORF135L | ATGAAGTGCACGGCGTTACT | TCCTCGGAGGTGCTGTAGGA | 400 |
| 19 | ORF140R | ATGCGCGTGCTCGAACTATT | TCAACATGTTTTTTGCACAC | 600 |
| 20 | ORF145R | CGTCACAGACGATGTATCCG | ACTCTAGGAGCACGACACGT | 900 |
| 21 | ORF151R | GCCTTTACCAGAGTGCACAC | TGCTCAAGGCCATCGCATGT | 800 |
| 22 | ORF156R | ATTCATTCGAGCCTGCCGGC | GGTGCCCTTACCGTAGAGCT | 300 |
| 23 | ORF161L | ATGGAGTCACAACTTGTCGC | TGTTGCACTGTGGATACGGC | 600 |
| 24 | ORF162R | ACGCAACCGAACAGGCGTGT | CACATGTACCGCAGTGGCGT | 1,200 |
| 25 | ORF171R | GCGTAGACCTACGCGACTGT | GTCCACAGCCATGTGGCACA | 1,000 |
| 26 | ORF179L | TAGGCTGCAGCTGGCCTACA | GGACACGAATGCGACGTCTA | 700 |
| 27 | ORF180R | CGACGTGTCGACCGAAGAGT | TGCGCCTGTGCACAGTAGCA | 1,200 |
| 28 | ORF186R | GACCTTGAAGGCTATGCAGT | GCACTTAGCAGAGCAGAGTC | 1,100 |
| 29 | ORF197L | GCGTCACAGGTACAAGCGCT | GAGAGCCACCTGGTAATGCG | 900 |
| 30 | ORF198R | GCATCCTGCAACACTGTGAG | CGTGTGCTGCACATAGTCGT | 800 |
| 31 | ORF224L | TGCGCATTACGAGCACCTGC | GTGCAGCTGTCGCACCTTGA | 1,200 |
| 32 | ORF226R | GCTCAGGATGTCCATGGCTC | TGGACGACGGCAGCGTCTTA | 800 |
| 33 | ORF234L | AAAGTCCAGCCTGCTGCGCA | CTGGCGCCTTACAGTGTCCA | 500 |
| 34 | ORF237L | ATGTTCCTCGTCAAATGCGG | TACGGCAATGGGTGTTTCTC | 400 |
| 35 | ORF239R | CGCGTCATCTGTCACTCGTG | CAACGCCAGCACAGTGTCCT | 1,100 |
| 36 | ORF256R | GTGTCACCGAAGAGTGGCTG | GCTGGCTTGTGTCGACAACC | 800 |
| 37 | ORF261R | AATTGCATTGCTGCGACCAC | TGAGTTGTACTCACTTTCGC | 300 |
| 38 | ORF268L | ATGTGCTCAGACCAAACGAC | TCAAGCAGGCGATCTGCAAC | 800 |
| 39 | ORF291L | ACGGTACCATCTTGTCTGTC | CACGGGCTATTGAATGGACA | 600 |
| 40 | ORF317L | AGTCGCGTGTTAGCAAGACG | GTTCGCAGCCTTCGTGCTCT | 1,100 |
| 41 | ORF321R | ATGCCGACTACCAAACACAC | CTCAAAGGCGCCGTGATACG | 400 |
| 42 | ORF324R | TCCGAGCTAACACTGTGGCG | GCGCATGAGCCATAATGGCC | 600 |
| 43 | ORF333L | GACCGTGTCGTGTCCATGCT | CGTCAACTCGGCCAATCTCG | 600 |
| 44 | ORF342L | ACTGGAGATTGGCGAAGTGC | TACAGTCGACTGTGCCACTG | 1,000 |
| 45 | ORF349L | CAGGGCAGATACTCTGTGAC | CCATGTCCACGACAGTCTGT | 1,300 |
| 46 | ORF351R | GGACTATATTGCTGAGCTAC | TACAGCTTGATTAGGGCGAC | 300 |
| 47 | ORF353R | GGCATCCTAACATCGCTGGC | CGTTGTCCGTATGCTGCCTG | 400 |
| 48 | ORF373L | CTGCTGACACTGCCCATTGG | GCAGAGCAGCATGTAGAGTC | 1,400 |
| 49 | ORF374R | ATGGGTGCTGCCCAGTCATT | TTATATATGATCGGCCTCGT | 1,400 |
| 50 | MCP (380) | GTACAGCAAGCTGCCCGTTA | ACGGTGACAGTCGATATGGC | 500 |
| 51 | ORF385R | ATGCGTTACGTACCATGCTG | TGCTGTTCGCTTGGCTGAAA | 800 |
| 52 | ORF390R | CACAACGGTGTAGTAACGCG | GACGGTTGCAGGGACCTACA | 800 |
| 53 | ORF394R | ATGCACACAGCGATGAACCT | CACTGTAGTACACACGGTGC | 600 |
| 54 | ORF396R | ATGGCACCGTGTGTACTACA | AGCCATGTGTACATCAGGGC | 1,000 |
| 55 | ORF401R | TGCACACTATGCTGAGTAGC | GGCCATGAGAAACCATGTCA | 700 |
| 56 | ORF407R | GTCTAGTGCCATTGCCATGG | GTAAGCAAGTGCACAGCACC | 400 |
| 57 | ORF412L | AGTTGTCCCTGACGGAGCTG | AAGCCCACATCGGCGTCTGT | 700 |
| 58 | ORF413R | GCCACGTTGATGCACTACAC | TGCGGTGACAAACTGCACGA | 400 |
| 59 | ORF420L | GCTTGCTCTGGGTGATAAGA | GCGCACAATATCATTGGCTG | 500 |
| 60 | ORF423L | GAGACCATCGCCAAGTATGC | CTGCGTATGCGGCCAGCATC | 300 |
| 61 | ORF424R | TTTGCTGCGACCGGAGATGC | CCGTGCTGCCATGGACCTAA | 1,300 |
| 62 | ORF426R | CCGCTTGTTGTCAAGACCAA | ATTGTGGCCTGTGACAACTC | 500 |
| 63 | ORF430L | GCAACTGATGCACGACCGTA | ATCAATGTTCATCGCTGCGC | 1,300 |
| 64 | ORF458L | GTCCGTGACGACCTGTGCAT | TCGACATTGACATGGCGTCG | 900 |
| 65 | ORF463R | ATGCGTCGCGACGGCATGTA | TGTCACCGTTGCCAGTACTG | 1,200 |
| 66 | ORF487L | TGAGCTGCTGTACGCCAACG | ATGCACCTCTACGGTGCCGA | 700 |
| 67 | ORF488R | GCTTGCTATGTGCTACGTGC | TCGCAGACGTGTACCGTGAC | 800 |
| 68 | ORF493R | CGGCTGATCGTCAGCAGTTT | TGTGCCTAGCGCACTGTACA | 1,200 |
| 69 | ORF502R | AGGCAGCCAGCACTGCGTTT | CGGCAGGCTCATGTCAACAC | 600 |
| 70 | ORF506R | CCGCTCAAGACATGTTCGGC | CCAAGGTGTACGCCATGTCG | 700 |
| 71 | ORF515R | AGCCTGTTCCAGCTGCCAGA | TGCGGTTCATTTGCTGTCCC | 700 |
| 72 | ORF522L | GAAGTCTTCCGCTCTGATGC | GGCGAATGTGTGATTGCCC | 700 |
| 73 | ORF534L | GCGCTCAAGGCTGTCTTGCT | AACTGGCGTGCGAACAGCGT | 1,000 |
| 74 | ORF535R | GATGCCTGCACATCTGTCGG | GCATGCTCAAGGGAGTGCAC | 500 |
| 75 | ORF543R | ACGAGACATGTGCCGTGTGC | CAACACAGCAACAGGTACAC | 300 |
| 76 | ORF550R | ACATCGTACGTGGACACCGC | CCCAGGTATGCAAACAGTGG | 700 |
| 77 | ORF554R | AGCGCTGATGAATGACTACC | GCCTCTTCAGCATGAGTGGT | 900 |
| 78 | ORF562R | ATGCATTGTGCCGTGCTCAC | GCATATGTGAGTGCCTCTCC | 500 |
| 79 | ORF569R | ATGATTTCAAGTGCACTGCT | CTAGGCCATAATGCCACGTC | 1,000 |
| 80 | ORF575R | ATGGCCCAGAAGCAGGATAA | CTAGGCAAATGCGGCAATAA | 1,000 |
| 81 | ORF586L | CACTGCAACAGAAGCTACCG | CGCTGCCACAACCGCAGTAT | 700 |
| 82 | ORF589L | TGCGCGGACTGTTCTTCACG | CTGGCAGCATATTCGAGCAT | 600 |
| 83 | ORF591R | GACCTGAGCAAGCTGCCAGT | CATGGTCCAGTCATGCGGCA | 400 |
| 84 | ORF596L | CCGCACGTTTGCGCAGACAA | CCGTCTGATGCTAGCTGTGC | 800 |
| 85 | ORF600L | CGCATGTCTGCAAGCGGTCT | CGCCTGATGTATGCCTCGAT | 500 |
| 86 | ORF606R | ACGCCATTGGACTGCCTGCA | GCGCAGTATAGCATCAAGCG | 1,000 |
| 87 | ORF617L | GTCAAAGTGTGGGCACCATT | CCTTGCATAACGCAGGATGT | 400 |
| 88 | ORF618R | CCGCAGGTTGTGCACTATGT | CGTCGACGGCATGAACGCAT | 1,000 |
| 89 | ORF628L | TATCCAAGGACACTGTGGCG | CACGGCAACAAGCAGTGGCT | 500 |
| 90 | ORF632L | GCGCATCACCGGTACTTGAC | GCACCAGCATACGGTGTTCC | 600 |
| 91 | ORF635L | GTCAGTTGCGCATGCACACC | GCAATAAGCTCTGCCTTGGC | 500 |
| 92 | ORF641L | ATGTCTGCATGGCGTGCAGT | CGTCCGTCTATCGGTGTAAC | 1,100 |
Microarray hybridization.
The Cy5/Cy3-dUTP-labeled cDNAs were resuspended in hybridization solution containing formamide, hybridized to the microarrays for 16 to 18 h at 42°C, and rinsed several times following the DNA microarray standard methods and previously published studies (3, 4, 32, 56): once with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at room temperature for 20 min, once with 0.2× SSC-0.1% SDS at room temperature for 20 min, twice with 0.2× SSC-0.1% SDS at 55°C for 20 min with slight shaking, and once with 0.2× SSC and 0.05× SSC at room temperature. Following the washing steps, the microarrays were dried by low-speed centrifugation (500 rpm for 5 min) (10), and immediately scanned using a GenePix 4000B array scanner (Axon Instruments, Inc.; www.axon.com).
Statistical data analysis.
Images obtained from scanning were analyzed by GenePix Pro 4.0 array analysis software (Axon Instruments, Inc.; www.axon.com). Normalization was performed to equilibrate Cy5 and Cy3 signal intensity with the spotted housekeeping gene (piscine β-actin genes) by adjusting the photomuliplier tube power and signal gain (4, 32). The median signal intensity, which was subtracted from the background signal, was used as an appropriate measure of absolute viral gene expression. Only those genes exhibiting a signal intensity greater than twofold changes in virus-infected samples compared with uninfected samples were used for statistical analysis (19, 34, 51, 63). Gene expression level measurements were statistically analyzed by using a t test (within Analyze-it for Microsoft Excel Ver. 1.71). The paired t test on replicated spots for each gene was performed to determine the significance at the 95% confidence level in the virus-infected samples versus those in the mock-infected samples or in the drug-treated samples versus those in the untreated samples (35). Viral genes were considered to be significantly expressed only if the following two criteria were met: (i) the median level of intensity in infected samples was at least twofold greater than that in the mock-infected samples and (ii) a paired t test for an increase in level of intensity under conditions of virus infection versus mock-infection was significant at a P value of less than 0.05.
The microarray data were also reported as the calibrated expression ratio, which was the ratio of the fluorescence intensity of an RSIV transcript in infected cells compared to that of the control transcripts (β-actin genes) (58). The expression ratio data were imported into Cluster Program 3.0 in conjunction with an average linkage hierarchical clustering algorithm, using Euclidian distance as the similarity metric. The cluster analysis allowed grouping of RSIV ORFs whose levels of expression show similar patterns. After clustering, the results were visualized in a tree structure by using Tree View program.
Reverse transcription-PCR.
To validate the microarray data, a reverse transcription (RT)-PCR assay was used to determine the expression of several RSIV ORFs. Twenty μl of cDNA was synthesized from 5 μg purified RNA of the same samples used for cDNA synthesis for the microarray experiments by using RSIV antisense-specific primer mixture and Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer's protocols. The reaction mixture contained 1 μl cDNA, Taq polymerase, and specific primers for each RSIV ORF in a volume of 30 μl. Cycling parameters consisted of an initial denaturation at 95°C for 3 min, followed by 27 to 30 cycles of denaturation at 95°C for 30 seconds, annealing at 56°C for 30 seconds, elongation at 72°C for 1 min, and a final elongation step at 72°C for 5 min.
Dilution RT-PCR.
To further reconfirm the microarray results, some RSIV ORFs selected for RT-PCR were also chosen for dilution RT-PCR. The cDNAs were serially diluted 10-fold (10−1 to 10−3) from the original cDNAs and used as templates. The reaction mixture contained 2 μl diluted cDNA, Taq polymerase, and specific primers for each RSIV ORF in a volume of 30 μl. The cycling parameters used were the same as those used for RT-PCR.
RESULTS
RSIV transcriptional program by viral DNA microarrays.
To investigate the transcriptional program for RSIV, the one-step growth conditions of infection were established in a cell culture system for time course sampling. At specified times, the cDNAs derived from virus-infected and mock-infected HINAE cells were labeled with fluorescence Cy5- or Cy3-aminoallyl-dUTP, respectively, and hybridized to the microarrays.
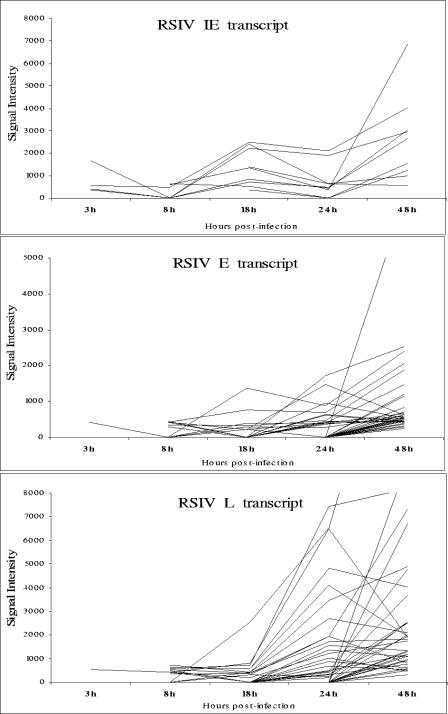
The paired t test was applied to the microarray data and determined only six viral ORFs that showed a significant change in expression at P < 0.05 in the early stages of infection at 3 h p.i., accounting for about 6.5% of total viral ORFs. However, as depicted in Table 2 and Fig. 1, the number of expressed RSIV ORFs increased significantly, up to 95.7% at 48 h p.i. (P < 0.0001). As shown in Fig. 1, little transcription was detected at 3 h p.i. but from 8 h p.i. onwards, signals for the majority of viral ORFs could be seen. In addition, the expression level of each ORF seemed to increase through the infection cycle, indicating a rapid escalation of RSIV gene expression. Various ORFs reached significant levels of expression at different time points, indicating that each ORF had its own expression kinetics. Taken together, these results suggest that RSIV's global transcriptional profile changed as it progressed through the life cycle.
TABLE 2.
Kinetic classes
| No. | ORF | Putative function | Accession no. | Signal intensity valuea at time (h p.i.):
|
Calibrated expression ratiob at time (h p.i.):
|
Kinetic classc | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 8 | 18 | 24 | 48 | 3 | 8 | 18 | 24 | 48 | |||||
| 1 | ORF016L | Hypothetical protein | 600 | 564 | 4,829 | 4,026 | 1.11 | 1.25 | 8.40 | 8.08 | L | |||
| 2 | ORF018R | Hypothetical protein | 919 | 1.85 | L | |||||||||
| 3 | ORF033R | Cytosine DNA methyltransferase | AAT71861 | 441 | 431 | 0.77 | 0.87 | E | ||||||
| 4 | ORF037R | Hypothetical protein | 560 | 4,083 | 1,925 | 1.24 | 7.10 | 3.87 | L | |||||
| 5 | ORF042R | Hypothetical protein | 512 | 377 | 1,923 | 7,316 | 0.95 | 0.83 | 3.38 | 14.69 | L | |||
| 6 | ORF049R | RING-finger-containing E3 ubiquitin ligase | AAT71876 | 696 | 481 | 2,659 | 1.54 | 0.84 | 5.34 | IE | ||||
| 7 | ORF054R | Putative RNA gianylytransferase | AAL98788 | 639 | 1.28 | E | ||||||||
| 8 | ORF063R | Largest subunit of DNA-dependent RNA polymerase | BAA82753 | 377 | 527 | 0.66 | 1.06 | E | ||||||
| 9 | ORF077R | Putative DNA-binding protein | AAT71873 | 412 | 215 | 1,476 | 614 | 0.76 | 0.48 | 2.57 | 1.23 | E | ||
| 10 | ORF092R | Putative replication factor | AAS18131 | 423 | 1,364 | 884 | 2,404 | 0.45 | 3.02 | 1.54 | 4.83 | E | ||
| 11 | ORF097R | Hypothetical protein | 576 | 1,355 | 346 | 6,839 | 1.07 | 3.00 | 0.60 | 13.73 | IE | |||
| 12 | ORF101R | Hypothetical protein | 185 | 2,510 | 0.32 | 5.04 | L | |||||||
| 13 | ORF106R | Hypothetical protein | 2,541 | 5.10 | L | |||||||||
| 14 | ORF111R | Hypothetical protein | 417 | 9,316 | 0.77 | 18.71 | L | |||||||
| 15 | ORF122R | Hypothetical protein | 509 | 1,023 | 471 | 0.94 | 1.78 | 0.95 | L | |||||
| 16 | ORF128R | Hypothetical protein | 434 | 946 | 551 | 0.80 | 1.65 | 1.11 | E | |||||
| 17 | ORF135L | Hypothetical protein | 533 | 427 | 189 | 1,196 | 1,726 | 0.56 | 0.79 | 0.42 | 2.08 | 3.47 | L | |
| 18 | ORF140R | Cytosine DNA methyltransferase | AAT71861 | 360 | 0.72 | ND | ||||||||
| 19 | ORF145R | Hypothetical protein | 638 | 1.28 | E | |||||||||
| 20 | ORF151R | Hypothetical protein | 302 | 0.61 | E | |||||||||
| 21 | ORF156R | Thiol oxidoreductase | AAP33193 | 327 | 0.66 | E | ||||||||
| 22 | ORF161L | Hypothetical protein | 514 | 1.03 | E | |||||||||
| 23 | ORF162R | Hypothetical protein | 399 | 0.80 | E | |||||||||
| 24 | ORF171R | Hypothetical protein | 565 | 1.13 | E | |||||||||
| 25 | ORF179L | Hypothetical protein | 435 | 0.87 | E | |||||||||
| 26 | ORF186R | Hypothetical protein | 478 | 0.96 | L | |||||||||
| 27 | ORF197L | Hypothetical protein | 913 | 1.83 | L | |||||||||
| 28 | ORF198R | Hypothetical protein | 647 | 452 | 1.13 | 0.91 | E | |||||||
| 29 | ORF224L | RNA polymerase beta subunit | AAT71848 | 966 | 1.94 | L | ||||||||
| 30 | ORF226R | Hypothetical protein | 585 | 727 | 7,413 | 8,175 | 1.08 | 1.61 | 12.89 | 16.42 | L | |||
| 31 | ORF234L | Deoxyribonucleoside kinase | AAT71846 | 296 | 0.59 | E | ||||||||
| 32 | ORF237L | Largest subunit of DNA-dependent RNA polymerase | AB018418 | 507 | 1.02 | E | ||||||||
| 33 | ORF239R | Largest subunit of DNA-dependent RNA polymerase | BAA82753 | 427 | 418 | 588 | 0.79 | 0.73 | 1.18 | E | ||||
| 34 | ORF256R | DNA repair protein RAD2 | BAA82754 | 413 | 415 | 2,554 | 0.76 | 0.72 | 5.13 | L | ||||
| 35 | ORF261R | Hypothetical protein | 315 | 0.63 | L | |||||||||
| 36 | ORF268L | Ribonucleotide reductase small subunit | BAA82755 | 840 | 1.69 | E | ||||||||
| 37 | ORF291L | Laminin-type epidermal growth factor-like domain | AAT71838 | 434 | 788 | 6,489 | 1,910 | 0.80 | 1.74 | 11.29 | 3.84 | L | ||
| 38 | ORF317L | DNA polymerase | O70736 | 408 | 755 | 686 | 2,055 | 0.76 | 1.67 | 1.19 | 4.13 | E | ||
| 39 | ORF321R | DNA polymerase | AB007366 | 361 | 0.72 | E | ||||||||
| 40 | ORF324R | DNA polymerase | AB007366 | 6,600 | 13.25 | E | ||||||||
| 41 | ORF333L | Hypothetical protein | 1,373 | 631 | 568 | 3.04 | 1.10 | 1.14 | IE | |||||
| 42 | ORF342L | Hypothetical protein | 1,637 | 2,203 | 1,901 | 2,967 | 1.72 | 4.87 | 3.31 | 5.96 | IE | |||
| 43 | ORF349L | Genine threonine protein kinase catalytic domain | AAT71828 | 1,289 | 2.59 | L | ||||||||
| 44 | ORF351R | Hypothetical protein | 379 | 0.76 | E | |||||||||
| 45 | ORF353R | Hypothetical protein | 646 | 515 | 1,535 | 1.20 | 1.14 | 3.08 | IE | |||||
| 46 | ORF373L | Hypothetical protein | 425 | 3,463 | 4,892 | 0.94 | 6.02 | 9.82 | L | |||||
| 47 | ORF374R | Hypothetical protein | 1,329 | 2.67 | L | |||||||||
| 48 | MCP(380R) | Major capsid protein | BAC66968 | 585 | 1.17 | L | ||||||||
| 49 | ORF385R | Catalytic domain of ctd-like phosphatase | AAT71821 | 722 | 1.45 | E | ||||||||
| 50 | ORF390R | Hypothetical protein | 476 | 1,166 | 0.83 | 2.34 | L | |||||||
| 51 | ORF394R | Hypothetical protein | 302 | 1,930 | 792 | 0.67 | 3.36 | 1.59 | L | |||||
| 52 | ORF396R | Transmembrane amino acid transporter | AAT71816 | 553 | 460 | 2,492 | 2,098 | 4,016 | 0.58 | 0.85 | 5.51 | 3.65 | 8.06 | IE |
| 53 | ORF401R | Hypothetical protein | 1,355 | 1,076 | 2.36 | 2.16 | L | |||||||
| 54 | ORF407R | ATPase | AB007367 | 1,706 | 2,523 | 2.97 | 5.07 | E | ||||||
| 55 | ORF412L | ATPase | AAO16492 | 227 | 383 | 1,478 | 0.50 | 0.67 | 2.97 | E | ||||
| 56 | ORF413R | ATPase | AB007367 | 398 | 664 | 0.69 | 1.33 | E | ||||||
| 57 | ORF420L | Hypothetical protein | 1,111 | 2.23 | L | |||||||||
| 58 | ORF423L | RING-finger domain-containing protein | AAT71906 | 882 | 595 | 1.53 | 1.19 | L | ||||||
| 59 | ORF424R | Putative ankyrin repeat protein | AAL98801 | 296 | 281 | 582 | 0.65 | 0.49 | 1.17 | E | ||||
| 60 | ORF426R | Hypothetical protein | 470 | 4,773 | 0.82 | 9.58 | L | |||||||
| 61 | ORF430L | Putative phosphatase | AAT71837 | 1,156 | 2.32 | E | ||||||||
| 62 | ORF458L | Hypothetical protein | 6,692 | 13.44 | L | |||||||||
| 63 | ORF463B | Hypothetical protein | 355 | 1,921 | 0.62 | 3.86 | L | |||||||
| 64 | ORF487L | Proliferating cell nuclear antigen | NP078615 | 433 | 0.87 | E | ||||||||
| 65 | ORF488R | Putative tumor necrosis factor receptor associated factor | AAL98835 | 697 | 539 | 1.21 | 1.08 | L | ||||||
| 66 | ORF493R | D5 family NTPase | AAS18067 | 316 | 743 | 0.55 | 1.49 | L | ||||||
| 67 | ORF502R | Hypothetical protein | 440 | 468 | 0.81 | 0.81 | ND | |||||||
| 68 | ORF506R | Hypothetical protein | 718 | 420 | 2,696 | 2,074 | 1.33 | 0.93 | 4.69 | 4.16 | L | |||
| 69 | ORF522L | Hypothetical protein | 352 | 391 | 1,709 | 1,829 | 0.65 | 0.87 | 2.97 | 3.67 | L | |||
| 70 | ORF534L | Ankyrin repeat-containing protein | AAT71909 | 287 | 616 | 410 | 0.53 | 1.07 | 0.82 | E | ||||
| 71 | ORF535R | Hypothetical protein | 356 | 836 | 423 | 3,046 | 0.37 | 1.85 | 0.74 | 6.12 | IE | |||
| 72 | ORF543R | RING-finger domain-containing protein | AAT71906 | 2,534 | 6,517 | 17,679 | 5.61 | 11.33 | 35.50 | L | ||||
| 73 | ORF550R | Hypothetical protein | 297 | 223 | 3,663 | 0.66 | 0.39 | 7.36 | L | |||||
| 74 | ORF554R | Hypothetical protein | 1,183 | 2.38 | L | |||||||||
| 75 | ORF562R | Hypothetical protein | 390 | 388 | 447 | 0.86 | 0.67 | 0.90 | E | |||||
| 76 | ORF569R | Hypothetical protein | 501 | 1.01 | E | |||||||||
| 77 | ORF575R | Hypothetical protein | 320 | 0.64 | L | |||||||||
| 78 | ORF586L | Hypothetical protein | 311 | 0.62 | E | |||||||||
| 79 | ORF589L | Hypothetical protein | 326 | 0.65 | E | |||||||||
| 80 | ORF591R | Hypothetical protein | 403 | 2,423 | 635 | 1,000 | 0.42 | 5.36 | 1.10 | 2.01 | IE | |||
| 81 | ORF596L | dsRNA-specific ribonuclease | AAT71898 | 1,996 | 4.01 | L | ||||||||
| 82 | ORF600L | Hypothetical protein | 463 | 0.93 | E | |||||||||
| 83 | ORF606R | Hypothetical protein | 1,539 | 1,342 | 2.68 | 2.69 | L | |||||||
| 84 | ORF617L | Hypothetical protein | 256 | 0.51 | E | |||||||||
| 85 | ORF618R | Hypothetical protein | 506 | 1.02 | E | |||||||||
| 86 | ORF628L | Hypothetical protein | 1,210 | 2.43 | E | |||||||||
| 87 | ORF632L | Hypothetical protein | 346 | 318 | 471 | 1,887 | 0.64 | 0.70 | 0.82 | 3.79 | E | |||
| 88 | ORF635L | Hypothetical protein | 357 | 1,225 | 0.79 | 2.46 | IE | |||||||
| 89 | ORF641L | Putative ankyrin repeat protein | AAL98801 | 226 | 550 | 0.50 | 1.10 | E | ||||||
Signal intensity value is the background-subtracted median value.
The calibrated expression ratio is the ratio of the expression of the RSIV ORF in infected cells compared to the β-actin control gene. The overall percentages of ORFs expressed at each time point were: 3 h p.i., 6.5% (P < 0.05); 8 h p.i., 21.7% (P < 0.05); 18 h p.i., 31.5% (P < 0.001); 24 h p.i., 50% (P < 0.001) and 48 h p.i., 95.7% (P < 0.0001).
Abbreviation: IE, immediate-early; E, early; L, late; ND, not detected.
FIG. 1.
Hierarchical cluster analysis of the expression data of RSIV transcripts throughout infection in vitro. Calibrated expression ratios for each ORF were categorized by an average linkage hierarchical clustering program. Each row represents the expression profile of a single ORF, and each column indicates time points after infection. The normalized expression levels across all the time points are color-coded. Green boxes indicate expression ratios lower than the mean. Red boxes indicate expression ratios greater than the mean. Black boxes indicate an intermediate level of expression and gray boxes indicate missing or not detected. The magnitude of up-regulation from the mean is shown by differing intensities of red, with deep red showing lower expression and bright red showing the highest levels of expression.
Based on the expression of some ORFs that have been predicted to be involved in viral DNA replication (e.g., 239R, encoding the largest submit of DNA-dependent RNA polymerase, and 317L, encoding DNA polymerase), we propose that DNA replication of RSIV began at about 8 h p.i. in infected HINAE cells in vitro. At 48 h p.i., most viral ORFs, including the ORF for the major capsid protein (MCP), were strongly expressed (Table 2 and Fig. 1). MCP is a structural protein that occupies up to 45% of total virion protein expression (55, 61). This indicates that most RSIV virions were completely assembled by about 2 days after infection.
Temporal kinetic classes of RSIV transcripts.
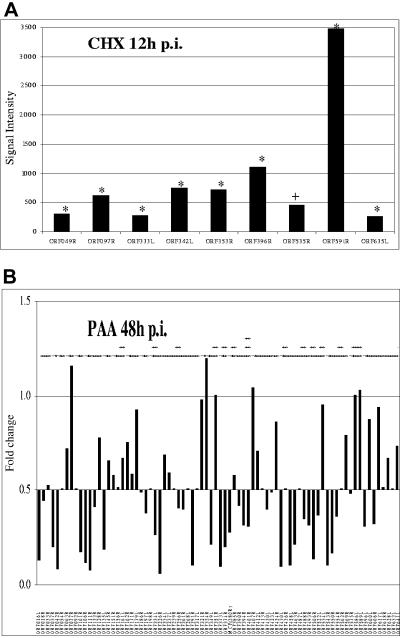
Drug inhibitors were used to map the RSIV transcripts into temporal kinetic classes during the infection in vitro as outlined in the Materials and Methods. CHX was used to investigate IE transcripts, while PAA was used to differentiate between E and L transcripts. A viral gene was classified to be in the IE expression class if it was statistically expressed under CHX treatment by using the same criteria described in Materials and Methods. On the basis of these criteria, nine RSIV transcripts were classified as IE (P < 0.05) (Table 2 and Fig. 2A). Under PAA treatment, a viral gene was considered to be in the L expression class if its median intensity ratio in the PAA-treated samples was at least twofold lower than in the untreated samples. A viral gene was considered to be in the E expression class if its expression level increased or remained within the twofold range after PAA treatment but was inhibited by CHX treatment (7, 15, 43, 50). The statistical analysis was also applied to the PAA data and identified 40 transcripts as E and 38 as L transcripts (Table 2 and Fig. 2B). As described in Fig. 3, most IE transcripts showed early expression at 3 h p.i. and were continuously expressed throughout the infection cycle. While both E and L transcripts were observed slightly later at 8 h p.i., the L transcripts peaked in expression levels at the late stages of the infection cycle between 24 to 48 h p.i.
FIG. 2.
Immediate-early, early, and late transcripts of RSIV. (A) Immediate-early transcripts of RSIV. HINAE cells were treated with CHX (50 μg/ml) 1 h prior to and throughout the RSIV infection and total RNA was isolated at 12 h p.i. The RSIV CHX-untreated and CHX-treated samples were labeled with Cy5 and Cy3-dUTP, respectively, and hybridized to the microarrays. RSIV IE transcripts were observed under CHX treatment. The signal intensity is calculated as the mean intensity of duplicate spots of each viral ORF minus the background signal. Statistical significance of changes in expression was assessed by paired t test (+, P > 0.05; *, P < 0.05) (B) Early and late transcripts of RSIV. HINAE cells were treated with PAA (100 μg/ml) 1 h prior to and throughout the RSIV infection and total RNA was isolated at 48 h p.i. The RSIV PAA-treated and RSIV-infected samples were labeled with Cy5- and Cy3-dUTP, respectively, and hybridized to the microarrays. The downward bars represent L genes whose expression levels were inhibited at least twofold (signal intensity ratios less than 0.5) by PAA treatment, while the upward bars represent E genes that were unaffected by PAA. The signal intensity ratio is calculated from the signal intensity of RSIV PAA-treated samples divided by the RSIV-infected samples. The experiment was performed in duplicate. The statistical significance of changes in expression was assessed by paired t test (+, P > 0.05; *, P < 0.05; **, P < 0.001; ***, P < 0.0001).
FIG. 3.
Time course expression analysis of three RSIV transcript classes.
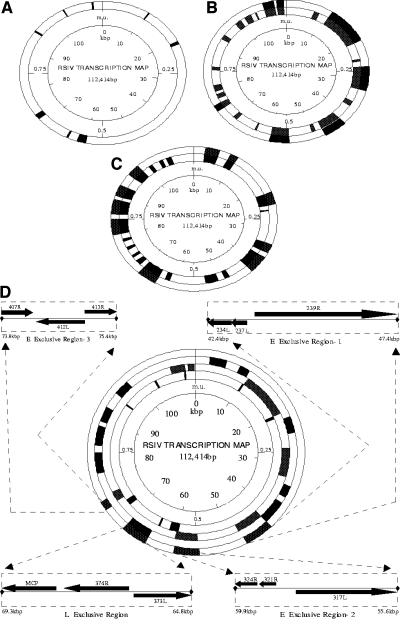
In order to determine the distribution of temporal classes in the RSIV genome, RSIV transcripts were localized on the viral physical map correlating with genome sequence data (31) (Fig. 4). Instead of clustering in any particular region of the genome, IE transcripts were distributed throughout the RSIV genome (Fig. 4A). E transcripts seemed to cluster in at least six discrete regions (Fig. 4B). Among them, at least three regions appeared to be exclusively devoted to E transcripts that encode enzymes associated with viral DNA replication (E1: bp 42,410 to 47,397; E2: bp 55,605 to 59,911, and E3: bp 73,774 to 75,409) (Fig. 4D). L transcripts were also observed throughout the genome, but appeared to originate from at least seven discrete regions (Fig. 4C).
FIG. 4.
Distribution of IE, E, and L transcripts in the RSIV genome and correlation with genomic sequence data. Transcriptionally active regions are shown in black, while inactive or undetected regions are shown in white. The innermost circle indicates map units (m.u.) and kilobases (kb) from map unit 0. (A) Distribution of IE transcripts in the RSIV genome. (B) Distribution of E transcripts in the RSIV genome. The inner solid circle shows regions of E transcription. The outer solid circle indicates major clusters of E transcription. (C) Distribution of L transcripts in the RSIV genome. The inner solid circle shows regions of L transcription. The outer solid circle indicates major clusters of L transcription. (D) Distribution of E and L exclusive regions in the RSIV genome. The three inner solid circles show major clusters of IE, E, and L transcription in the innermost, middle, and outermost circles, respectively. The outer solid circle indicates three regions exclusive to E transcripts encoding enzymes associated with viral DNA replication and an L region containing MCP. Potential ORFs are derived from genomic sequence data of Kurita et al. (28).
Analysis of toxicity of CHX and PAA.
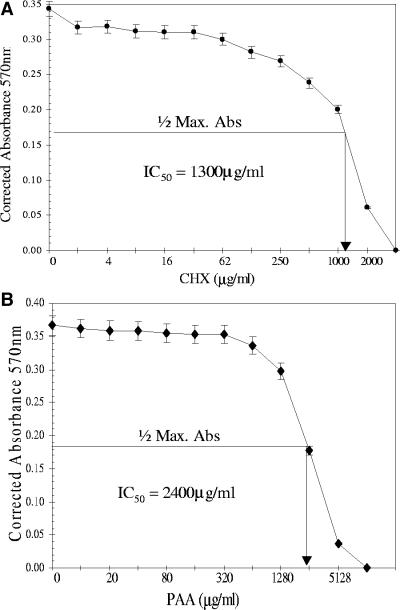
Growth of HINAE cells in various concentrations of either CHX or PAA was measured to determine the IC50 of these drug inhibitors. The assay analysis resulted in detection of IC50 at about 1,300 and 2,400 μg/ml for CHX and PAA, respectively (Fig. 5A and B). As seen in Fig. 5A, CHX concentrations less than 100 μg/ml had no significant effect on cell growth, however, concentrations greater than 500 μg/ml induced a drastic decrease in the number of living cells that was directly proportional to the 570-nm absorbance values. Similarly, no significant differences were found in the numbers of living cells during PAA treatment at concentrations up to 640 μg/ml, yet higher doses (from concentrations of 1,280 μg/ml) affected the growth of cells significantly (Fig. 5B).
FIG. 5.
IC50 detection of CHX and PAA. The assays were performed with HINAE cells following the CellTiter 96 nonradioactive cell proliferation assay kit's protocol. IC50 values were determined by locating the drug treatment value corresponding to one-half the maximum absorbance (Max. Abs.) value. The y axis represents corrected absorbance values at 570 nm that were subtracted from the absorbance value of the positive control (100% lysed cells). The x axis represents concentrations of drug inhibitors on a log scale to plot against the corrected absorbance values. The concentrations were twofold dilutions, and each concentration was tested in quadruplicate. The assay of each drug was performed in duplicate. (A) IC50 detection of CHX. (B) IC50 detection of PAA.
Confirmation of array results by RT-PCR analysis.
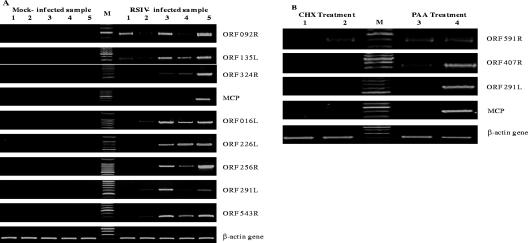
The microarray results for several ORFs whose expression changed during the viral multiplication cycle were checked by RT-PCR (Fig. 6A). These ORFs consisted of 092R (putative replication factor) and 135L, which were expressed in the early stages of infection; 324R (DNA polymerase), which has been predicted to be involved in viral DNA replication; MCP, which has been used to detect and measure the virus (5, 31); and 016L, 226R, 256R (DNA repair protein RAD2), 291L (laminin-type epidermal growth factor-like domain), and 543R (ring-finger domain-containing protein), which were strongly expressed throughout the viral infection. As expected, no viral band was observed in the mock-infected samples. For the virus-infected samples, the expression patterns and peak expression times produced by the microarray analysis and RT-PCR assay were basically the same and hence validated the microarray findings.
FIG. 6.
RT-PCR analyses for RSIV transcripts. cDNAs were synthesized from 5 μg total RNA taken from the same samples used for the microarray hybridizations. One μl cDNA was used for 30-μl RT-PCR with cycling conditions as follows: an initial denaturation at 95°C for 3 min, followed by 27 to 30 cycles of denaturation at 95°C for 30 seconds, annealing at 56°C for 30 seconds, and elongation at 72°C for 1 min, and a final elongation step at 72°C for 5 min. (A) RT-PCR detection of RSIV transcripts at different time points after infection. Lane 1, 3 h p.i.; lane 2, 8 h p.i.; lane 3, 18 h p.i.; lane 4, 24 h p.i.; lane 5, 48 h p.i.; and lane M, 100-bp DNA ladder. (B) RT-PCR detection of RSIV transcripts under drug treatment. Lane 1, CHX-treated uninfected at 12 h p.i.; lane 2, CHX-treated RSIV-infected at 12 h p.i.; lane 3, PAA-treated and RSIV-infected at 48 h p.i.; lane 4, RSIV-infected at 48 h p.i.; and lane M, 100-bp DNA ladder.
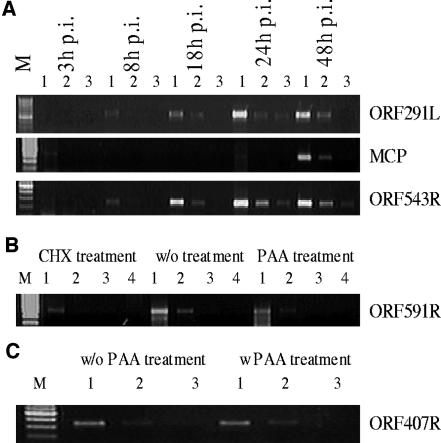
Similarly, to verify the microarray data for classification of RSIV genes into temporal kinetic expression classes, several RSIV ORFs including IE gene ORF 591R, E gene ORF 407R (ATPase), and L genes ORFs 291L (laminin-type epidermal growth factor-like domain) and MCP were selected for RT-PCR (Fig. 6B). The RT-PCR data confirmed the microarray results showing the same relative regulation of transcription of the selected genes. ORF 591R was expressed during viral infection in the presence of CHX or PAA treatment, indicating that its accumulation was not dependent on de novo protein or DNA synthesis. This confirms the microarray data which categorized this ORF as an IE gene. The band representing E gene ORF 407R was detected in the PAA-treated sample, but not the CHX-treated sample. The expression of ORF 291L and MCP was inhibited in the presence of CHX and PAA, indicating that their transcription requires de novo protein synthesis and viral DNA replication. These ORFs are therefore designated as L genes, confirming the microarray results.
Confirmation of array results by dilution RT-PCR analysis.
Some RSIV ORFs selected for RT-PCR were also chosen for dilution RT-PCR to further reconfirm the microarray data. These ORFs included three randomly selected ORFs, 291L, 543R, and MCP, which were used to confirm expression of RSIV ORFs during the viral infection cycle (Fig. 7A), and IE gene ORF 591R and E gene ORF 407R, which were used to confirm expression of RSIV ORFs under drug treatments (Fig. 7B and C). As expected, at the earlier stages of infection, PCR signals of ORFs 291L and 543R were detected after only one dilution (at 8 h p.i.) or two dilutions (at 18 h p.i.). In contrast, at the later stages of infection (at 24 h and 48 h p.i.), the signals could be seen until the third dilution (Fig. 7A). Therefore, the dilution RT-PCR results indicated increase in abundance levels of RSIV transcripts throughout the multiplication cycle, confirming the microarray results.
FIG. 7.
Dilution RT-PCR analyses for RSIV transcripts. cDNAs were serially 10-fold diluted from the original cDNAs which were used as templates for RT-PCR amplification. Two microliters of diluted cDNA was used in a 30-μl dilution RT-PCR volume. Dilution RT-PCR conditions were the same as those used for RT-PCR. (A) Dilution RT-PCR analysis of RSIV transcripts at different time points after infection. Lane 1, 10−1 diluted cDNA; lane 2, 10−2 diluted cDNA; lane 3, 10−3 diluted cDNA; and lane M, 100-bp DNA ladder. (B) Dilution RT-PCR analysis of RSIV IE transcripts. Lane 1, 100 diluted cDNA; lane 2, 10−1 diluted cDNA; lane 3, 10−2 diluted cDNA; lane 4, 10−3 diluted cDNA; and lane M, 100-bp DNA ladder. (C) Dilution RT-PCR analysis of RSIV E transcripts. Lane 1, 100 diluted cDNA; lane 2, 10−1 diluted cDNA; lane 3, 10−2 diluted cDNA; and lane M, 100-bp DNA ladder).
IE gene ORF 591R was observed with slightly different bands between RSIV-infected samples with and without drug treatments, however, its expression under the drug treatments indicated that it was independent on de novo protein synthesis and viral DNA replication (Fig. 7B). Signals of E gene ORF 407R (ATPase) were detected in both RSIV-infected samples with and without PAA at the same dilutions (10−1 dilution), indicating its expression was not affected by viral DNA replication inhibitor (Fig. 7C). Thus, the dilution RT-PCR data also strongly reconfirmed the microarray data in the case of drug treatments.
DISCUSSION
Evaluation of the RSIV DNA microarrays.
We have used DNA microarray technology, which has been successfully used to study the gene transcriptional profiles and temporal kinetics of several viruses (1, 7, 15, 30, 41, 53, 62) and knowledge of the RSIV genome to characterize the viral replication cycle in vitro on a transcriptional basis. In addition, this technology has also used to classify RSIV transcripts into temporal kinetic classes based on their dependence on de novo protein synthesis and viral DNA replication. An RSIV DNA microarray contained 92 putative viral ORFs as well as negative controls and internal controls for normalization. Every RSIV ORFs was spotted in duplicate and the duplicate spots were split between different parts of the microarrays to assess the consistency of hybridization, following the DNA microarray standard methods and published results (3, 4, 23, 32).
By using this approach, hybridization of each RSIV ORF was observed in duplicate on the individual microarray, resulting in confirming the consistent hybridization results. Our results indeed show that the signal intensities were identical between the duplicate spots of each RSIV ORF on the individual array. The microarray results were statistically analyzed by using the paired t test. The data were also imported into the cluster program that uses standard statistical algorithms to arrange genes according to similarity in pattern of gene expression (16). Furthermore, the microarray data were well confirmed by more quantitative assays, RT-PCR and dilution RT-PCR.
For normalizing genes, the microarray data showed that the median intensities of the β-actin genes were statistically unchanged (standard deviation <2-fold changes). Also, the β-actin transcript levels, as determined by RT-PCR, were similar between virus-infected and mock-infected samples, drug-treated and untreated samples as well as time course samples. The β-actin genes were therefore used to normalize viral gene expression results across the microarrays.
Taken together, the RSIV microarray is a useful tool for rapid analysis of the viral gene transcriptional profile over time course of the in vitro infection in infected cells and for grouping genes into temporal kinetic classes, providing a global picture of transcription and kinetics of RSIV genes during the replication cycle. Furthermore, the microarray analysis can predict the function of the viral genes. The coexpression of uncharacterized genes with those of known function under the same conditions or at the same developmental stages, together with their classification into the same kinetic groups may help to understand the functions of these genes (23, 63). This is the first viral DNA microarray that has been designed to study gene expression at the molecular level for piscine viruses in general and iridoviruses in particular.
RSIV transcription program over the course of an in vitro infection.
The time course experiments have allowed us to monitor the expression of each RISV ORF through an infection in vitro. It is clear that different putative ORFs generally have different expression profiles at various time points of the infection cycle. Some ORFs showed fluctuating expression patterns through the infection cycle while others were constantly expressed (Table 2 and Fig. 1). Similar results have been observed in MHV-68, HHV-8, and white spot syndrome virus infections (1, 46, 58). Our microarray results also revealed that expression of some ORFs was not continuously detected throughout the infection. For example, ORF 092R (putative replication factor) was observed at 3 h, 18 h, 24 h, and 48 h p.i., except for 8 h p.i.; however, a faint band could be seen at 8 h p.i. by RT-PCR (Fig. 6A).
According to Jenner et al. (23) the relative times at which gene expression is detected by any method are intrinsically linked to the sensitivity of the detection methods used. Therefore it may be beyond the threshold of sensitivity of the microarray method to detect low levels of expressed ORFs. Interestingly, there have been similar problems in studies of the MHV-68 transcription program. According to Ahn et al. (1) only a MHV-68 IE gene, ORF73, was detected in the absence of protein synthesis based on their microarray analysis, while three other MHV-68 genes, including ORFs 50, 57, and K3, were also determined as IE genes by other studies. Thus, more sensitive and dedicated techniques should be developed to elucidate these gene expression events.
Although the patterns and time course events of replication vary with conditions such as cell type, virus, multiplicity of infection, and temperature (61), our virus infection was carried out at MOI of about 5 throughout all experiments, following one-step growth conditions to ensure that almost all cells receive at least one infectious unit. The microarray analysis of RSIV infection in infected HIANE cells suggests that transcription of RSIV ORFs begins as early as 3 h p.i. and the viral DNA replication process begins at around 8 h p.i. in infected cells. Transcription was observed a little earlier in cells infected by Chilo iridescent virus (CIV), a virus from the family Iridoviridae. CIV transcripts were detectable by 0.5 h p.i. and the DNA replication process began at around 6 h p.i (11). However, in FHM cells infected with frog virus 3 (FV3), another iridovirus, FV3 DNA fragmentation was first detected at 6 to 7 h p.i. by Northern blot assay (8).
In this study, the expression of MCP was detected from about 48 h after infection in HINAE cells. Similarly, in RSIV-infected GF cells, MCP was detected 2 days after the first RSIV DNA fragments were observed. Many RSIV virions at distinct stages of development were also seen in the enlarged cells by electron microscope at this time (20). Taken together, these results indicate that the time required to completely assemble RSIV virions is about 2 days after infection in infected cells in vitro.
Among the genes that were consistently expressed upon viral infection, some encode enzymes associated with viral DNA replication (e.g., ORFs 239R, 317L, and 412L). Some have not been assigned functions, but they are predicted to code for proteins containing special motifs or transactivators (e.g., ORFs 291L, 396R, and 424R) (Table 2). ORF 291L is homologous to a protein containing a laminin-type epidermal growth factor-like domain of rock bream iridovirus (RBIV). It has been reported that the nidogen-binding site of laminin plays a role in cell-to-cell adhesion and may support the rapid spread of progeny virus from infected cells to uninfected cells while protecting the virus from host enzymes and antibodies (14, 63).
ORF 396R is homologous to a protein containing the transmembrane amino acid transporter of RBIV. ORF 424R is homologous to a putative ankyrin repeat protein of infectious spleen and kidney necrosis virus. CHOhr, an ankyrin repeat gene of vaccinia virus, has been reported to prevent virus-induced apoptosis in CHO cells, which thus provides sufficient time for replication of the virus before cell death (21). Another ankyrin repeat gene of African swine virus also has a role in escaping the immune response during viral infection (49). Similarly, it is possible that RSIV ORFs 291L, 396R, and 424R also play a crucial role during virus multiplication, hence their constant expression throughout the virus infection. However, some other strongly expressed ORFs (e.g., 097R, 135L, 226R, 342L, 506R, and 522L) are of unknown function. This group of genes, therefore, will be interesting subjects for further studies.
Temporal kinetic expression of RSIV transcripts.
RSIV transcripts are defined as IE, E, and L by their independence of de novo protein synthesis and viral DNA replication. IE transcripts normally function as regulatory trans-acting factors; therefore, they are expressed in the presence of a de novo protein synthesis inhibitor, which prevents de novo protein synthesis by preventing translation but not transcription. Most E transcripts are involved in viral nucleic acid metabolism, while L transcripts function as structural proteins and are expressed after the onset of viral DNA replication. Therefore, inhibition of viral DNA synthesis blocks the expression of L transcripts, allowing them to be distinguished from IE and E transcripts (15, 33, 53). In this study, CHX (50 μg/ml) was used to identify the kinetic class of IE, while PAA (100 μg/ml) was used to identify E and L transcripts. The concentrations of both of CHX and PAA used here were less than at least 20 times the IC50, and were within the range of concentrations that had no significant effects on the growth of HINAE cells (Fig. 5A and B). Moreover, HINAE cells were also checked for drug toxicity by β-actin gene expression observed using RT-PCR. The results revealed that there was no difference in expression in the drug-treated and untreated cells (Fig. 6B), indicating minimal toxicity to the cells by these drugs.
RSIV IE transcripts were expressed under CHX treatment. L transcripts were inhibited under PAA treatment, whereas E transcripts showed increased or unchanged expression after PAA treatment compared to RSIV-infected cells without PAA but were blocked under CHX treatment (Fig. 2A and B). Three temporal kinetic classes of transcripts have also been identified in FV3, CIV, and Wiseana iridescent virus infections (8, 12, 36, 61). In these studies, the second class is called delayed-early. For example, in CIV infection, 37 transcripts were classified as IE, 35 transcripts as DE, and 65 transcripts as L (12). Thus, the temporal expression kinetics observed in RSIV are similar to those observed in other iridoviruses, suggesting that they are a common feature of the family Iridoviridae.
The expression of RSIV genes upon infection occurred in a temporal manner: IE genes were expressed first, then followed by E and L genes (Fig. 3). These observations are similar to those detected in transcription profiles of other DNA viruses, including CIV and HHV-8. CIV gene expression also occurred in a temporal cascade with three kinetic classes (IE, DE, and L). CIV IE transcripts were first observed during 0 to 3 h p.i. while DE transcripts were first detected at 3 h p.i. and showed peak levels between 3 h to 12 h p.i. L transcripts were detected either at 6 h, 12 h, or 24 h p.i (11, 12). In the transcription program of HHV-8, genes involved in gene regulation or IE genes showed the earliest increase in expression. Genes involved in DNA replication or E genes showed increase in expression slightly later (46).
IE transcripts were not clustered in any specific region, whereas 6 discrete regions of E and 7 discrete regions of L transcription were detected in the RSIV genome (Fig. 4A, B, and C). These results are similar to those observed in other members of the family Iridoviridae. IE transcripts of CIV were also not clustered in any specific regions in the CIV genome, DE transcripts were clustered in at least three DE discrete regions, and L transcripts were distributed throughout the genome (12). IE transcripts originated from at least one third of the genome in FV3 (57) and from seven different regions in Wiseana iridescent virus (36). In two other large DNA viruses, IE and E transcripts are distributed throughout the genome. At least eight IE discrete regions have been observed in the baculovirus Autographa californica nucleopolyhedrovirus (12) and a half of the vaccinia virus genome is devoted to E transcription (45).
At least three E regions are exclusive to E transcripts that encode potential enzymes associated with viral DNA replication. The exclusive region E1 contains a cluster of three ORFs (234L, 237L, and 239R). ORF 234L is homologous to a deoxyribonucleoside kinase of RBIV. ORFs 237L and 239R encode the largest submit of the DNA-dependent RNA polymerase. Region E2 includes three ORFs (317L, 321R, and 324R) coding for DNA polymerase, while region E3 contains three ORFs (407R, 412L, and 413R) coding for ATPase (Table 2 and Fig. 4D). The presence of these enzymes suggests that the three E-exclusive regions seem to play a significant role in the DNA replication of RSIV. Similarly, the L region (bp 64,820 to 69,270) (Fig. 4D) contains three ORFs, including MCP, which accounts for up to 45% of total virion protein, suggesting that this L region would play a significant role in the viral assembly process. However, the significance of other E and L discrete regions is not known because most ORFs in these regions are of unknown functions. Further studies are required to identify more functionally defined genes and to assess the roles of these regions.
In conclusion, a DNA microarray for RSIV has been developed and used to monitor the in vitro transcriptional profile of the RSIV in infected cells and to classify its genes into IE, E, and L kinetic classes. RSIV DNA replication began at around 8 h p.i. and the time required to completely assemble RSIV virions was about 2 days after infection in vitro. The RSIV gene expression strategies occurred in a temporal manner with three stages (IE, E, and L), a finding consistent with a classical virus gene expression cascade. These studies should impart a greater understanding of the replication mechanism, gene regulation strategies, and pathogenesis of RSIV in particular and piscine iridoviruses in general. This viral microarray may also be used as a tool to examine the RSIV transcription pattern in vivo in order to better understand the pathogenesis of RSIV. Further studies are needed to understand how host responses contribute to the complex host-pathogen interaction during infection. For this purpose, a combined viral and host gene microarray would be useful.
Acknowledgments
We thank Jun Kurita of the National Research Institute of Aquaculture, Fisheries Research Agency, for providing genome information on RSIV.
This research was supported in part by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Sciences, and Technology of Japan.
REFERENCES
- 1.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowtell, D., and J. Sambrook (ed.). 2002. DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 4.Byon, J. Y., T. Ohira, I. Hirono, and T. Aoki. 2005. Use of a cDNA microarray to study immunity against viral hemorrhagic septicemia (VHS) in Japanese flounder (Paralichthys olivaceus) following DNA vaccination. Fish Shellfish Immunol. 18:135-147. [DOI] [PubMed] [Google Scholar]
- 5.Caipang, C. M., I. Hirono, and T. Aoki. 2003. Development of a real-time PCR assay for the detection and quantification of red sea bream Iridovirus (RSIV). Fish Pathol. 38:1-7. [Google Scholar]
- 6.Carter, K. L., E. Cahir-McFarland, and E. Kieff. 2002. Epstein-barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76:10427-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinchar, V. G., L. Bryan, J. Wang, S. Long, and G. D. Chinchar. 2003. Induction of apoptosis in frog virus 3-infected cells. Virology 306:303-312. [DOI] [PubMed] [Google Scholar]
- 9.Clem, L. W., L. Moewus, and M. M. Sigel. 1961. Studies with cells from marine fish in tissue culture. Proc. Soc. Exp. Biol. Med. 108:762-766. [DOI] [PubMed] [Google Scholar]
- 10.Cuadras, M. A., D. A. Feigelstock, S. An, and H. B. Greenberg. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J. Virol. 76:4467-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Costa, S. M., H. Yao, and S. L. Bilimoria. 2001. Transcription and temporal cascade in Chilo iridescent virus infected cells. Arch. Virol. 146:2165-2178. [DOI] [PubMed] [Google Scholar]
- 12.D'Costa, S. M., H. J. Yao, and S. L. Bilimoria. 2004. Transcriptional mapping in Chilo iridescent virus infections. Arch. Virol. 149:723-742. [DOI] [PubMed] [Google Scholar]
- 13.DeFilippis, V., C. Raggo, A. Moses, and K. Fruh. 2003. Functional genomics in virology and antiviral drug discovery. Trends Biotechnol. 21:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do, J. W., C. H. Moon, H. J. Kim, M. S. Ko, S. B. Kim, J. H. Son, J. S. Kim, E. J. An, M. K. Kim, S. K. Lee, M. S. Han, S. J. Cha, M. S. Park, M. A. Park, Y. C. Kim, J. W. Kim, and J. W. Park. 2004. Complete genomic DNA sequence of rock bream iridovirus. Virology 325:351-363. [DOI] [PubMed] [Google Scholar]
- 15.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiss, G. K., R. E. Bumgarner, M. C. An, M. B. Agy, A. B. van't Wout, E. Hammersmark, V. S. Carter, D. Upchurch, J. I. Mullins, and M. G. Katze. 2000. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology 266:8-16. [DOI] [PubMed] [Google Scholar]
- 18.Huang, C., X. Zhang, K. Y. Gin, and Q. W. Qin. 2004. In situ hybridization of a marine fish virus, Singapore grouper iridovirus with a nucleic acid probe of major capsid protein. J. Virol. Methods 117:123-128. [DOI] [PubMed] [Google Scholar]
- 19.Hung, S. P., P. Baldi, and G. W. Hatfield. 2002. Global gene expression profiling in Escherichia coli K12. The effects of leucine-responsive regulatory protein. J. Biol. Chem. 277:40309-40323. [DOI] [PubMed] [Google Scholar]
- 20.Imajoh, M., H. Sugiura, and S. Oshima. 2004. Morphological changes contribute to apoptotic cell death and are affected by caspase-3 and caspase-6 inhibitors during red sea bream iridovirus permissive replication. Virology 322:220-230. [DOI] [PubMed] [Google Scholar]
- 21.Ink, B. S., C. S. Gilbert, and G. I. Evan. 1995. Delay of vaccinia virus-induced apoptosis in nonpermissive Chinese hamster ovary cells by the cowpox virus CHOhr and adenovirus E1B 19K genes. J. Virol. 69:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inouye, K., K. Yamano, Y. Maeno, K. Nakajima, M. Matsuoka, Y. Wada, and M. Sorimachi. 1992. Iridovirus infection of cultured red sea bream, Pargus major. Fish Pathol. 27:19-27. [Google Scholar]
- 23.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, J. B., L. J. Jun, M. H. Yoo, M. S. Kim, J. L. Komisar, and H. D. Jeong. 2003. Characterization of the DNA nucleotide sequences in the genome of red sea bream iridoviruses isolated in Korea. Aquaculture 220:119-133. [Google Scholar]
- 25.Jeong, J. B., K. H. Park, H. Y. Kim, S. H. Hong, K. H. Kim, J. K. Chung. J. L. Komisar, and H. D. Jeong. 2004. Multiplex PCR for the diagnosis of red sea bream iridoviruses isolated in Korea. Aquaculture Article in Press.
- 26.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karaca, G., J. Anobile, D. Downs, J. Burnside, and C. J. Schmidt. 2004. Herpesvirus of turkeys: microarray analysis of host gene responses to infection. Virology 318:102-111. [DOI] [PubMed] [Google Scholar]
- 28.Kasai, H., and M. Yoshimizu. 2001. Establishment of two Japanese flounder embryo cell lines. Bull. Fisheries Sci. Hokkaido Univ. 52:67-70. [Google Scholar]
- 29.Kawakami, H., and K. Nakajima. 2002. Cultured fish species affected by red sea bream iridoviral disease from 1996-2000. Fish Pathol. 37:45-47. [Google Scholar]
- 30.Khadijah, S., S. Y. Neo, M. S. Hossain, L. D. Miller, S. Mathavan, and J. Kwang. 2003. Identification of white spot syndrome virus latency-related genes in specific-pathogen-free shrimps by use of a microarray. J. Virol. 77:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurita, J., K. Nakajima, I. Hirono, and T. Aoki. 2002. Complete genome sequencing of red sea bream Iridovirus (RSIV). Fisheries Sci. 68:1113-1115. [Google Scholar]
- 32.Kurobe, T., M. Yasuike, T. Kimura, I. Hirono, and T. Aoki. 2005. Expression profiling of immune-related genes from Japanese flounder Paralichthys olivaceus kidney cells using cDNA microarrays. Dev. Comp. Immunol. 29:515-523. [DOI] [PubMed] [Google Scholar]
- 33.Liu, W. J., Y. S. Chang, C. H. Wang, G. H. Kou, and C. F. Lo. 2005. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 334:327-341. [DOI] [PubMed] [Google Scholar]
- 34.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937-19944. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMillan, N. A., and J. Kalmakoff. 1994. RNA transcript mapping of the Wiseana iridescent virus genome. Virus Res. 32:343-352. [DOI] [PubMed] [Google Scholar]
- 37.Morgan, R. W., L. Sofer, A. S. Anderson, E. L. Bernberg, J. Cui, and J. Burnside. 2001. Induction of host gene expression following infection of chicken embryo fibroblasts with oncogenic Marek's disease virus. J. Virol. 75:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima, K., T. Ito, J. Kurita, H. Kawakami, T. Itano, Y. Fukuda, Y. Aoi, T. Tooriyama, and S. Manabe. 2002. Effectiveness of a vaccine against red sea bream irodoviral disease in various cultured marine fish under laboratory conditions. Fish Pathol. 37:90-91. [Google Scholar]
- 39.Nakajima, K., and M. Sorimachi. 1994. Biological and physio-chemical properties of the iridovirus isolated from cultured red sea bream, Pagrus major. Fish Pathol. 29:29-33. [Google Scholar]
- 40.Nakajima, K., Y. Maeno, M. Fukudome, Y. Fukuda, S. Tanaka, and M. Sorimachi. 1995. Immunofluorescence test for the rapid diagnosis of red sea bream iridovirus infection using monoclonal antibody. Fish Pathol. 30:115-119. [Google Scholar]
- 41.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nalcacioglu, R., H. Marks, J. M. Vlak, Z. Demirbag, and M. M. van Oers. 2003. Promoter analysis of the Chilo iridescent virus DNA polymerase and major capsid protein genes. Virology 317:321-329. [DOI] [PubMed] [Google Scholar]
- 43.Oster, B., and P. Hollsberg. 2002. Viral gene expression patterns in human herpesvirus 6B-infected T cells. J. Virol. 76:7578-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsuka, M., H. Aizaki, N. Kato, T. Suzuki, T. Miyamura, M. Omata, and N. Seki. 2003. Differential cellular gene expression induced by hepatitis B and C viruses. Biochem. Biophys. Res. Commun. 300:443-447. [DOI] [PubMed] [Google Scholar]
- 45.Paoletti, E., and L. J. Grady. 1977. Transcriptional complexity of vaccinia virus in vivo and in vitro. J. Virol. 23:608-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pietiainen, V., P. Huttunen, and T. Hyypia. 2000. Effects of echovirus 1 infection on cellular gene expression. Virology 276:243-250. [DOI] [PubMed] [Google Scholar]
- 48.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent end points. Am. J. Hyg. 27:493-497. [Google Scholar]
- 49.Revilla, Y., M. Callejo, J. M. Rodriguez, E. Culebras, M. L. Nogal, M. L. Salas, E. Vinuela, and M. Fresno. 1998. Inhibition of nuclear factor kappaB activation by a virus-encoded IkappaB-like protein. J. Biol. Chem. 273:5405-5411. [DOI] [PubMed] [Google Scholar]
- 50.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 52.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 53.Stingley, S. W., J. J. Ramirez, S. A. Aguilar, K. Simmen, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2000. Global analysis of herpes simplex virus type 1 transcription using an oligonucleotide-based DNA microarray. J. Virol. 74:9916-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, L. A., C. M. Carthy, D. Yang, K. Saad, D. Wong, G. Schreiner, L. W. Stanton, and B. M. McManus. 2000. Host gene regulation during coxsackievirus B3 infection in mice: assessment by microarrays. Circ. Res. 87:328-334. [DOI] [PubMed] [Google Scholar]
- 55.Tidona, C. A., P. Schnitzler, R. Kehm, and G. Darai. 1998. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 16:59-66. [DOI] [PubMed] [Google Scholar]
- 56.Ton, C., D. Stamatiou, V. J. Dzau, and C. C. Liew. 2002. Construction of a zebrafish cDNA microarray: gene expression profiling of the zebrafish during development. Biochem. Biophys. Res. Commun. 296:1134-1142. [DOI] [PubMed] [Google Scholar]
- 57.Tondre, L., T. N. Tham, P. H. Mutin, and A. M. Aubertin. 1988. Molecular cloning and physical and translational mapping of the frog virus 3 genome. Virology 162:108-117. [DOI] [PubMed] [Google Scholar]
- 58.Tsai, J. M., H. C. Wang, J. H. Leu, H. H. Hsiao, A. H. Wang, G. H. Kou, and C. F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van 't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, C. S., H. H. Shih, C. C. Ku, and S. N. Chen. 2003. Studies on epizootic iridovirus infection among red sea bream, Pagrus major (Temminck & Schlegel), cultured in Taiwan. J. Fish Dis. 26:127-133. [DOI] [PubMed] [Google Scholar]
- 61.Williams, T. 1996. The iridoviruses. Adv. Virus Res. 46:345-412. [DOI] [PubMed] [Google Scholar]
- 62.Yamagishi, J., R. Isobe, T. Takebuchi, and H. Bando. 2003. DNA microarrays of baculovirus genomes: differential expression of viral genes in two susceptible insect cell lines. Arch. Virol. 148:587-597. [DOI] [PubMed] [Google Scholar]
- 63.Ye, R. W., T. Wang, L. Bedzyk, and K. M. Croker. 2001. Applications of DNA microarrays in microbial systems. J. Microbiol. Methods 47:257-272. [DOI] [PubMed] [Google Scholar]