Abstract
Recently, a histone H3 variant in Drosophila and humans, the H3.3 protein, was shown to replace canonical H3 in active chromatin in a replication-independent (RI) manner. In the fission yeast Schizosaccharomyces pombe, there exists a single form of H3, which is equivalent to H3.3 and is thought to participate in both replication-independent (RI) and replication-coupled (RC) nucleosome assembly. In this study, we show that RI deposition of H3 at heterochromatic regions is consistently lower than that at a gene-free euchromatic region, and deletion of the conserved heterochromatin-specific proteins Swi6 or Clr4 markedly increases RI deposition at heterochromatic regions such as the silent mating-type loci or centromeres. These results clearly show that RI deposition of H3 occurs preferentially in euchromatic regions. We also observed that RI deposition of H3 could be increased at the thi3+ gene when transcription is induced, indicating transcription further facilitates RI deposition of H3. Taken together, these observations demonstrate that selective deposition of histone H3.3 at transcriptionally active chromatin by the RI assembly pathway is conserved in fission yeast and, thus, our data support an essential role of histone H3 replacement in maintaining active chromatin among diverse eukaryotic organisms ranging from fission yeast to humans.
INTRODUCTION
Nucleosomes, which are composed of 146 bp of DNA wrapped around an octamer of four core histone proteins, are fundamental elements of chromatin (1). Deposition of histone proteins primarily occurs during DNA replication to complete chromatin duplication (2). However, several studies indicate that histone proteins can be deposited outside of S phase in a replication-independent (RI) manner (3–5). One of the best-characterized histones is the H3.3 protein, which is an H3 variant, found in Drosophila and humans that is constitutively expressed throughout the cell cycle (5–8).
H3.3 differs from the canonical H3 by only four amino acids but uses a completely different histone chaperone than H3 (HIRA for H3.3 and CAF-I for H3), indicating that cells have a specialized pathway to assemble H3.3-containing chromatin (9). In Drosophila KC cells, H3.3 is enriched at transcriptionally active ribosomal gene arrays on metaphase chromosomes, and in human U2OS cells, H3.3 is enriched at artificially constructed transgenes after transcriptional activation (5,6). These results strongly suggest that transcription may be a critical determinant for H3.3 deposition. However, both active ribosomal gene arrays in Drosophila KC cells and activated transgene arrays in human U2OS cells undergo alterations in their chromatin structures during transcriptional activation, and it is therefore not possible to determine from these data whether transcription itself or the alterations in chromatin formation associated with transcription is the basis for RI deposition of H3.3 at active loci.
In the fission yeast, Schizosaccharomyces pombe, histone H3 exists as a single subtype, which is the equivalent of H3.3 in higher eukaryotes, suggesting that H3.3 is an essential component of eukaryotic systems. Therefore, it is likely that the fission yeast H3 may be involved in both bulk chromatin assembly and RI assembly (10).
In this study, to gain further insight into the mechanism of RI deposition of H3.3, we developed an assay to monitor RI deposition of histone H3 and H4 in fission yeast using a chromatin immunoprecipitation (ChIP) assay. Using this assay, we show that RI deposition of fission yeast H3 occurs preferentially at euchromatic regions. RI deposition of H3 in heterochromatic regions was significantly lower than deposition at a non-transcribed euchromatic locus. Moreover, RI deposition of H3 at the heterochromatic mating-type region was elevated by deletion of the heterochromatin-specific proteins Swi6 or Clr4, indicating that heterochromatin formation can block RI deposition of H3. In addition, we observed that RI deposition of H3 can be enhanced by transcriptional activation at promoter and coding regions of an inducible gene, thi3+, indicating the importance of transcription in RI deposition. Thus, we conclude that RI deposition of H3.3 is linked to transcriptionally active chromatin in a wide range of eukaryotic organisms.
MATERIALS AND METHODS
Plasmids
pINV1-H3-HA or pINV1-H4-HA was generated by PCR cloning of hht1+ or hhf1+ into the NdeI–NotI sites of the pINV1-spc1+-HA6his vector [(11) kindly provided by Dr. P. Russell]. The correct in-frame fusion and absence of PCR errors were confirmed by DNA sequencing (data not shown).
Cell culture and medium
Culture of fission yeast cells followed standard protocols (12). Glucose medium for repressing the INV1 promoter was made by adding 8% glucose to standard EMM medium (11). Sucrose medium for inducing the invertase promoter INV1 was glucose depleted EMM medium to which 4% sucrose was added (11).
Block of DNA replication by HU treatment
Cells transformed with pIVN1-H3-HA or pINV1-H4-HA were grown in 8% glucose medium at 30°C to a density of 2.5 × 106 cells/ml. The cultures were treated with 25 mM hydroxyurea (HU) for 4 h at 30°C to block DNA replication. The cultures were then washed with distilled water containing 25 mM HU and split into two aliquots. One aliquot was transferred to 4% sucrose medium with 25 mM HU and grown at 30°C for 2 h. The other aliquot was grown in 8% glucose medium with 25 mM HU at 30°C for 2 h.
Sample preparation for FACS analysis
For flow cytometry, cells were fixed in 70% ethanol at 4°C. FACS analysis was performed by staining the fixed cells with propidium iodide. Samples were briefly sonicated before analysis. FACS analysis was performed according to the manufacturer's instructions (BD FACS Calibur).
ChIP assay
ChIP assays were performed as described previously (13). For cross-linking, 200 ml of cells at a density of 5 × 106 cells/ml were treated with 1% formaldehyde for 20 min at room temperature. After purification of the chromatin fraction by ultracentrifugation, chromatin was fragmented to an average of 500 bp in size by sonication. The resulting extracts were incubated with an antibody to hemagglutinin (HA) (12CA5, Roche), an RNA polymerase II antibody (CTD4H8, upstate) or an H3 antibody (AB1791, Abcam) and collected with protein A (for 12CA5) or protein G (CTD4H8) Sepharose. Then, DNA was purified from the immunoprecipitates and amplified by PCR. For PCR, 2 µl of the DNA recovered by ChIP and 2 µl of a 1/100 dilution of the whole cell extract (WCE) were used as the template and subjected to 27 amplification cycles that were in the linear range. For amplification of the reference region (a gene free euchromatic region), two primers, gfr2-upper, 5′ CCC AAC ATC CAA AAA TGA GG 3′ and gfr2-lower, 5′ AAA CCG TAA AGC GTC AAA CG 3′, were used. The reference region corresponds to the nucleotides 13023–13188 of the cosmid SPBC409 which is located in the intergenic sequence between the wis1 gene and the open reading frame of SPBC409.08.
RESULTS
Uncoupling H3 or H4 expression from DNA replication in fission yeast
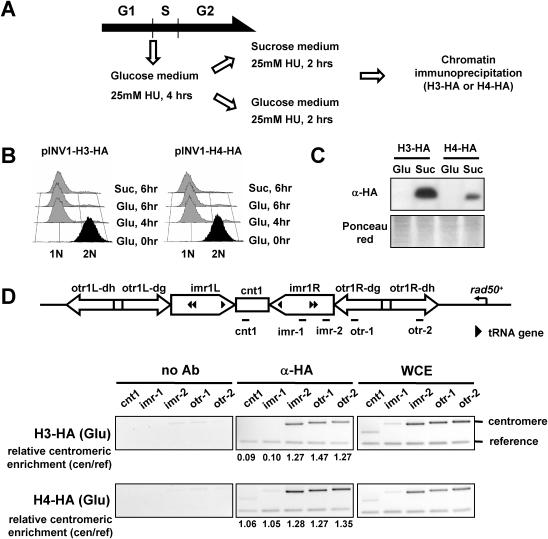
The fission yeast S.pombe has only one subtype of histone H3 which is equivalent to the histone H3 variant, H3.3, in animals (10). Thus, it is expected that fission yeast H3 should be involved in both bulk chromatin assembly and RI assembly. To specifically monitor RI deposition of fission yeast H3 or H4 in vivo, we generated a vector expressing H3 or H4 whose C-terminus is tagged with HA under the control of an invertase promoter (INV1). The invertase promoter is repressed when cells are grown in glucose medium but can be rapidly (within 2 h) induced when cells are grown in sucrose medium (11). To uncouple the expression of HA-tagged histones from DNA replication, cells transformed with pINV1-H3-HA or pINV1-H4-HA and grown in glucose medium were treated with 25 mM hydroxyurea (HU), which permanently blocks DNA replication in fission yeast (14), for 4 h (Figure 1A). The cultures were then split into two aliquots, one of which was transferred to sucrose medium to induce H3-HA or H4-HA. The other aliquot was continuously grown in glucose medium as a control (see below). After medium exchange, each aliquot was incubated for an additional 2 h in the presence of 25 mM HU to induce histone expression. Each sample was then cross-linked with formaldehyde and subjected to ChIP with an HA antibody to monitor the incorporation of H3-HA or H4-HA into chromatin regions.
Figure 1.
Exogenously expressed H3-HA and H4-HA are functional, as determined by their incorporation into centromeric regions. (A) Experimental strategy for incorporation of exogenous H3-HA or H4-HA in fission yeast (see text for details). (B) FACS analysis to monitor DNA content of cells treated with 25 mM HU at the indicated time points. Cells were fixed in 70% ethanol and analyzed by flow cytometry. (C) Induction of exogenous H3-HA or H4-HA in sucrose medium was analyzed by western blot with an HA antibody. Staining of the membrane with Ponceau red solution shows equal loading. (D) A diagram of centromere 1 and locations of primer pairs are shown on top. Primer pairs designated cnt1 and imr-1 are located within the central core region. Primer pairs designated imr-2, otr-1 and otr-2 are located within centromeric heterochromatin. Cells containing pINV1-H3-HA or pINV1-H4-HA grown in glucose medium with 25 mM HU for 6 h were subjected to ChIP with protein A Sepharose only (no Ab, control) or protein A Sepharose and HA antibody (α-HA). PCR with the designated primers was performed from the no Ab sample, α-HA sample, or WCE to detect incorporation of basally expressed H3-HA or H4-HA grown in glucose medium. The reference region is a gene-free euchromatic region of chromosome 2 (13023–13188). Relative centromeric enrichment shown beneath each lane was calculated by dividing the ratio of the centromeric loci/reference PCR products in the ChIP sample with that in the WCE sample. The experiments above were repeated at least twice, to ensure reproducibility.
We confirmed the HU-induced cell cycle arrest by flow cytometry (Figure 1B). S phase arrest was attained after 4 h incubation with HU and was maintained at 6 h, by when histone genes are induced. H3-HA or H4-HA protein induction in sucrose medium was monitored by western blot analysis with an HA antibody (Figure 1C). Prolonged exposure showed that there are weak signals under glucose-repressed conditions, due to leaky expression of H3-HA or H4-HA grown in glucose medium (data not shown). We estimated that the HA-tagged H3 or H4 represent <10% of the total H3 or H4 in the cell by western blot analysis with an H3 or H4 antibody (data not shown). Before investigating RI deposition of H3-HA or H4-HA, we tested whether the epitope-tagged H3 and H4 are functional and whether cells containing the tagged histones can grow normally. We confirmed that the induction of tagged histones in sucrose medium does not affect cell growth or viability (data not shown) and we then monitored the chromatin incorporation properties of basally expressed H3-HA and H4-HA under glucose-repressed conditions. We used the centromeric region as an indicator since the central core region (cnt1 and inner parts of imr1 delimited by tandem tRNA genes, Figure 1D) is occupied by the centromere-specific H3 variant Cnp1 instead of canonical H3 (15). Because histone H4 can pair with both H3 and Cnp1, it is expected that H4 can be incorporated into the centromeric regions. When we monitored the incorporation of basally expressed histones, we found that H3-HA was preferentially incorporated outside of the central core region (imr-2, otr-1 and otr-2) while H4-HA was incorporated into all the centromeric regions. By this criterion, we concluded that the tagged H3 and H4 could functionally interact with chromatin as effectively as their wild-type counterparts.
RI deposition of H3 or H4 is correlated with chromatin structure
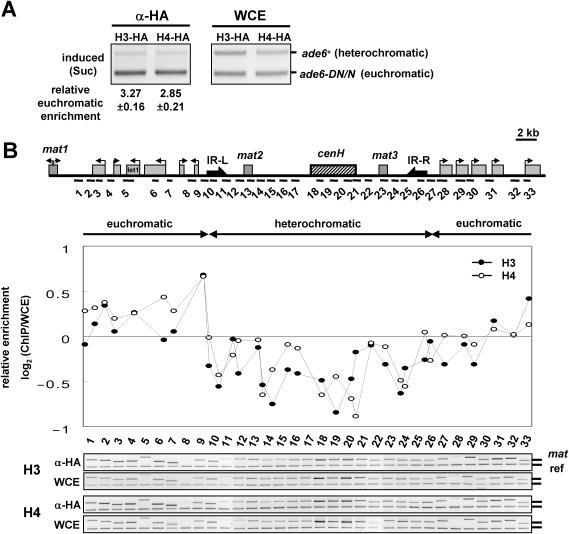
Next, we asked whether RI deposition of H3 or H4 occurs at heterochromatic regions or euchromatic regions. For this experiment, we used a strain containing an ade6+ reporter gene inserted near the heterochromatic mat3 locus and a truncated minigene, ade6-DN/N, at the endogenous euchromatic locus. By using a primer pair that can simultaneously amplify both alleles, we compared RI deposition of H3 and H4 between these alleles in anti-HA-ChIPed samples. When we monitored RI deposition of H3 or H4 in sucrose-induced samples, a relative enrichment of ∼3-fold (3.27 for H3-HA and 2.85 for H4-HA) at the euchromatic reporter was observed, showing that RI deposition is more active at euchromatic sites (Figure 2A). To address whether heterochromatin indeed suppresses RI deposition of H3, we monitored RI deposition of H3 or H4 at 33 loci across the silent mating-type loci in fission yeast (Figure 2B). The fission yeast silent mating-type loci form a heterochromatin-like structure whose boundaries are delimited by insulator elements called IR-L and IR-R (16). RI deposition of H3 at regions outside of these boundaries, which are euchromatic, was generally equivalent to levels of deposition at the reference region. In contrast, RI deposition of H3 at regions inside the boundaries, which are heterochromatic, was consistently lower than the reference region. The RI deposition pattern of H4 is essentially the same as that of H3, indicating their pairwise incorporation into chromatin. The reference region is a euchromatic but gene-free region located on chromosome 2 which should not be transcribed and we did not detect any RNA polymerase II localization at this locus (data not shown). However, we cannot exclude the possibility that some undetectable transcriptional activity may allow RI deposition of H3 at the reference region to occur more efficiently than at heterochromatic regions. Alternatively, repressive chromatin structure itself may somehow inhibit RI deposition of H3 in heterochromatic regions.
Figure 2.
RI deposition of H3 and H4 occurs predominantly at euchromatic regions. (A) Deposition of H3 or H4 at the reporters ade6+, located in the heterochromatic mat3 locus, and ade6-DN/N, located in an endogenous euchromatic locus, in S.pombe grown in sucrose medium. The relative euchromatic enrichment shown beneath each lane was calculated by dividing the ratio of the ade6-DN/N/ade6+ PCR products in the ChIP sample with that in the WCE sample and the data are presented as the mean of triplicate measurements ± SD. (B) A diagram of the mat locus and the locations of primers used are shown at the top. IR-L and IR-R represent the left and right heterochromatin boundaries, respectively. ChIP analysis with an HA antibody was performed using cells expressing H3-HA or H4-HA in sucrose medium to measure their incorporation throughout the mat locus (1–33). Relative enrichment of H3-HA or H4-HA at each mat locus was calculated as the ratio of the PCR products in the ChIP sample to that in the WCE sample for each site (1–33). The ratio is plotted as a log scale and aligned with the map of the mat locus. The experiments above were repeated at least twice, to ensure reproducibility.
Heterochromatin structure blocks RI deposition of H3
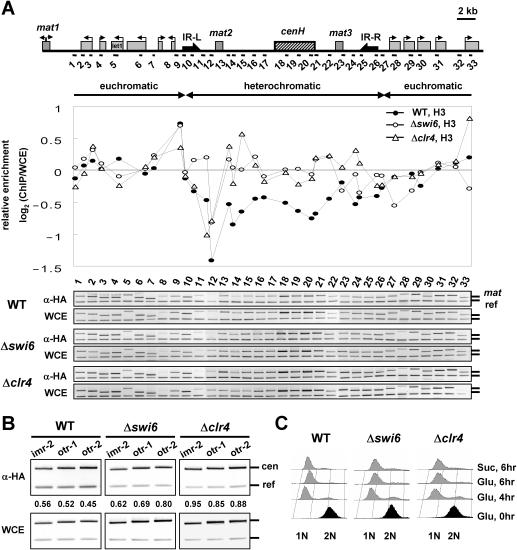
To confirm whether heterochromatin structure is required to block RI assembly at the silent mating-type loci, we monitored RI deposition of H3 in cells lacking Swi6 or Clr4, conserved heterochromatin proteins equivalent to HP1 or Suv39h, respectively (17,18). We found that deletion of Swi6 markedly increases RI deposition of H3 at silent mating-type loci while not affecting the RI deposition at euchromatic regions (Figure 3A). Similar results were observed in cells lacking Clr4, indicating that the heterochromatin structure formed by Swi6 and Clr4 blocks RI deposition of H3 at silent mating-type loci. Silent mating-type loci contain genes or transcribable elements including mat2, mat3 and cenH where siRNA precursors are transcribed (19,20) and their transcription is subject to chromatin structure modifications. Thus, these experiments cannot distinguish whether alterations in chromatin structure or in transcriptional activity are responsible for the increased RI deposition of H3 in the absence of Swi6 or Clr4. Next, to confirm the inhibitory role of heterochromatin on RI assembly, we monitored RI deposition of H3 at centromeric heterochromatin. Consistent with the silent mat locus data, we observed that disruption of heterochromatin structure by deletion of swi6+ or clr4+ increases RI deposition of H3 at centromeric heterochromatin (Figure 3B). We also noted a greater increase in RI deposition of H3 at this region by deletion of clr4+ than by deletion of swi6+. Previous data have shown that deletion of clr4+ weakens centromeric heterochromatin more profoundly than deletion of swi6+, unlike the silent mating-type loci where deletion of either gene displays similar effects (21). Thus, the greater effect of Δclr4 on RI deposition at centromeric heterochromatin may be due to the greater disruption of heterochromatin structure or more robust transcription in Δclr4 cells than in Δswi6 cells. We confirmed that the cell cycle arrest by HU was not affected by deletion of swi6+ or clr4+ by flow cytometry (Figure 3C).
Figure 3.
RI deposition of H3 at silent mat loci is increased by deletion of swi6+ or clr4+. (A) ChIP analysis with an HA antibody was performed with wild-type (WT), Δswi6, or Δclr4 cells expressing H3-HA in sucrose medium to measure its incorporation throughout the mat locus (sites 1–33). Relative enrichment of H3-HA in each strain was calculated and plotted as a log scale as described in Figure 2C. (B) The same ChIP samples used in (A) were subjected to PCR with primer pairs corresponding to centromeric heterochromatin (imr-2, otr-1 and otr-2), as shown in Figure 1C. (C) FACS analysis was performed to monitor DNA content of WT, Δswi6, or Δclr4 cells treated with 25 mM HU at the designated time points. The experiments above were repeated at least twice, to ensure reproducibility.
Heterochromatin structure does not alter histone H3 occupancy
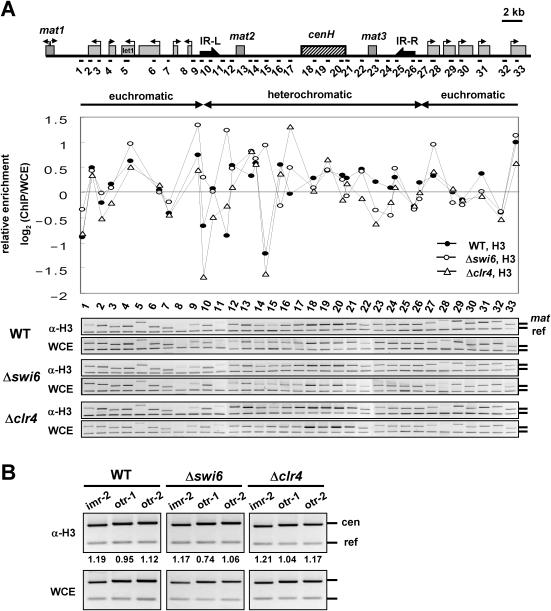
Heterochromatin can block access of proteins such as the Escherichia coli dam methylase (22,23). Thus, one can argue that apparent changes in RI deposition at heterochromatic regions by Δswi6 or Δclr4 might reflect alterations in antibody accessibility or histone-DNA interactions that affect cross-linking efficiency. If this is the case, then ChIP with an antibody recognizing endogenous histone H3 would be expected to produce results similar to the data presented above. To test this possibility, we performed ChIP analysis with an antibody recognizing the C-terminus of histone H3 in wild-type, Δswi6 or Δclr4 cells (Figure 4A). In wild-type cells, we found no gross differences in H3 occupancy in heterochromatin and euchromatin regions of the mating-type loci. In addition, we could not find any marked changes in H3 occupancy at heterochromatic regions in cells lacking Swi6 or Clr4. The occupancy of histone H3 was also measured at centromeric heterochromatin, where Δswi6 or Δclr4 cells showed increased RI deposition of histone H3-HA (Figure 4B). Again, we could not observe any difference in H3 occupancy among the strains, supporting the conclusion that inhibition of RI deposition at heterochromatic regions is not due to lower cross-linking efficiency or histone occupancy in heterochromatic regions.
Figure 4.
The total occupancy of histone H3 at the silent mat loci is not altered by deletion of swi6+ or clr4+. (A) ChIP analysis with an H3 antibody was performed with the same samples used in Figure 3 to measure H3 occupancy throughout the mat locus (sites 1–33). (B) The same ChIP samples were subjected to PCR with primer pairs corresponding to centromeric heterochromatin (imr-2, otr-1 and otr-2), as shown in Figure 1C. The experiments above were repeated at least twice, to ensure reproducibility.
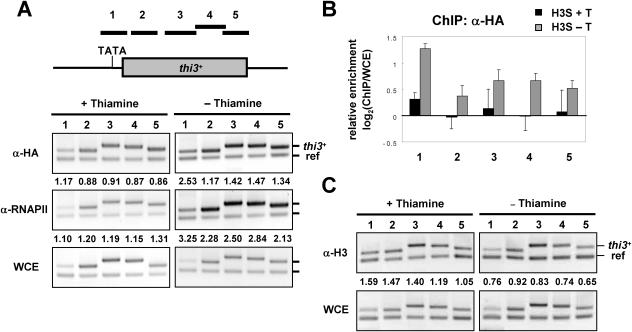
Transcription by RNA polymerase II can enhance RI deposition of H3 at promoter and coding regions of thi3+
Recently, it has been reported in Drosophila and human that H3.3 becomes enriched in active chromatin when transcription is induced (5,6). It has also been reported in budding yeast that histone occupancies decrease in proportion to transcriptional activity (24–28). Thus, it is possible that transcription may enhance RI deposition of histone H3 in fission yeast as demonstrated in other organisms. To test this hypothesis, we monitored transcription-dependent RI deposition of H3 at an inducible gene thi3+ (Figure 5A). This gene is repressed when thiamine is present in culture medium but is highly induced when thiamine is absent in the medium (29). When transcription is repressed (+Thiamine), RI deposition of H3 at the promoter (primer set 1) and coding (primer sets 2–5) regions is similar to the reference region. However, when transcription is induced (−Thiamine), RI deposition of H3 is significantly increased at the promoter and coding regions of thi3+. Notably, RI deposition of H3 at the promoter region is consistently higher than at coding regions, indicating that H3 exchange may be most active at promoter regions where nucleosome remodeling is expected to occur (Figure 5B). Consistent with this observation, a recent study shows that H3.3 preferentially localizes to promoter regions in mouse (30). To confirm whether the difference in RI deposition in these samples is indeed due to the difference in transcription by RNA polymerase II, we analyzed binding of RNA polymerase II in each sample using ChIP (Figure 5A). As expected, RNA polymerase II is localized to the promoter and coding regions only when transcription is induced (−Thiamine). Next, to determine whether increased RI deposition by transcription is due to conformational changes affecting cross-linking efficiency, we examined histone H3 densities in the same samples (Figure 5C). In contrast to the increased RI deposition of H3-HA by transcriptional induction, we found that the endogenous histone H3 occupancy was rather decreased at the promoter and coding regions of thi3+ in transcriptionally active cells. This result clearly shows that the enhanced RI deposition of histone H3 by transcriptional activation at thi3+ is genuine and suggests that the increase in RI deposition might be more robust than previously measured. At loci where transcription is highly active such as the induced thi3+ gene, histone displacement may be more efficient than reassembly, resulting in the net loss of histone proteins at those loci. The most robust loss of histone H3 was detected at the promoter where RI deposition of H3-HA occurred most actively. Based on these results, we propose that transcription enhances RI deposition of H3 at RNA polymerase II-transcribed genes such as thi3+, especially at their promoter regions.
Figure 5.
Transcription enhances RI deposition of H3 at promoter and coding regions of thi3+. (A) A diagram showing the promoter (TATA) and coding region of the thi3+ gene. The location of each primer pair is shown on top. Samples grown in sucrose medium in the presence (+T) or absence of thiamine (−T) were subjected to ChIP analysis with an HA antibody or an RNA polymerase II (RNAP II) antibody. (B) The H3-HA enrichment at thi3+ in each sample was quantified and the relative enrichment of RI deposited H3-HA at each locus is shown as the mean ± SD of triplicate samples. (C) The total occupancy of histone H3 at thi3+ in each sample. The same extracts used in (A) were subjected to ChIP analysis with an anti H3 antibody to assess the level of total histone H3 bound to each locus. The experiments above were repeated at least twice, to ensure reproducibility.
DISCUSSION
In this study, we investigated RI incorporation of histone H3 into chromatin in fission yeast. As fission yeast possesses only one subtype of histone H3, which is equivalent to the metazoan H3.3, we monitored exogenously expressed, HA-tagged H3 in DNA replication-blocked cells to assess RI deposition of histone H3. To block DNA replication, we used hydroxyurea whose treatment arrests cells early in S phase. To arrest cells before entering S phase, we examined the use of a temperature-sensitive cell division cycle mutant (cdc10ts). However, to arrest cells at G1, cdc10ts mutants require incubation at high temperature, which is known to affect higher-order chromatin structure in fission yeast (31,32). Therefore, we used HU throughout these experiments since it does not require any temperature shift. Nevertheless, we found that RI deposition using cdc10ts was very similar to that using HU (E.S. Choi, Y.K. Jang, unpublished data).
The central conclusion of our study is that RI deposition of histone H3 (and probably H4 as well) occurs predominantly at euchromatic regions. As RI deposition of H3 or H4 implies the displacement of pre-existing histones, we propose that loss of H3-H4 tetramers (or nucleosome octamers) may occur to some degree at open chromatin even in the absence of transcription. As we observed both robust loss of total histone H3 and increased RI deposition of exogenously expressed H3 at thi3+ gene upon transcriptional induction, we concluded that transcription at active chromatin may induce H3 displacement and reassembly by the RI pathway. In fact, previous studies in Drosophila revealed that RI deposition of H3.3 occurs primarily at euchromatic regions where most transcriptionally active genes reside (5,7,8). Although the exact mechanism by which transcription induces histone replacement is largely unknown, it may involve the action of chromatin remodeling machinery associated with transcription.
Previous studies (5–8) have suggested that RI deposition of histone H3.3 is blocked at heterochromatic regions. In the present study, we showed this is the case in fission yeast as well. However, the mechanism by which heterochromatin inhibits H3/H3.3 replacement is largely unknown. As fission yeast heterochromatic regions contain transcribed elements including siRNA precursors, heterochromatin may inhibit RI deposition by simply blocking transcription of such elements. Alternative possibilities assume more direct roles of heterochromatin.
Although little is known about internucleosomal forces within heterochromatin, several possibilities can be inferred from known properties of heterochromatin. First, the chromodomain of HP1 binds the H3 tail methylated at lysine 9 and the chromoshadow domain of HP1 mediates associations between HP1 proteins (33–35). Thus, multimeric associations of HP1 can cross-link neighboring nucleosomes within heterochromatin, limiting their mobility. Second, heterochromatin blocks access of proteins such as E.coli dam methylases (22,23). Thus, it is possible that factors required for histone exchange such as ATP-dependent remodeling factors and/or histone-modifying enzymes cannot access nucleosomes within heterochromatin. An example of such remodeling factors (and histone modifying enzymes) globally working at euchromatic regions was identified in budding yeast (36–38).
In summary, our study demonstrates that RI deposition of H3 in fission yeast occurs at active chromatin where genes reside and transcription is high. Thus, it is likely that histone H3 replacement is intimately linked to transcription in fission yeast as well as other eukaryotic organisms. This conservation suggests that H3.3 replacement may be fundamental to maintain transcriptionally active chromatin, in eukaryotes ranging from yeast to human. Considering the relative ease of genetic manipulations in fission yeast, this evolutionary conservation would make our experimental system a valuable tool for further delineating roles and mechanisms of RI deposition of H3.3 in eukaryotic cells, e.g. elucidation of the precise role of chromatin structure in regulating RI deposition, the mechanism of histone replacement by transcription and identification of factors involved in these processes.
Acknowledgments
We thank Dr Paul Russell for plasmids and helpful information on the plasmids. This work was supported by Korea Research Foundation Grant (KRF-2004-015-C00401) and in part by the National Cancer Center Research Grant (0210110 and 0510050) to Y.K.J. Funding to pay the Open Access publication charges for this article was provided by Korea Research Foundation Grant (KRF-2004-015-C00401) and the National Cancer Center Grant (0210110 and 0510050).
Conflict of interest statement. None declared.
REFERENCES
- 1.Workman J.L., Kingston R.E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Mello J.A., Almouzni G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 3.Altheim B.A., Schultz M.C. Histone modification governs the cell cycle regulation of a replication-independent chromatin assembly pathway in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1999;96:1345–1350. doi: 10.1073/pnas.96.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray-Gallet D., Quivy J.P., Scamps C., Martini E.M., Lipinski M., Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad K., Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 6.Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz B.E., Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mito Y., Henikoff J.G., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 9.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad K., Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc. Natl Acad. Sci. USA. 2002;99(Suppl. 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iacovoni J.S., Russell P., Gaits F. A new inducible protein expression system in fission yeast based on the glucose-repressed inv1 promoter. Gene. 1999;232:53–58. doi: 10.1016/s0378-1119(99)00116-x. [DOI] [PubMed] [Google Scholar]
- 12.Moreno S., Klar A., Nurse P. Methods. Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 13.Choi E.S., Kim H.S., Jang Y.K., Hong S.H., Park S.D. Two ubiquitin-conjugating enzymes, Rhp6 and UbcX, regulate heterochromatin silencing in Schizosaccharomyces pombe. Mol. Cell. Biol. 2002;22:8366–8374. doi: 10.1128/MCB.22.23.8366-8374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enoch T., Carr A.M., Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K., Chen E.S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 16.Noma K., Allis C.D., Grewal S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 17.Lorentz A., Ostermann K., Fleck O., Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 18.Ivanova A.V., Bonaduce M.J., Ivanov S.V., Klar A.J. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nature Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 19.Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 20.Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nature Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 21.Partridge J.F., Borgstrom B., Allshire R.C. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschling D.E. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh J., Klar A.J.S. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- 24.Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 25.Reinke H., Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee C.K., Shibata Y., Rao B., Strahl B.D., Lieb J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 27.Schwabish M.A., Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristjuhan A., Svejstrup J.Q. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 2004;23:4243–4252. doi: 10.1038/sj.emboj.7600433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweingruber A.M., Fankhauser H., Dlugonski J., Steinmann-Loss C., Schweingruber M.E. Isolation and characterization of regulatory mutants from Schizosaccharomyces pombe involved in thiamine-regulated gene expression. Genetics. 1992;130:445–449. doi: 10.1093/genetics/130.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow C.M., Georgiou A., Szutorisz H., Maia e Silva A., Pombo A., Barahona I., Dargelos E., Canzonetta C., Dillon N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allshire R.C., Javerzat J.P., Redhead N.J., Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 32.Ayoub N., Goldshmidt I., Cohen A. Position effect variegation at the mating-type locus of fission yeast: a cis-acting element inhibits covariegated expression of genes in the silent and expressed domains. Genetics. 1999;152:495–508. doi: 10.1093/genetics/152.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 34.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 35.Ye Q., Callebaut I., Pezhman A., Courvalin J.-C., Worman H.J. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi G., Shen X., Landry J., Wu W.H., Sen S., Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 37.Krogan N.J., Keogh M.C., Datta N., Sawa C., Ryan O.W., Ding H., Haw R.A., Pootoolal J., Tong A., Canadien V., et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 38.Kobor M.S., Venkatasubrahmanyam S., Meneghini M.D., Gin J.W., Jennings J.L., Link A.J., Madhani H.D., Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:587–599. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]