Abstract
DNA ligases are important enzymes which catalyze the joining of nicks between adjacent bases of double-stranded DNA. NAD+-dependent DNA ligases (LigA) are essential in bacteria and are absent in humans. They have therefore been identified as novel, validated and attractive drug targets. Using virtual screening against an in-house database of compounds and our recently determined crystal structure of the NAD+ binding domain of the Mycobacterium tuberculosis LigA, we have identified N1, Nn-bis-(5-deoxy-α-d-xylofuranosylated) diamines as a novel class of inhibitors for this enzyme. Assays involving M.tuberculosis LigA, T4 ligase and human DNA ligase I show that these compounds specifically inhibit LigA from M.tuberculosis. In vitro kinetic and inhibition assays demonstrate that the compounds compete with NAD+ for binding and inhibit enzyme activity with IC50 values in the µM range. Docking studies rationalize the observed specificities and show that among several glycofuranosylated diamines, bis xylofuranosylated diamines with aminoalkyl and 1, 3-phenylene carbamoyl spacers mimic the binding modes of NAD+ with the enzyme. Assays involving LigA-deficient bacterial strains show that in vivo inhibition of ligase by the compounds causes the observed antibacterial activities. They also demonstrate that the compounds exhibit in vivo specificity for LigA over ATP-dependent ligase. This class of inhibitors holds out the promise of rational development of new anti-tubercular agents.
INTRODUCTION
DNA ligases are important enzymes, vital for replication and repair, which catalyze the joining of nicks between adjacent bases of double-stranded DNA. These enzymes are classified as NAD+ or ATP-dependent based on the respective co-factor specificities. NAD+-dependent ligases (also called LigA) are found exclusively in eubacteria and some viruses (1–3) while their ATP-dependent counterparts are found in all kingdoms of life (1). Gene knockout and other studies have shown that NAD+-dependent ligases are essential in several bacteria including Escherichia coli, Staphylococcus aureus and Bacillus subtilis (4–6). Consistent with LigA being essential, it was not possible to isolate bacteria with the gene deleted in Mycobacterium tuberculosis (7,8). Additionally, LigA is also not found in humans and are therefore attractive drug targets.
Both NAD+- and ATP-dependent DNA ligases are highly modular proteins with distinct domain architectures. Their mechanistic steps involve large conformational changes, among other things (9–12), and the respective enzyme mechanisms are also broadly conserved. Briefly, the respective enzymes form an enzyme-adenylate intermediate in the first step after attacking the α-phosphorous of ATP or NAD+. A DNA adenylate intermediate is formed in the second step where the bound AMP is transferred to the 5′ end of DNA. The respective enzymes then catalyze the joining of the 3′ nicked DNA to the intermediate and release AMP in the final step.
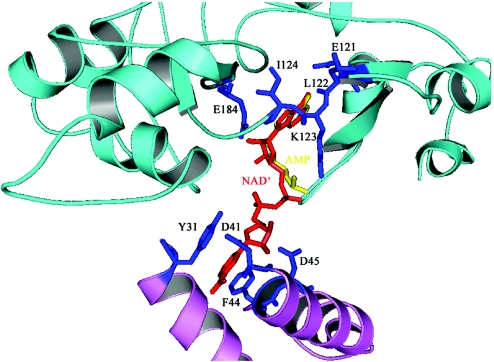
A crystal structure of the full-length LigA with bound AMP (Adenosine mono phosphate) is available from T.filiformis (12), while structures of the adenylation domain are available from Bacillus stearothermophilus (no co-factor) and Enterococcus faecalis (with NAD+) (10,11). We have recently reported the crystal structure of the adenylation domain of LigA from M.tuberculosis bound to AMP (13). The adenylation domain contains five out of six conserved sequence motifs in NAD+-dependent ligases (14) and they mainly line the NAD+ binding pocket (Figure 1). The active site lysine (K123), which forms the covalent ligase-adenylate intermediate, and a co-factor ‘conformation discriminating’ Glu (E184) are part of motifs I and III, respectively (13,15).
Figure 1.
Co-factor binding site in NAD+-dependent DNA ligase from M.tuberculosis. Parts from sub-domains 1a and 1b which make up the binding site are depicted in lavender and cyan, respectively. Interacting residues with NAD+ are indicated in blue and labeled for clarity, while the co-factor itself is shown in red. Bound AMP from the crystal structure (PDB: 1ZAU) is indicated in yellow. This and the next figure was made using PyMOL (41) (http://www.pymol.org).
Despite the availability of crystal structures, efforts to find LigA-specific inhibitors have resulted in the identification of only three classes of compounds, namely arylamino compounds, pyridochromanones and glycosyl ureides (16,17,13). This could partly be due to the inherent difficulties in finding molecules capable of distinguishing between related binding sites in NAD+ and ATP ligases. Arylamino compounds and derivatives of chloroquine inhibit the enzyme in the low µM range but exhibit some DNA binding properties which affect the potency but not the specificity of the tested inhibitors (16). Pyridochromanones and glycosyl ureides, on the other hand, are competitive inhibitors and bind to the NAD+ binding site. These compounds have up to nanomolar and low micromolar affinities for the enzyme, respectively. They are also able to distinguish between the NAD+ and ATP binding sites as supported by both inhibition assays and docking studies (17,13).
M.tuberculosis, the etiological agent of the disease, kills more than 2 million people every year worldwide in concurrence with HIV-related infections (18). Moreover, appearance of multi drug resistant (MDR) strains of M.tuberculosis to many, if not all, of the existing drugs has been noted. This has necessitated more urgent and new approaches to find novel therapies based on different mechanisms of action (19).
As part of a long range goal we are using virtual screening strategies to identify novel classes of inhibitory molecules which bind competitively to the co-factor binding site and to develop them as potential anti-tubercular entities. We had earlier identified glycosyl ureides as potent, competitive LigA-specific inhibitors (13). Here, we report a series of 5-deoxy-xylofuranosylated amines active against the M.tuberculosis NAD+-dependent DNA ligase with IC50 values in the µM range and able to discriminate between the human (ATP-dependent) and pathogen enzymes. This class of compounds had earlier been reported by us as having anti-tubercular activity (20). In vitro inhibition assays show specificity of the compounds for MtuLigA over ATP-dependent ligases from human and bacteriophage sources and also that they compete with NAD+ for binding. In vivo inhibition/antibacterial assays involving LigA-deficient bacterial strains rescued with MtuLigA suggests that inhibition of MtuLigA is the cause of the earlier reported anti-tubercular activity of the compounds. These results pave the way for rational optimization and synthesis of second generation inhibitors with potent anti-tubercular activity.
MATERIALS AND METHODS
Virtual screening protocol
Docking calculations were carried out using AUTODOCK v3.0.5 (21). A PERL/PerlTk/Python-based script was used to add the capability of automated docking against a ligand database to AUTODOCK. A computer cluster consisting of SGI ORIGIN350 servers and SGI OCTANES were used for the computation and analysis of docked complexes. Two models each of well-characterized NAD+- and ATP-dependent DNA ligases were selected as targets for in silico screening calculations.
Preparation of templates
MtuLigANAD. The crystal structure of MtuLigA (PDB: 1ZAU) was used as the basis for generating the NAD+ binding site by superposing the individual sub-domains in the structure onto the E.faecalis LigA-NAD+ co-crystal structure (PDB: 1TAE).
E.faecalis ligase
The NAD+-dependent ligase from E.faecalis (PDB: 1TAE) in which the NAD+ binding pocket is well defined was also chosen.
ATP-dependent DNA ligases
To compare docking results and to identify compounds with specificity for LigA, well-characterized ATP ligases from two different sources, viral (T4) and human ATP-dependent ligase I (PDB: 1X9N) were also chosen for docking studies. Selection of human ligase was based on the fact that M.tuberculosis is a major human pathogen. A homology model for T4Lig was generated using MODELLER6v2 (22) where T7 DNA ligase (23) (PDB: 1A0I) was the template. The model was refined by subjecting it to a few rounds of minimization using the DISCOVER_3 module in InsightII (24). The stereo-chemical quality of the model was verified using PROCHECK (25) and WHAT IF (26). Prior to docking studies, crystallographic waters and heteroatoms were removed from the crystal structures. Polar hydrogens were added and also Kollman charges were assigned to all atoms (http://www.scripps.edu/mb/olson/dock/autodock/tools.html).
Ligand preparation
An in-house database consisting of over 15 000 compounds was used. This database can be filtered for specific properties, such as anti-tubercular activity, etc. based on prior experiments and synthesis expertise is also available. The 3D structures of the ligands were built and optimized using the BUILDER module in InsightII. The ligand's translation, rotation and internal torsions were assigned for AUTODOCK runs.
Autodock run parameters
The grid for docking calculations was centered on Lys123 for 1ZAU, Lys159 for T4, Lys120 for 1TAE and Lys568 for 1X9N in the docking studies. An 80 × 80 × 80 3D affinity grid with 0.375 Å spacing was calculated, respectively, for each of the following atom types C, A (aromatic C), N, O, S, H, F, Cl, Br and e by Autogrid 3.0.
The Lamarckian genetic algorithm implemented in AUTODOCK v3.0.5 was used. Docking parameters were as follows: 100 docking trials, population size of 150, random starting position and conformation translation step ranges of 1.5 Å, rotation step ranges 35, elitism of 1, mutation rate of 0.02, cross-over rate of 0.8, local search rate of 0.06 and 10 million energy evaluations. The jobs were distributed to the computer cluster. The docked conformations were clustered by the use of a tolerance of 1.5 Å root-mean-square deviation. The complexes were sorted based on the scoring function and fitness scores implemented in the program. Control docking experiments were carried out to reproduce the AMP/NAD+ complexes in the crystal structures. Selected compounds from the best 10% docked complexes (as observed from the AUTODOCK scoring function) were taken up further for in vitro and in vivo ligase assays.
Synthesis of compounds 1–5
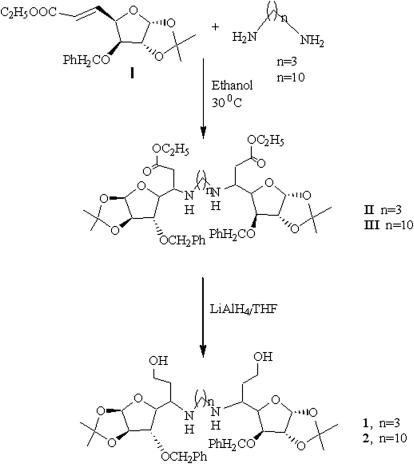
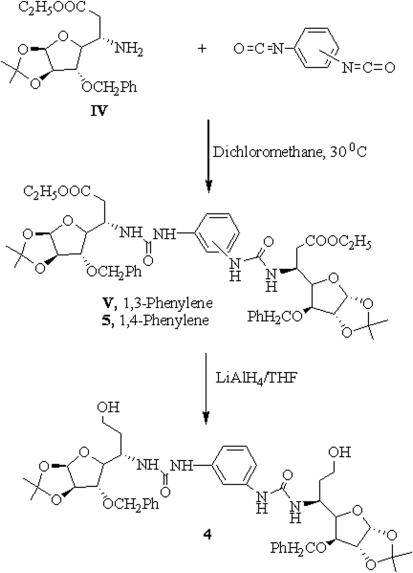
5-Deoxy-xylofuranosylated diamines 1 and 2 were synthesized by conjugate addition of 1 mole equivalent of diamines with glycosyl olefinic ester I followed by reduction of the intermediate glycosyl amino esters II and III by the protocol already reported by us (20,27) and compounds were isolated as diastereochemically pure by column chromatography (Scheme 1). Compound 3 was prepared by reductive aminoalkylation of xylofuranose dialdose with 1, 12-dodecyl amine as already reported by us (27). Compound 4 was prepared by reaction of xylofuranosylated amino ester IV (28) with 1, 3-phenylene diisocyanate followed by reduction of the intermediate amino ester V with LiAlH4. However, synthesis of compound 5 was achieved simply by reacting glycosyl amino ester IV with 1, 4-phenylene diisocyanate following our earlier protocol (29,30) (Scheme 2).
Scheme 1.
Synthesis of glycosyl amines with aminoalkyl spacers.
Scheme 2.
Synthesis of glycosyl amines with phenylene carbamoyl spacers.
In vitro assay
In vitro assays for ligase activity were performed using a double-stranded 40 bp DNA substrate carrying a single-strand nick between bases 22 and 23 as reported earlier (13). Briefly, the substrate was created in TE buffer by annealing 22 and 18mer DNA complementary strands to a 40mer (5′-ATG TCC AGT GAT CCA GCT AAG GTA CGA GTC TAT GTC CAG G-3′). At the 5′ end, the 18mer was radiolabeled with [γ-32P]ATP (3000 Ci/mmol, Board of Radiation and Isotope Technology, Mumbai). This labeled, nicked 40 bp DNA substrate was used to assay the in vitro inhibitory activity of different compounds against MtuLigA, T4Lig and HuLigI. Amounts of the respective enzymes were optimized for similar ligation extents in the absence of any inhibitor under assay conditions.
Full-length MtuLigA was cloned, expressed and purified as reported earlier (13). The assays were carried out using 2 ng of the purified protein. Reaction mixtures (15 µl) containing 50 mM, Tris–HCl, pH 8.0, 5 mM DTT, 10 mM MgCl2, 10% dimethyl sulfoxide (Me2SO), 50 µM NAD+, 2 pmol of 32P-labeled nicked duplex DNA substrate and different concentration of compounds were incubated for 1 h at 25°C. Subsequently, they were quenched with formamide and EDTA. The products were resolved electrophoretically on a 15% polyacrylamide gel containing 8 M urea in TBE (90 mM Tris-borate and 2.5 mM EDTA). Autoradiograms were developed and ligation extents were measured using Image Master 1D Elite software (Amersham). All compounds were dissolved in 100% Me2SO. The compound solutions comprised one-tenth volume of the ligation reaction mixture; thus, 10% Me2SO was included in all the control reactions.
The same procedure was followed for T4 DNA ligase also. Briefly, T4 DNA ligase assay was carried out in a volume of 15 µl containing 0.05 U of enzyme (Amersham), 2 pmol of labeled template and 66 µM ATP in 66 mM Tris–HCl, pH 7.6, 6.6 mM MgCl2, and 10 mM DTT and 10% Me2SO. The Human DNA ligase I expression plasmid was transformed into E.coli BL21 (DE3) and purified as described previously (31). Purified protein was concentrated to 2 mg/ml. An aliquot of 2 µg protein was used for assay in 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 5 mM DTT, 50 µg/ml BSA and 1 mM ATP as described above.
Calculation of IC50 values
The IC50 values were determined by plotting the relative ligation activity versus inhibitor concentration and fitting to the equation:
using GraphPad Prism®. V0 and Vi represent rates of ligation in the absence and presence of inhibitor, respectively, and [I] refers to the inhibitor concentration.
DNA-inhibitor interactions
Fluorescence assay
DNA intercalating properties of the compounds, if any, were measured by attempting to displace ethidium bromide from DNA. Detection of its displacement from DNA is based on the strong loss in fluorescence that should occur upon its detachment from DNA (32). The assay mixture contained 5 µg of calf thymus DNA, 5 µM ethidium bromide, 25 mM Tris–HCl, pH 8.0, 50 mM NaCl and 1 mM EDTA in a total volume of 100 µl.
Change in ethidium bromide fluorescence was followed on addition of increasing inhibitor concentrations at excitation and emission wavelengths of 485 and 612 nm, respectively.
Gel shift assay
An aliquot of 100 ng of plasmid DNA (pUC 18) was incubated with increasing compound concentration in TE at 25°C for 1 h. Subsequently, the DNA was analyzed in a 1% agarose gel.
Mode of inhibition
Using Michaelis–Menten kinetics, saturating substrate concentration for MtuLigA was determined by increasing the NAD+ concentration from 0.2 to 50 µM. Km for NAD+ was determined in 10% Me2SO using the assay procedure. Kinetics for different amount of compounds were determined using varying concentrations of NAD+ from 0 up to 50 µM under standard assay conditions as described earlier.
Rate of the ligation reaction was determined based on the extents of ligation by scanning the gel using ImageMaster 1D Elite software (Amersham). Data were plotted using Michaelis–Menten kinetics in Graph Pad Prism®. Similarly, Ki values were determined by plotting the apparent Km values against the respective compound concentrations. Mode of inhibition was determined through standard analysis of Lineweaver–Burk kinetics.
Antibacterial activity
The recombinant plasmid pRBL (33) containing the gene for T4Lig in pTrc99A was transformed into E.coli GR501 ligAts mutant (34). In order to have the same genetic background, the MtuLigA clone in pTrc99A (13) was also transformed into E.coli GR501. In growth experiments the strains expressing MtuLigA or T4Lig were compared with a control GR501 strain carrying empty pTrc99A without any gene insertions at 37°C. As reported earlier (17) and reproduced by us, the temperature-sensitive E.coli GR501 ligAts strain grows well at 30°C while it is strongly delayed at 37°C. Complementation with either MtuLigA or T4Lig restores the growth of the mutant strain.
Minimum inhibitory concentration (MIC) values for the inhibitors were determined for MtuLigA and T4Lig in E.coli GR501 ligAts mutant along with Salmonella typhimurium LT2 and its DNA ligase minus (null) mutant derivative which had been rescued with a plasmid (pBR313/598/8/1b) encoding the T4Lig gene (35) in order to check the specificity of compounds for NAD+-dependent ligases from other sources as well. Antimicrobial activity was monitored in microtiter plates using microdilution assay technique in a volume of 200 µl.
Approximately 105 CFU/ml in the case of E.coli LigAts mutant, 106 CFU/ml in the case of S.typhimurium LT2 and its mutant LigA− strain, rescued with T4Lig, were incubated with different compound concentrations under ambient conditions for 20 h and MIC were determined on the basis of the presence of any visible growth. E.coli mutant strain was grown in Luria–Bertani (LB) medium while nutrient broth was used for S.typhimurium strains. The media contained 20 µg/ml polymyxin B nonapeptide to facilitate passage of the inhibitors across the outer membrane.
RESULTS
Virtual screening
Choice of models and NAD+ binding site
Available crystal structures of NAD+-dependent DNA ligases include those with NAD+ (PDB: 1TAE) (11) and AMP (PDB: 1ZAU) (13). The AMP binding site consists only of residues in subdomain 1b. On the other hand, the NAD+ binding site is generated when the mobile subdomain 1a comes into close proximity of subdomain 1b (Figure 1). Mutational analysis of corresponding conserved residues in this class of enzymes has shown, among other things, that the tyrosine corresponding to Y31 of MtuLigA is essential for activity while mutating those corresponding to F44 diminishes activity (36). This is in addition to other residues in subdomain 1b, such as K123 and E184, which were earlier identified as essential for activity (37). The use of the complete NAD+ binding site, as opposed to only subdomain 1b which binds AMP, in virtual screening experiments would be expected to pick up molecules that compete with NAD+. We therefore generated the NAD+ binding site in MtuLigA based on the MtuLigA and EfaLigA crystal structures as reported earlier (13,11) (Figure 1) and used it for virtual screening. We were also interested in compounds that could potentially distinguish between NAD+ and ATP binding sites in the respective enzymes as compounds having less affinity for the ATP binding site would be more interesting as expectedly they were likely to have less affinity for the major human DNA Ligase I enzyme. To improve the chances of identifying potential inhibitors with such distinguishing properties, four models (Table 1) were used for virtual screening experiments. Two structures each, namely EfaLigA (PDB: 1TAE) and MtuLigA (PDB: 1ZAU), were used in screening for affinity for NAD+-dependent enzymes while T4 DNA ligase (a homology model) and Human DNA ligase I (PDB: 1X9N) were used for ATP-dependent enzymes.
Table 1.
In silico screening using AUTODOCK v3.0.5
| Compounds | MtuLigANAD | EfaLigA | T4Lig | HuLigI |
|---|---|---|---|---|
| NAD | −14.7 | −14.9 | — | — |
| AMP | −10.7 | −11.9 | −9.3 | −10.8 |
| Chloroquine | −10.7 | −10.8 | −9.3 | −10.6 |
| Doxorubicin | −18.6 | −19.7 | −13.7 | −17.0 |
| Pyridochromanone 3 | −16.7 | −17.7 | −13.5 | −17.7 |
| 1 | −15.8 | −16.9 | −12.4 | −12.6 |
| 2 | −11.2 | −12.2 | −11.7 | −11.0 |
| 3 | −13.8 | −11.8 | −10.7 | −12.0 |
| 4 | −14.3 | −15.2 | −13.0 | −14.9 |
| 5 | −11.2 | −12.1 | −13.3 | −14.4 |
The docking energies are in kcal/mol. MtuLigANAD, EfaLigA, T4Lig and HuLigI refer to the respective ligases from M.tuberculosis, E.faecalis, T4 bacteriophage and human sources mentioned in the text.
Database used and in silico screening
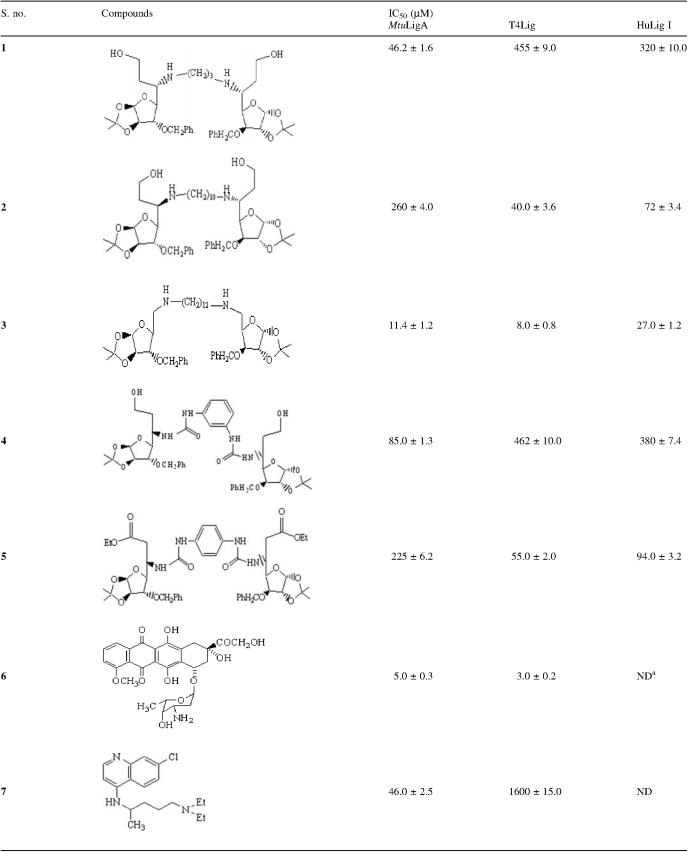
The top 10% of the docked complexes were sorted based on the scoring function and fitness scores as implemented in the AUTODOCK v3.0.5 program. Control docking experiments were able to reproduce the AMP/NAD+ complexes in their crystal structures with respective enzymes (13) and were used to optimize the docking parameters. The docking energy of chloroquine was much less than NAD+ in line with earlier reports (16) that chloroquine does not interact with the co-factor binding site in LigA (Table 1). The other selected compounds had comparable predicted binding affinities to NAD+. We then focused our efforts on the glycosylamines for further analysis as earlier reports had suggested that certain compounds exhibited anti-tubercular activity (20) although their mode of action is unknown. The selected glycofuranosylated diamines could be divided further into two subclasses (Table 2), namely bis xylofuranosylated diamines with aminoalkyl spacers (compounds 1, 2 and 3) and those with phenylene carbamoyl-based spacers (compounds 4 and 5, respectively).
Table 2.
In vitro inhibition of M.tuberculosis NAD+-dependent DNA ligase, T4 DNA ligase and human DNA ligase I by the respective compounds
aND refers to ‘not determined’.
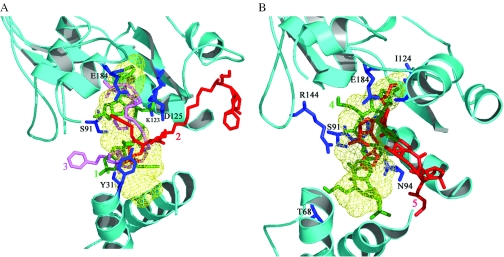
An analysis of the AUTODOCK predicted docking modes of the compounds suggests that they interact with several essential residues in the NAD+ binding site (Figure 2). Among compounds with aminoalkyl spacers (1, 2 and 3), 1 and 3 have electrostatic interactions with K123 and E184. They also interact with essential residues, such as Y31 and D45, from subdomain 1a which binds to the NMN moiety of NAD+ (Figure 2A). Compound 2, however, exhibits polar interactions only with Y31, D41 and D45 in the NAD+ binding site although it has van der Waals contacts with E184 also. These are in addition to other interactions with conserved residues not directly interacting with bound NAD+.
Figure 2.
(A) Interactions of compounds 1, 2 and 3 (those with aminoalkyl spacers) in the NAD+ binding site of the enzyme from M.tuberculosis. The compounds are shown in green, red and pink respectively while the NAD+ binding region is depicted as a light green wire mesh. Key interacting residues are shown as blue sticks and labeled for clarity. (B) Interactions of compounds 4 and 5 (those with phenylene carbamoyl spacers) in the NAD+ binding site of the M.tuberculosis enzyme. The compounds are depicted in green and red, respectively. The color scheme is similar to (A).
Among the compounds with phenylene carbamoyl spacers, namely compounds 4 and 5, respectively, the former interacts more extensively with residues directly and indirectly involved in NAD+ recognition. The interactions include those with the essential E184 and D45. Compound 5 on the other hand has less extensive interactions with residues in the NAD+ binding site, although it also interacts with E184 and D45 (Figure 2B). These compounds were then synthesized and assayed as described.
Synthesis
The identified compounds were synthesized as per our earlier reported protocols (20,27–30). Schematic representations of the synthesis of glycosylamines with aminoalkyl and phenylene carbamoyl spacers are shown in Schemes 1 and 2, respectively. The purity and integrity of the compounds were verified according to the standard procedures (data not shown).
In vitro DNA ligase inhibition assays
We assayed for the in vitro inhibitory potency of the compounds against the full-length NAD+-dependent enzyme from M.tuberculosis, the major human DNA ligase I and bacteriophage T4 DNA ligase, respectively. The latter two, as mentioned before, are ATP-dependent enzymes and we were interested in compounds which could potentially distinguish between NAD+- and ATP-dependent ligases. Chloroquine and doxorubicin (compounds 6 and 7, Table 2) were also used as control inhibitors as reported by us earlier (13). The results are summarized in Table 2.
Of the three bis xylofuranosylated diamines with 3, 10 and 12 carbon spacers, respectively, compound 1 with 3-carbon spacer was able to distinguish between the two classes of enzymes by up to 9-fold and exhibited specificity for the M.tuberculosis enzyme. Compound 3 could also distinguish between the human and M.tuberculosis enzymes by a factor of 2 and has higher affinity for the latter with IC50 values in the low micromolar range. Compound 2, on the other hand, bound to the ATP-dependent enzymes from human and bacteriophage sources with higher affinities.
Of the two compounds 4 and 5 with phenylene carbamoyl-based spacers, compound 4 with the 1,3-linker could distinguish between the human and tuberculosis enzymes exquisitely and exhibited a higher affinity for the latter. Compound 5 with the 1,4-linker had higher affinity for the ATP-dependent enzymes in the assays.
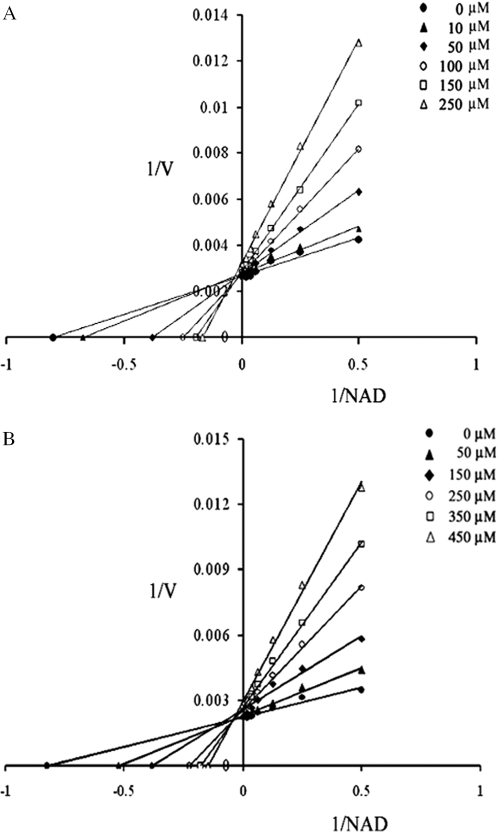
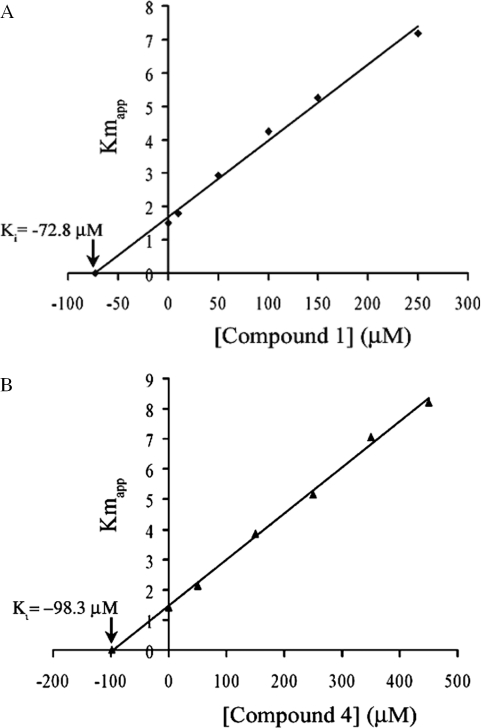
In silico docking analysis suggested an overlap of the binding sites of NAD+ and glycosylamines. We therefore chose compounds 1 and 4 to evaluate by standard kinetics if the compounds act competitively with NAD+ in the overall nick sealing reaction in vitro. In the absence of the inhibitor we determined a Km of 1.56 µM for NAD+ in the presence of 10% Me2SO in the assay mixture, which agrees well with previously reported data (8,13). In our inhibition studies, when the amount of NAD+ was increased up to 50 µM in the presence of rising concentrations of compound 1 (0–250 µM) and compound 4 (0–450 µM) under saturating DNA concentration (0.85 µM), the kinetics clearly indicated competitive inhibition of NAD+ by the compounds as also visualized in double-reciprocal plots (Figure 3). The linear regression using the apparent Km values leads to a Ki of 72.8 µM for compound 1 and 98.3 µM for compound 4 (Figure 4). These results strongly suggest that the binding sites of the glycosylamines and NAD+ overlap with each other.
Figure 3.
Competitive inhibition of MtuLigA with respect to NAD+ by the bis xylofuranosylated diamine with three carbon spacer (Compound 1) and with 1, 3-phenylene carbamoyl spacer (Compound 4). Data were fitted using standard linear regression. The double-reciprocal plots clearly indicate competitive binding between NAD+ and compounds 1 and 4. (A) Activity of MtuLigA measured in the presence of rising concentrations of compound 1 (0–250 µM) and NAD+ (0–50 µM). (B) Activity of MtuLigA measured in the presence of rising concentrations of compound 4 (0–450 µM) and NAD+ (0–50 µM).
Figure 4.
Linear regression plots of the inhibitor concentrations versus the Kmapp. The Ki values are marked with an arrow. The plots correspond to (A) compound 1 and (B) compound 4, respectively.
The in vitro inhibition assay results correlate well with the docking results and are rationalized in ‘Discussion’.
DNA binding assay
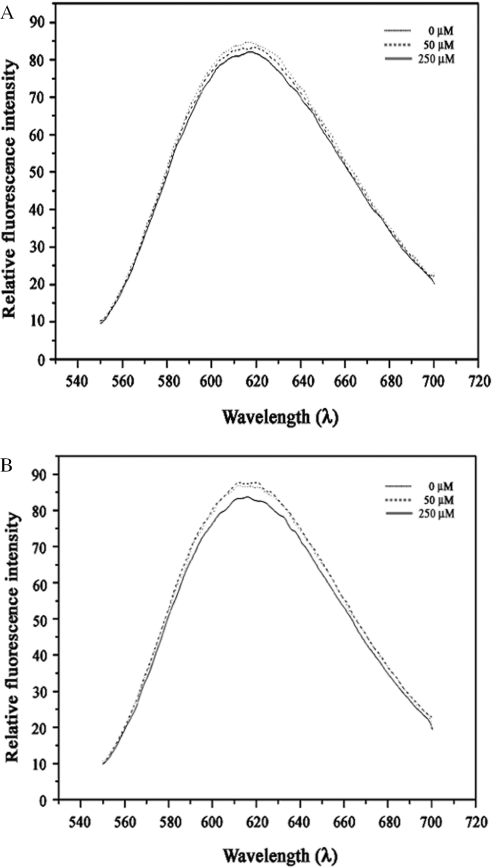
We carried out ethidium bromide displacement assays for the selected inhibitors in the current study to probe for interactions with DNA, if any. It was reported earlier that aryl amino compounds, a class of DNA ligase inhibitors, generally intercalate with DNA and this might influence their inhibitory potencies (16). Compounds were added to a maximum concentration of 250 µM which represents a 50-fold excess over ethidium bromide. We could not find any evidence for general interaction of the compounds with DNA by examining the fluorescence (Figure 5). We also carried out gel shift assays where the electrophoretic mobility of DNA was checked in the presence of increasing inhibitor concentrations. These experiments also did not support any general interaction of glycosylamines with DNA. Subsequently, the compounds were assayed to examine whether they inhibit the action of the enzyme in bacteria also and whether that would be inimical to their growth.
Figure 5.
Ethidium bromide displacement assay. Relative fluorescence intensity measured in the presence of 10 (dark dotted lines) and 50 times (continuous lines) increasing concentrations. The control (light dotted lines) is in the absence of the respective inhibitors. (A) and (B) correspond to compounds 1 and 4, respectively.
Antibacterial/in vivo assays
We used two bacterial systems to evaluate the in vivo inhibition of NAD+ ligases. In the first instance we chose the E.coli GR501 strain which harbors a temperature-sensitive lig251 mutation in its LigA. Its growth is strongly impaired at physiological temperatures while at 30°C it grows well. This defect is overcome when it is rescued by NAD+ or ATP-dependent ligases (17,34,38). This strain has been used therefore to demonstrate the in vivo specificity of inhibitors for LigA. We accordingly used pTrc99A-based systems involving MtuLigA and T4Lig (33) in this strain and also reproduced earlier results (34,38).
As a second system we used a strain of the prominent human pathogen S.typhimurium LT2 (39) where again NAD+ ligase has been shown to be essential. We also used its DNA ligase null derivative TT15151 (35) that had been rescued with T4 DNA ligase. These two systems give a handle on not only whether the compounds inhibit LigA in vivo but also allows for probing differences in efficacy against ATP-dependent ligases in similar bacterial systems.
Results of these assays are tabulated in Table 3 and results of the growth inhibition studies involving the S.typhimurium strains are shown in Figures 6 and 7. There is a low residual ligase activity in the mutant E.coli GR501 strain as compared with the corresponding growth rescued versions (growth curves not shown) because the latter possesses a high amount of expressed ligase. This leads to the much higher sensitivity of the mutant strain for the compounds (Table 3). The in vivo results are consistent with the in vitro inhibition assays. For both the tested compounds, namely 1 and 4, the MICs are less for the strain rescued by MtuLigA and higher for the T4 DNA ligase rescued version. The same trend is observed in the S.typhimurium system also where the compounds exhibit higher sensitivity for the wild-type salmonella strain (harboring its natural NAD+-dependent DNA ligase) as compared with its ligase-deficient variant rescued by T4Lig.
Table 3.
Antibacterial activity of glycosylamines
| Compounds | MIC (µg/ml) | ||||
|---|---|---|---|---|---|
| E.coli GR 501+ pTrc99A | E.coli GR 501+ Mtu NAD+ligase | E.coli GR 501+ T4 DNA ligase | S.typhimurium LT2 | S.typhimurium TT15151 | |
| 1 | 0.2 | 12 | 54 | 10.5 | 52 |
| 4 | 0.1 | 18 | 76 | 20 | 80 |
The MIC values were determined by broth microdilution for the bacterial strains E.coli GR501 and S.typhimurium LT2 (which contains its NAD+ ligase) and the DNA ligase-minus (null) derivative TT15151 (lig-2::Mu dJ/pBR313/598/8/1b [T4Lig+] AMPr) (35) rescued with T4Lig+ plasmid. E.coli GR501 ligAts is a strain containing a temperature-sensitive ligase mutant (34); a defect that is restored by the overexpression of MtuLigA (13,38) or T4Lig (34). Polymyxin B nonapeptide (20 µg/ml) was added to the growth medium to facilitate passage of the inhibitors across the outer membrane of the cell.
Figure 6.
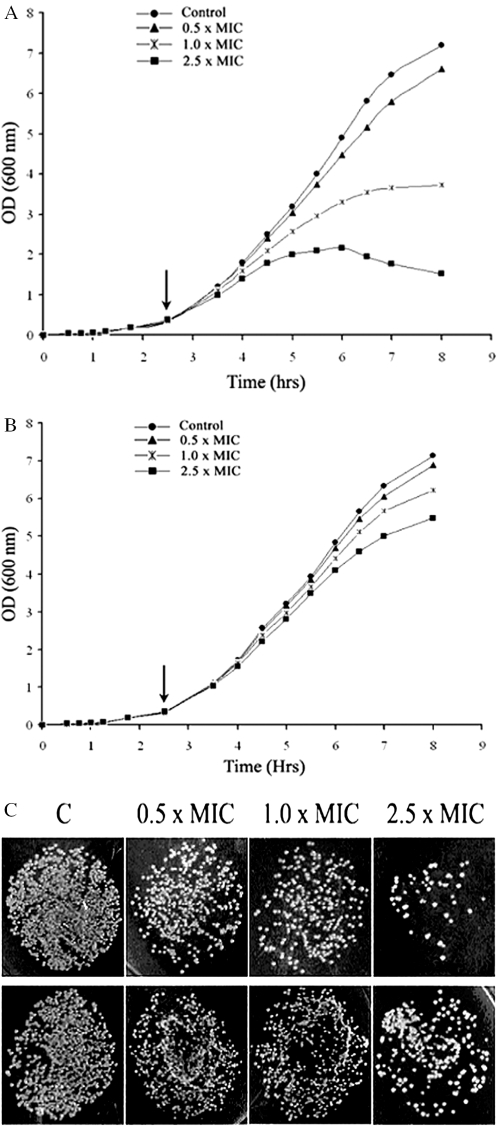
Bactericidal activity of compound 1. Effect on growth as reflected in changes of the optical density at 600 nm of (A) S.typhimurium LT2 and (B) its DNA ligase minus (null) derivative TT15151[Lig−/T4 Lig+] on their respective exposures to compound 1 at 5.25–26.25 µg/ml representing 0.5–2.5 times the MIC value. The arrow indicates the point at which the compound was added. (C) Comparative viability between S.typhimurium LT2 (top row) and its corresponding DNA ligase minus (null) derivative TT15151[Lig−/T4 Lig+] (bottom row) rescued with a plasmid containing the gene for T4 Ligase as shown by surviving CFU, 5 h after addition of compound 1 to the growth medium. The cells were plated at dilution ratios of 10−4 on nutrient agar and the indicated MIC values correspond to that of S.typhimurium LT2 against the inhibitor. The control (marked ‘C’) and the amount of added inhibitor as multiples of MIC are also indicated.
Figure 7.
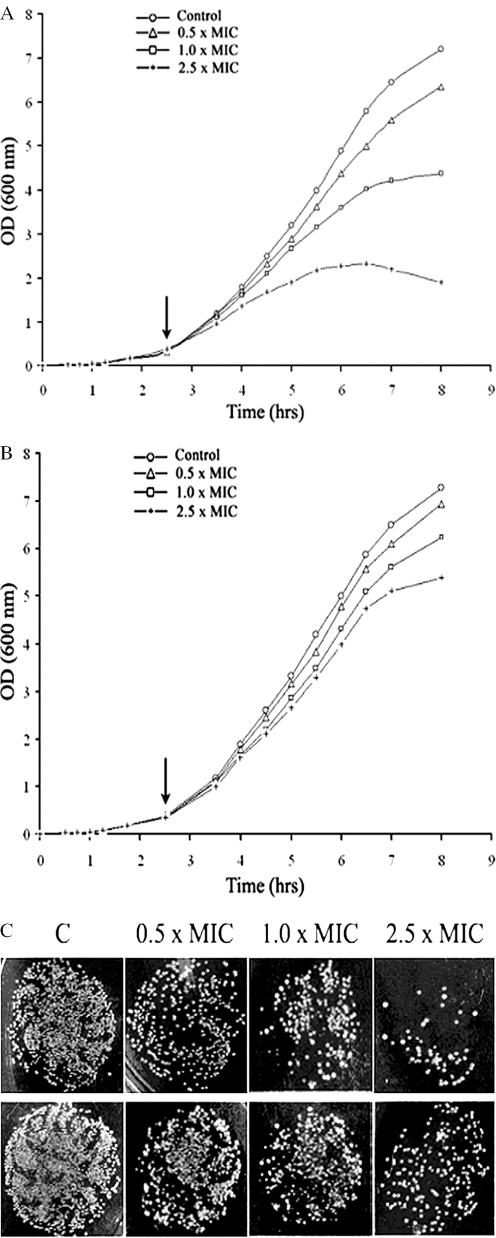
Bactericidal activity of compound 4. Effect on growth as reflected in changes of the optical density at 600 nm of (A) S.typhimurium LT2 and (B) its corresponding DNA ligase minus (null) derivative TT15151[Lig−/T4 Lig+] on their respective exposures to compound 4 at 10–50 µg/ml representing 0.5–2.5 times the MIC value. The arrow indicates the point at which the compound was added. (C) Comparative viability between S.typhimurium LT2 (top row) and its corresponding DNA ligase minus (null) derivative TT15151[Lig−/T4 Lig+](bottom row) rescued with a plasmid containing the gene for T4 Ligase as shown by surviving CFU, 5 h after addition of compound 4 to the growth medium. The cells were plated at dilution ratios of 10−4 on nutrient agar and the indicated MIC values correspond to that of S.typhimurium LT2 against the inhibitor. The control (marked ‘C’) and the amount of added inhibitor as multiples of MIC are also indicated.
Correspondingly, growth inhibition studies performed using the compounds are in line with the above results. Increasing the inhibitor concentration leads to more bactericidal activity against the S.typhimurium LT2 strain compared with its ligase null derivative (rescued by T4Lig) (Figure 6A and B and Figure 7A and B) at the same compound concentration.
Cell viability which was tested using the compounds again show that the wild-type S.typhimurium LT2 strain is less viable at same compound concentrations compared with the viability of the ligase-deficient variant rescued with T4Lig (Figures 6C and 7C).
The in vivo assay results demonstrate that compounds 1 and 4 have higher specificity for NAD+-dependent ligases and strongly suggest that the observed antibacterial activities are due to in vivo inhibition of LigA.
DISCUSSION
One of the main differences in the sequences of NAD+- and ATP-dependent ligases in the co-factor binding domain is that several residues lining the NAD+ binding site are conserved across species while they are not in ATP-dependent ligases. Subdomain 1a occurs only in LigA and is absent in ATP-dependent ligases. Such differences can be potentially exploited in designing LigA-specific inhibitors. It is instructive in this context to examine the docking modes of the inhibitors and compare them with the in vitro inhibition assay results against NAD+- and ATP-dependent ligases. Among the compounds 1, 2 and 3 (those with aminoalkyl spacers, Table 2), compound 2 has higher affinity for human DNA ligase I. On the other hand, compounds 1 and 3 can distinguish between MtuLigA and the human enzyme and are more specific for the former. These results correlate well with the observed docking modes of compounds 1 and 3, respectively (Figure 2A). Both compounds seem to mimic the binding modes of NAD+ and have more interactions within the binding site of the co-factor. This seems to be the basis for their ability to distinguish between MtuLigA and the human enzyme. Compound 2, on the other hand, has more interactions outside of the NAD+ binding site. This supposedly decreases its specificity for the NAD+-dependent ligase.
The same analogy extends to the cases of compounds 4 and 5 (those with phenylene carbamoyl spacers). Compound 4 which exhibits higher specificity for the M.tuberculosis enzyme (Figure 2B and Table 2) has more extensive contacts within the NAD+ binding site compared with compound 5. The latter compound has more interactions outside the NAD+ binding site. The in vitro assays are in agreement and show that compound 5 has less affinity for MtuLigA. The present results may therefore be used as a guiding rule in selecting inhibitors with the ability to distinguish between NAD+- and ATP-dependent DNA ligases.
The compounds described here are able to discriminate between the two classes of DNA ligases. The in vivo results are in good consonance with the in vitro assay results and demonstrate that the inhibitors have higher affinity for NAD+-dependent ligases in vivo also. We used two different LigA-deficient strains for the in vivo assays. While the E.coli-based strain was rescued with LigA from M.tuberculosis, the other strain was a S.typhimurium one containing its own LigA. The efficacy of compounds 1 and 4 against both strains point to their potential development as broad antibacterials also. This is also the case with pyridochromanones, another class of LigA-specific inhibitors, which have also been found to inhibit LigA from at least E.coli and M.tuberculosis (17,8).
The similar binding site architecture in NAD+- and ATP-dependent ligases makes it difficult to identify inhibitors capable of distinguishing between them. However, this is an important and necessary step for exploiting the enzyme's potential as a drug target. Several sequence motifs lining the co-factor binding site are conserved across bacteria (40), and it is therefore naturally expected that inhibitors of LigA will act as general antibacterials also. A further challenge, however, is to tweak the specificity such that the inhibitors are more potent against specific pathogens, such as M.tuberculosis. In the present work, we have identified glycofuranosylated diamine-based inhibitors which are more specific for LigA and which were known from earlier work to possess some anti-tubercular activity [(20); our unpublished data]. The in vitro assays against MtuLigA and in vivo experiments with the LigA-deficient strains reported here suggest that inhibition of MtuLigA is the basis of the observed anti-tubercular activity of the compounds although an alternate mechanism in the pathogen cannot be ruled out. These compounds also do not appear to exhibit any unwanted general interactions with DNA. Indeed, ongoing experiments for the next generation of more potent anti-tubercular compounds include structural studies on the enzyme–inhibitor complexes to support our docking and optimization strategies.
Acknowledgments
S.typhimurium LT2 wild-type strain and its DNA ligase-minus (null) derivative containing T4 Lig+ plasmid pBR313/598/8/1b TT15151 were kindly provided by Dr J. R. Roth (University of Utah, Utah). pTrc99A, E.coli GR501 ligAts mutant and T4-pTrc99A (pRBL) construct were kind gifts from Dr Richard Bowater (University of East Anglia, Norwich, UK). Full-length human DNA ligase I construct was kindly gifted by Dr Deborah Barnes (Clare Hall laboratories, Herfordshire, UK). S.K.S., D.D., N.T. and N.D. acknowledge fellowships from Council of Scientific and Industrial Research (CSIR), India. The work was funded by CSIR network grants CMM0017 and SMM003. This is communication number 6784 from CDRI. Funding to pay the Open Access publication charges for this article was provided by CSIR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilkinson A., Day J., Bowater R. Bacterial DNA ligases. Mol. Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- 2.Sriskanda V., Moyer R.W., Shuman S. NAD+-dependent DNA ligase encoded by a eukaryotic virus. J. Biol. Chem. 2001;276:36100–36109. doi: 10.1074/jbc.M105643200. [DOI] [PubMed] [Google Scholar]
- 3.Lu J., Tong J., Feng H., Huang J., Afonso C.L., Rock D.L., Barany F., Cao W. Unique ligation properties of eukaryotic NAD+-dependent DNA ligase from Melanoplus sanguinipes entomopoxvirus. Biochimica et Biophysica Acta. 2004;1701:37–48. doi: 10.1016/j.bbapap.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Dermody J.J., Robinson G.T., Sternglanz R. Conditional-lethal deoxyribonucleic acid ligase mutant of Escherichia coli. J. Bacteriol. 1979;139:701–704. doi: 10.1128/jb.139.2.701-704.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaczmarek F.S., Zaniewski R.P., Gootz T.D., Danley D.E., Mansour M.N., Griffor M., Kamath A.V., Cronan M., Mueller J., Sun D., et al. Cloning and functional characterization of an NAD+-dependent DNA ligase from Staphylococcus aureus. J. Bacteriol. 2001;183:3016–3024. doi: 10.1128/JB.183.10.3016-3024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petit M.A., Ehrlich S.D. The NAD+-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis. Nucleic Acids Res. 2000;28:4642–4648. doi: 10.1093/nar/28.23.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 8.Gong C., Martins A., Bongiorno P., Glickman M., Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 9.Pascal J.M., O'Brien P.J., Tomkinson A.E., Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 10.Singleton M.R., Hakansson K., Timson D.J., Wigley D.B. Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure. 1999;7:35–42. doi: 10.1016/s0969-2126(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 11.Gajiwala K.C., Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004;12:1449–1459. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.Y., Chang C., Song H.K., Moon J., Yang J.K., Kim H.-K., Kwon S.T., Suh S.W. Crystal structure of NAD+-dependent DNA ligase: modular architecture and functional implications. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava S.K., Tripathi R.P., Ramachandran R. NAD+-dependent ligase (Rv3014c) from M.tuberculosis H37Rv: crystal structure of the adenylation domain and identification of novel inhibitors. J. Biol. Chem. 2005;280:30273–30281. doi: 10.1074/jbc.M503780200. [DOI] [PubMed] [Google Scholar]
- 14.Shuman S., Schwer B. RNA capping enzyme and DNA ligase—a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhu H., Shuman S. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli NAD+-dependent DNA ligase (LigA) J. Biol. Chem. 2005;280:12137–12144. doi: 10.1074/jbc.M413685200. [DOI] [PubMed] [Google Scholar]
- 16.Ciarrocchi G., MacPhee D.G., Deady L.W., Tilley L. Specific inhibition of the eubacterial DNA ligase by arylamino compounds. Antimicrob. Agents Chemother. 1999;43:2766–2772. doi: 10.1128/aac.43.11.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brőtz-Oesterhelt H., Knezevic I., Bartel S., Lampe T., Warnecke-Eberz U., Ziegelbauer K., Habich D., Labischiinski H. Specific and potent inhibition of NAD+-dependent DNA ligase by pyridochromanones. J. Biol. Chem. 2003;278:39435–39442. doi: 10.1074/jbc.M306479200. [DOI] [PubMed] [Google Scholar]
- 18.Sander P., Rezwan M., Walker B., Rampini S.K., Kroppenstedt R.M., Ehlers S., Keller C., Keeble J.R., Hagemeier M., Colston M.J., et al. Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2004;52:1543–1552. doi: 10.1111/j.1365-2958.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 19.Duncan K. The impact of genomics on the search for novel tuberculosis drugs. Novartis Found. Symp. 1998;217:228–237. doi: 10.1002/0470846526.ch17. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi R.P., Tiwari V.K., Tewari N., Katiyar D., Saxena N., Sinha S., Gaikwad A., Srivastava A., Chaturvedi V., Manju Y.K., et al. Synthesis and antitubercular activities of bis-glycosylated diamino alcohols. Bioorg. Med. Chem. 2005;13:5668–5679. doi: 10.1016/j.bmc.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 22.Marti-Renom M.A., Stuart A., Fiser A., Sánchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 23.Subramanya H.S., Doherty A.J., Ashford S.R., Wigley D.B. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 24.ACCELRYS ver. 11. San Diego, CA: Accelrys Inc.; 2000. pp. 92121–92152. [Google Scholar]
- 25.Laskowski R.A., Mac Arthur M.W., Moss D.S., Thornton J.M. A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;6:283–291. [Google Scholar]
- 26.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 27.Tiwari V.K., Tewari N., Katiyar D., Tripathi R.P., Arora K., Gupta S., Ahmad R., Srivastava A.K., Khan M.A., Murthy P.K., et al. Synthesis and antifilarial evaluation of N1,Nn- xylofuranosylated diaminoalkanes. Bioorg. Med. Chem. 2003;11:1789–1800. doi: 10.1016/s0968-0896(03)00033-6. [DOI] [PubMed] [Google Scholar]
- 28.Tripathi R.P., Tripathi R., Tiwari V.K., Bala L., Sinha S., Srivastava A., Srivastava R., Srivastava B.S. Synthesis of glycosylated β-amino acids as new class of anti tubercular agents. Eur. J. Med. Chem. 2002;37:773–781. doi: 10.1016/s0223-5234(02)01398-3. [DOI] [PubMed] [Google Scholar]
- 29.Tewari N., Ramesh, Mishra R.C., Tripathi R.P., Srivastava V.M.L., Gupta S. Leishmanicidal activity of phenylene bridged C2 symmetric glycosyl ureides. Bioorg. Med. Chem. Lett. 2004;14:4055–4059. doi: 10.1016/j.bmcl.2004.05.035. [DOI] [PubMed] [Google Scholar]
- 30.Tewari N., Mishra R.C., Tiwari V.K., Tripathi R.P. DBU catalysed cyclatic amidation reaction: a convenient synthesis of C-nucleoside analogs. Synlett. 2002;11:1779–1782. [Google Scholar]
- 31.Mackenney V.J., Barnes D.E., Lindahl T. Specific function of DNA ligase I in simian virus 40DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J. Biol. Chem. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 32.Le Pecq J.-B., Paoletti C.A. Fluorescent complex between ethidium bromide and nucleic acids. J. Mol. Biol. 1967;27:87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 33.Ren Z.J., Baumann R.G., Black L.W. Cloning of linear DNAs in vivo by overexpressed T4 DNA ligase: construction of a T4 phage hoc gene display vector. Gene. 1997;195:303–311. doi: 10.1016/s0378-1119(97)00186-8. [DOI] [PubMed] [Google Scholar]
- 34.Lavesa-Curto L., Sayer H., Bullard D., MacDonald A., Wilkinson A., Smith A., Bowater L., Hemmings A., Bowater R.P. Characterization of a temperature-sensitive DNA ligase from Escherichia coli. Microbiology. 2004;150:4171–4180. doi: 10.1099/mic.0.27287-0. [DOI] [PubMed] [Google Scholar]
- 35.Wilson G.G., Murray N.E. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterization of the recombinants. J. Mol. Biol. 1979;132:471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- 36.Sriskanda V., Shuman S. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+ J. Biol. Chem. 2002;277:9685–9700. doi: 10.1074/jbc.M111164200. [DOI] [PubMed] [Google Scholar]
- 37.Sriskanda V., Schwer B., Ho C.K., Shuman S. Mutational analysis of E.coli DNA ligase identifies amino acids required for nick-ligation in vitro and for in vivo complementation of the growth of yeast cells deleted for CDC9 and LIG4. Nucleic Acids Res. 1999;27:3953–3963. doi: 10.1093/nar/27.20.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson A., Sayer H., Bullard D., Smith A., Day J., Kieser T., Bowater R. NAD+-dependent DNA ligases of Mycobacterium tuberculosis and Streptomyces coelicolor. Proteins. 2003;51:321–326. doi: 10.1002/prot.10361. [DOI] [PubMed] [Google Scholar]
- 39.Park U.E., Olivera B.M., Hughes K.T., Roth J.R., Hillyard D.R. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J. Bacteriol. 1989;171:2173–2180. doi: 10.1128/jb.171.4.2173-2180.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 41.DeLano W.L. The PyMOL molecular graphics system. 2002.