Abstract
Repression of poly(A)-binding protein (PABP) mRNA translation involves the binding of PABP to the adenine-rich autoregulatory sequence (ARS) in the 5′-untranslated region of its own mRNA. In this report, we show that the ARS forms a complex in vitro with PABP, and two additional polypeptides of 63 and 105 kDa. The 63 and 105 kDa polypeptides were identified, as IMP1, an ortholog of chicken zip-code binding polypeptide, and UNR, a PABP binding polypeptide, respectively, by mass spectrometry of the ARS RNA affinity purified samples. Using a modified ribonucleoprotein (RNP) immunoprecipitation procedure we further show that indeed, both IMP1 and UNR bind to the ARS containing reporter RNA in vivo. Although both IMP1 and UNR could bind independently to the ARS RNA in vitro, their RNA-binding ability was stimulated by PABP. Mutational analyses of the ARS show that the presence of four of the six oligo(A) regions of the ARS was sufficient to repress translation and the length of the conserved pyrimidine spacers between the oligo(A) sequences was important for ARS function. The ability of mutant ARS RNAs to form the PABP, IMP1 and UNR containing RNP complex correlates well with the translational repressor activity of the ARS. There is also a direct relationship between the length of the poly(A) RNAs and their ability to form a trimeric complex with PABP, and to repress mRNA translation. UV crosslinking studies suggest that the ARS is less efficient than a poly(A) RNA of similar length, to bind to PABP. We show here that the ARS cannot efficiently form a trimeric complex with PABP; therefore, additional interactions with IMP1 and UNR to form a heteromeric RNP complex may be required for maximal repression of PABP mRNA translation under physiological conditions.
INTRODUCTION
Poly(A)-binding protein (PABP) is a highly conserved, abundant and ubiquitous RNA-binding protein which preferentially associates with the 3′-poly(A) tail of eukaryotic mRNA. PABP also interacts with other cytosolic proteins such as Rna15 (1), eIF4B (2), eIF4G (3), eRF3 (4), Paip1 (5), Paip2 (6), TcUBP-1 (7), UNR (8) and poly(C)-binding proteins (9). Almost all known functions of PABP are attributed to its ability to interact with the 3′-poly(A) tract and to act as a scaffold for protein–protein interactions. PABP contains four highly conserved RNA-binding domains (RBDs I–IV) arranged in tandem at the N-terminus, followed by a less conserved proline-rich auxiliary domain of variable length at its C-terminus (10). The first two RBDs towards the N-terminus (RBD-I/II) exhibit preferential affinity for poly(A) RNA, whereas the RBDs towards the C-terminus (RBD-III/IV) show tropism for poly(U) RNA (11–13). The C-terminal region, which contains a highly conserved 74 amino acid-long PABC domain (14), does not bind RNA but interacts with other polypeptides as well as promotes oligomerization of PABP on poly(A) RNA (15).
PABP has been implicated in global gene regulation both by stimulating translation initiation (16) and by enhancing mRNA stability (17). A popular hypothesis addressing the mechanism of translation stimulation focuses on the connection between the 3′-poly(A) tail bound PABP and the 5′-cap bound eIF4F holoenzyme. eIF4F is a ternary complex consisting of RNA-helicase eIF4A and 5′-cap binding protein eIF4E, connected together by a central scaffold protein eIF4G (18). According to this model, interaction between PABP and eIF4G results in mRNA circularization, thereby promoting recycling of terminating ribosomes from the 3′ end to the 5′ end for another round of translation initiation. The interaction between PABP and eIF4G has been reconstituted using purified components (19), and has been demonstrated in yeast (3) and wheat germ (20) cell-free extracts. However, direct evidence that this interaction stimulates translation is lacking (21). The essential role of PABP in mRNA translation has been confirmed recently by using a PABP depleted cell-free system (22).
Considering the significance of PABP in gene expression, controlling its cellular level may be critical. Available evidence suggests that the cellular level of PABP is regulated in a developmental or growth-dependent manner at the translational level. PABP mRNA is a member of the terminal oligopyrimidine track (TOP) containing mRNA family, which includes mRNAs that encode components of the protein synthesizing machinery. The long 5′-untranslated region (5′-UTR) of PABP mRNA consists of at least two cis-acting translation control elements. The first control element TOP at the 5′-terminus (23) is followed by a highly conserved A-rich autoregulatory sequence (ARS) (24). The role of these cis-acting elements in regulation of PABP expression is well established but not fully understood. Several mRNAs containing a 5′-TOP sequence exhibit growth dependent translational control (25,26). Thus, the 5′-TOP probably confers regulation of PABP mRNA translation in a growth-dependent manner to coordinate expression of components of the protein synthesizing machinery, whereas the ARS may be involved in constitutive control of the PABP level under all circumstances (23,24).
Results of both in vitro and in vivo studies suggest that the ARS of PABP mRNA is able to confer autoregulation of PABP expression by a negative feedback mechanism (23,24,27). Previous studies have shown that PABP interacts with the ARS region and addition of purified PABP to a cell-free system repressed translation of mRNAs containing the A-rich sequence in their 5′-UTR (25,27). Deletion of the ARS region from PABP mRNA abolishes its autoregulation, whereas addition of this region at the 5′-UTR of a reporter gene represses its translation (24). According to the current understanding, idling PABP represses translation of its own mRNA by binding to the ARS region, thereby stalling the movement of the 40S pre-initiation complex within the 5′-UTR (28,29). Recent studies have shown that the C-terminal region of PABP, which is involved in protein–protein interaction, is necessary for autoregulation of PABP mRNA translation (15). This implies that interaction between ARS bound PABP and other polypeptides may be an important aspect of translational control.
In the present study we have investigated whether other polypeptides besides PABP bind to the ARS region. The results presented here suggest that the ARS region specifically interacts with at least three core polypeptides, IMP1, PABP and UNR, to form a multi subunit autoregulatory ribonucleoprotein complex (ARC). IMP1 and UNR can bind independently to the ARS but their RNA-binding activity was stimulated by PABP. Therefore, the formation of a heteromeric complex may be responsible for the previously observed stalling of the 40S pre-initiation complex at a site near the ARS (29).
MATERIALS AND METHODS
Plasmid construction
The 5′-UTR region (nt 71–131) of the human PABP cDNA (GeneBank ID Y00345) corresponding to the ARS was cloned into appropriate plasmid vectors by using synthetic oligonucleotides. Double-stranded oligodeoxynucleotides encoding either oligo(A) of different length, wild-type or mutant ARS sequences were generated by annealing complementary synthetic oligonucleotide sets (Table 1, only sense sequences are given). The annealed double-stranded DNA products contained an EcoRI restriction site followed by a T7 RNA polymerase promoter sequence at the 5′ end, and a BamHI restriction site at the 3′ end. The annealed products were digested with respective restriction enzymes (MBI, Fermentas), purified from a 2.5% agarose gel using the QIAquick gel extraction kit (Qiagen), cloned into pUC18 (MBI, Fermentas) and pEGFP-N3 (Clontech, BD Biosciences) plasmid vectors.
Table 1.
Sense oligonucleotides used to create oligo(A) and various ARS constructs
| Primer | Sequence |
|---|---|
| ARS | agaattc gtaatacgactcactataggg aaaaaatccaaaaaaaatctaaaaaaatcttttaaaaaaccccaaaaaaatttacaaaaaa ggatcca |
| ΔARS-4 | agaattc gtaatacgactcactataggg tccaaaaaaaatctaaaaaaatcttttaaaaaaccccaaaaaaattt ggatcca |
| ΔARS-2L | agaattc gtaatacgactcactataggg tccacctctactctactcctatcttttaaaaaaccccaaaaaaattt ggatcca |
| ΔARS-2R | agaattc gtaatacgactcactataggg tccaaaaaaaatctaaaaaaatcttttatcctaccccactcctattt ggatcca |
| ΔARS-0 | agaattc gtaatacgactcactataggg tccacctctactctactcctatctttatcctaccccactcctattt ggatcca |
| LS-ARS | agaattc gtaatacgactcactataggg aaaaaaaatctcctcttctccaaaaaaatccctctccctctaaaaaacttccctccctctaaaaaaa ggatcca |
| Poly(A)50 | agaattc gtaatacgactcactataggg aaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaggatcca |
| Poly(A)20 | agaattc gtaatacgactcactataggg aaaaaaaaaaaaaaaaaaaa ggatcca |
| Poly(A)13 | agaattc gtaatacgactcactataggg aaaaaaaaaaaaa ggatcca |
For the synthesis of N-terminus 6× His tag epitope containing protein expression vectors, the IMP1 (GenBank ID: NM006546), PABP (GeneBank ID: Y00345) and UNR (GenBank ID: BC032446) cDNAs were amplified by PCR using respective pQE-primers sets (Table 2). The forward and reverse primer contained BamHI and HindIII restriction sites, respectively. The PCR product was digested with appropriate restriction enzymes (MBI, Fermentas), purified from a 1% agarose gel by QIAquick gel extraction kit (Qiagen), and cloned into pQE80L (Qiagen) prokaryotic expression vector. All plasmid constructs were propagated into Escherichia coli DH5α (Invitrogene) employing standard recombinant techniques (30), isolated using GenElute plasmid maxi-prep kit (Sigma) and confirmed to be correct by DNA sequencing.
Table 2.
Primers used to create Protein expression vectors
| Primer | Sequence |
|---|---|
| pQE-IMP1(s) | aaaggatccaacaagctttacatcgg |
| pQE-PABP(s) | aaaggatccaaccccagtgccccc |
| pQE-UNR(s) | aaaggatccagctttgatccaaaccttc |
| pQE-IMP1(as) | ctaaagcttcacttcctccgtgcctg |
| pQE-PABP(as) | ctaaagcttaaacagttggaacacc |
| pQE-UNR(as) | ctaaagcttagtcaatgacaccagc |
In vitro RNA synthesis
The pUC18 or pEGFP-N3 plasmids containing either oligo(A) of different length, wild-type or mutant ARS region under the control of the T7 RNA polymerase sequence were linearized with BamHI, and pGEM-T vector (Promega) was linearized with SalI restriction enzyme for in vitro run-off transcription. Transcription reaction was usually performed at 37°C for 3 h in a final volume of 100 µl containing 10 µg of a DNA template, 2.5 mM of each NTP and 100 U of T7 RNA polymerase (Promega) in the N4 buffer [20 mM MgCl2, 1 mM spermidine, 0.01% Triton X-100, 20 mM DTT and 40 mM Tris–HCl (pH 8.1)]. Uniformly radiolabeled RNA was synthesized under similar conditions in a final reaction volume of 25 µl containing 150 µCi [α-32P]ATP (MP Biomedicals) and the final concentration of cold ATP reduced to 25 µmol. The contaminating nucleotides, incompletely transcribed products and the DNA template were removed by fractionating transcription reaction mixtures on 8% polyacrylamide gels under denaturing conditions (31). The amount of RNA and its specific radioactivity were determined using a spectrophotometer and scintillation counter, respectively.
Preparation of cytoplasmic extracts
Cytoplasmic protein extracts were prepared as described previously (32). HeLa, HEK 293, NIH 3T3 and C2 myoblasts were grown to 90% confluency in DMEM (Sigma) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Gibco). Cells were rinsed twice with ice-cold phosphate-buffered saline (PBS) [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 (pH 7.4)] and harvested by scraping in a hypotonic buffer CEB [10 mM HEPES–KOH (pH 7.5), 3 mM MgCl2, 14 mM KCl, 5% glycerol, 1 mM DTT, 0.02% Igepal CA-630, 0.5 mM phenylmethlysulfonyl fluoride (PMSF), 10 µg/ml leupeptin and 2 µg/ml aprotinin (Sigma)]. The cells were lysed by repeated passage through a 28-guage needle, centrifuged for 2 min at 4°C to remove the nuclei and cell fragments and the supernatants were stored at −85°C in small aliquots. The protein concentration was estimated to be ≈20 µg/µl by the BioRad protein assay kit.
Expression and purification of 6× His-tag fusion protein
E.coli DH5α containing the IMP1, PABP or UNR open reading frames cloned into the pQE80L expression vector and E.coli BL21(DE3) containing pET28b-β-galactosidase plasmid were grown to an early log phase and induced for 4 h with isopropyl-β-d-thiogalactoside. The bacterial cells were harvested by centrifugation and lysed by incubating with 1 mg/ml of lysozyme in a lysis buffer [50 mM NaH2PO4, 500 mM NaCl, 30 mM imidizole, 13 mM β-mercaptoethanol, 2 mM MgCl2, 1 mM PMSF, 0.5% IgepalCA-630 and 5% glycerol (pH 8.0)] at 0°C for 30 min. The lysate was cleared by centrifugation and the supernatant was mixed with Ni-NTA agarose beads (Qiagen). After shaking at 0°C for 30 min, the beads were washed extensively with a washing buffer [50 mM NaH2PO4, 500 mM NaCl, 50 mM imidizole, 13 mM β-mercaptoethanol, 2 mM MgCl2, 1 mM PMSF, 0.5% IgepalCA-630 and 5% glycerol (pH 8.0)] and the bound proteins were eluted in the elution buffer [50 mM NaH2PO4, 500 mM NaCl, 300 mM imidizole, 13 mM β-mercaptoethanol, 2 mM MgCl2, 1 mM PMSF, 0.5% IgepalCA-630 and 5% glycerol (pH 8.0)]. The eluted fraction was equilibrated with a storage buffer [10 mM HEPES–KOH (pH 7.5), 3 mM MgCl2, 140 mM KCl, 5% glycerol, 1 mM DTT, 0.02% Igepal CA-630, 0.5 mM PMSF, 10 µg/ml leupeptin and 2 µg/ml aprotinin] using the microcon YM-30 concentration column (Millipore) and stored at −80°C in small aliquots.
For in vivo protein labeling, the E.coli were grown for 24 h at 37°C in 50 ml of minimal media [6.8 g Na2HPO4, 3.0 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 3 mg CaCl2, 0.25 g MgSO4.7H2O, 3 g sucrose in one liter H2O (pH 7.4)]. The cells were collected by centrifugation and resuspended in 1 ml of the same media supplemented with 1 mCi of Trans [35S]-label (MP Biomedicals) metabolic labeling reagent. Protein expression was induced and proteins were purified as described above.
RNA electrophoretic mobility shift assay (REMSA)
REMSA was performed as described previously (32). In short, 20 µg of the cytoplasmic extract was incubated with ≈1 ng (1 × 105 c.p.m.) of radiolabeled RNA for 10 min at 22°C in a total reaction volume of 18 µl in the binding buffer [10 mM HEPES–KOH (pH 7.5), 3 mM MgCl2, 140 mM KCl, 5% glycerol, 1 mM DTT, 0.02% Igepal CA-630, 10 µg of E.coli tRNA and 0.02% bromophenol blue]. Subsequently, heparin was added to a final concentration of 10 µg/µl and incubation was resumed for another 5 min. The sample was treated with RNase T1 (25 U) for 5 min and analyzed by electrophoresis in a 2% agarose gel in 0.5× TBE buffer [45 mM Tris–borate, 1 mM EDTA (pH 8.0)], 100 V, at 4°C. The gel was then vacuum-dried and autoradiographed.
For competition assays, the cell extract was preincubated with 20–200 ng of an unlabeled RNA for 5 min prior to the addition of the radiolabeled RNA. For supershift assays, the cell extract was preincubated with 1 µg of the indicated antibody on ice for 10 min prior to the addition of the radiolabeled RNA. A PABP deficient cell extract was prepared by incubating 5 µl of the human PABP antibody with 25 µl of HeLa cell extract at 4°C for 2 h in CEB buffer followed by clearing the extract through a protein A–Sepharose column (Sigma).
UV crosslinking assay
For UV-induced crosslinking assays, 60 µg of cytoplasmic extract was incubated with ≈3 ng (3 × 105 c.p.m.) of radiolabeled RNA at 22°C for 10 min in a total reaction volume of 27 µl in the binding buffer. Heparin was added to the final concentration of 10 µg/µl and the sample was irradiated by UV-light (254 nm, 4000 µw/cm2) at 4°C for 5 min. The non-crosslinked RNA was cleaved by RNase T1 (25 U) and RNase A (1 µg) treatment at 37°C for 5 min. Finally, the sample was boiled in a protein sample loading buffer [6% glycerol, 2% SDS, 100 mM DTT and 0.02% bromophenol blue in 60 mM Tris–HCl (pH 6.6)] for 5 min and analyzed by 10% SDS–PAGE. For competition assays, the cell extract was preincubated with 50–500 ng of an unlabeled RNA for 5 min prior to the addition of the radiolabeled RNA. The gels were vacuum-dried and autoradiographed.
Isolation of RNA–protein complexes from agarose gels
To isolate the RNA–protein complex, 100 µg of HeLa extract was incubated with ≈5 ng (5 × 105 c.p.m.) of radiolabeled ARS RNA for 10 min at 22°C in a total reaction volume of 40 µl in the binding buffer. Heparin was added to a final concentration of 10 µg/µl and the sample was irradiated under UV light for 5 min at 4°C. The ribonucleoprotein (RNP) complex was resolved on a 2% agarose gel as described and visualized by wet gel autoradiography. The gel portion corresponding to the RNP complex was excised, homogenized and treated with RNase A (2 µg) and RNase T1 (50 units) for 10 min at 37°C. Finally, the sample was analyzed by 10% SDS–PAGE followed by autoradiography.
RNA-affinity chromatography
The ARS binding proteins were purified by RNA-affinity chromatography as described previously (33). Fifty micrograms of in vitro synthesized and gel purified RNA were oxidized by sodium periodate (10 mM) in 200 µl of coupling buffer (100 mM sodium acetate, pH 5.0) at 0°C in the dark for 2 h. The oxidized RNA was precipitated twice with 2.5 vol of ethanol. Two milliliters (50% slurry) of adipic acid dihydrazide agarose beads (Sigma) were equilibrated with the coupling buffer, mixed with the oxidized RNA, and incubated with gentle shaking at 4°C for 4 h. The unbound RNA was removed by washing the beads twice with 2 ml of 2 M NaCl followed by equilibrating the beads with the chromatography buffer [10 mM HEPES–KOH (pH 7.5), 3 mM MgCl2, 140 mM NaCl, 5% glycerol, 1 mM DTT and 0.01% Triton X-100].
HeLa cell extract (250 µl) was precleared by incubating with 2 ml (50% slurry) of adipic acid hydrazide agarose beads in the chromatography buffer in presence of protease inhibitors (0.5 mM PMSF, 10 µg/ml leupeptin and 2 µg/ml aprotinin) for 1 h at 4°C. Finally, the RNA conjugated beads were incubated with the precleared HeLa cell extract at 4°C for 1 h. The unbound proteins were removed by washing the beads extensively with the chromatography buffer. The affinity-bound proteins were eluted by an increasing salt gradient (0.5, 1.0 and 2.0 M NaCl) in the chromatography buffer. The eluted protein samples were analyzed by 10% SDS–PAGE and visualized by silver staining (34).
Peptide analysis
Protein bands from silver nitrate stained gels were excised and digested in gel with trypsin as described previously (34). The samples were analyzed by LC/MS/MS using a Thermo Finnigan LCQ DECA XP mass spectrometer and the raw mass data was cross-correlated to a theoretical database using TurboSequest (Thermo) to identify the proteins.
Transfection of cells
Approximately 3 × 105 subconfluent HeLa cells grown on a 35 mm dish in DMEM medium with 10% FBS were used for transfection. Plasmid DNA (1 µg) was incubated with 3 µg of LipofectAMINE in 100 µl of Opti-MEM (Invitrogen) for 30 min at 25°C before adding to the cells. Cells were incubated for 5 h at 37°C with the DNA/liposome mixture in 1 ml of Opti-MEM. Following the incubation, 1 ml of growth medium containing 20% FBS was added to the culture. After 12 h of incubation, the medium was replaced by fresh, complete DMEM medium (Sigma) containing 10% FBS.
Isolation of RNA and measurement of mRNA levels
The total cellular RNA was isolated using the High Pure RNA Isolation Kit (Roche Biochemical). The quality and quantity of the RNA was determined by 1.5% agarose gel electrophoresis and spectrophotometry, respectively. The level of a specific mRNA in the sample was determined by comparative real time RT–PCR using Rotor-gene 3000 (Corbett Research, NSW, Australia) (35). An aliquot of total RNA (1 µg) was reverse transcribed at 50°C for 1 h in a total reaction volume of 25 µl by SuperScript II reverse transcriptase and 150 ng random primers according to manufacturer's instruction (Invitrogen). After the reaction, the cDNA sample was amplified by PCR with platinum SYBR green qPCR supermix-UDG kit (Invitrogen) with primers specific for β-Gal, GFP or β-actin cDNAs (Table 3). Specificity of the amplified PCR products were examined after the final cycle by generating a melting curve with a heating rate of 1°C/s between 72 and 99°C. The data were analyzed using Rotor-gene 3000 software with 2−ΔΔCt method. The relative expression values of β-Gal and GFP mRNAs were normalized to that of β-actin mRNA.
Table 3.
Primers used for the real-time RT-PCR
| MRNA | Nucleotide sequence | GenBank ID |
|---|---|---|
| Human β-actin (s) | ctcttccagccttccttcct (780–799) | BC013835 |
| Human β-actin (as) | caccttcaccgttccagttt (963–982) | |
| GFP(s) | taccagcggtggtttgtttg (4091–4110) | U57609 |
| GFP(as) | gcagagcgaggtatgtaggc (4252–4271) | |
| β-Gal (s) | gctggataacgacattg (2435–2453) | U02451 |
| β-Gal (as) | cagcaccgcatcagcaag (2560–2577) |
Measurement of protein levels
Cells were washed twice with PBS and lysed by boiling in 400 µl protein sample loading buffer, 24 h after transfection. The samples were subjected to 10% SDS–PAGE, and the separated polypeptides were electrophoretically transferred to a nitrocellulose membrane for western blotting (30). Membrane was treated with GFP (BD Biosciences) and β-actin (Sigma) primary antibodies, washed, and further incubation with the alkaline phosphatase conjugated secondary antibody. Detection was carried out using the NBT/BCIP reagents and quantitative analysis was performed using ImageJ (NIH) gel band densitometry program.
Immunoprecipitation of RNA–protein complex
The in vivo RNA–protein crosslinking and immunoprecipitation assay was performed as described previously (36). HeLa cells (≈3×106 cells) were treated with 1% formaldehyde (Electron Microscopy Sciences) in PBS for 10 min at room temperature to crosslink the RNA and proteins of RNP complexes. Following the crosslinking, the cells were lysed in 0.5 ml of RIPA buffer [50 mM Tris–HCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl, 0.5 mM PMSF, 10 µg/ml leupeptin, 2 µg/ml aprotinin, 100 U RNasin (Promega) and 100 µg of E.coli tRNA (pH 7.5)]. Antibody (10 µg) coated protein A–Sepharose beads (30 µl, packed volume) was incubated with the cell lysate (250 µl) in RIPA buffer with rotations at room temperature for 2 h. The beads were washed extensively with high-stringency RIPA buffer [50 mM Tris–HCl, 1% Igepal CA-630, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 M NaCl, 2.5 M Urea and 0.5 mM PMSF (pH 7.5)]. Following the washing step the RNA–protein crosslink was reversed by incubation at 70°C for 1 h in 200 µl of elution buffer [50 mM Tris–HCl, 5 mM EDTA, 10 mM DTT and 1% SDS (pH 7.0)]. The sample was extracted once with phenol and once with chloroform followed by precipitation with ethanol. Contaminating DNA in the sample was removed by RQ1 RNase-free DNase (Promega) treatment and the sample was reverse transcribed using random primers followed by the PCR amplification using GFP-specific primers (sense, 5′-ccgtcagatccgctagcgc; antisense, 5′-gtcgagctggacggcgac). The samples were analyzed by electrophoresis in a 2% agarose gel.
RESULTS
Formation of ARS RNA–protein complex
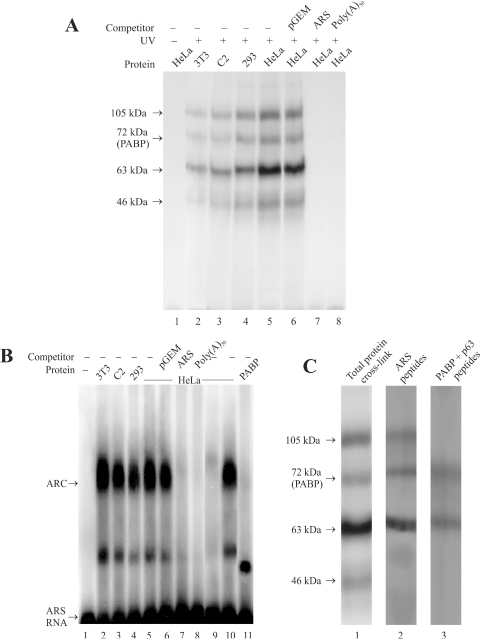
PABP is known to interact with several polypeptides to accomplish its function in the cell (1–9). We have, therefore, examined whether PABP can act as a protein scaffold to form a multisubunit complex with the ARS region of its mRNA. The results of crosslinking of the ARS RNA and cellular proteins from HeLa cells by UV light show that at least four polypeptides of Mr 105, 72, 63 and 46 kDa bind to the ARS RNA (Figure 1A, lane 5). Binding of these polypeptides was not seen in samples without the UV treatment (Figure 1A, lane 1). To test the specificity of this interaction unlabeled competitor RNAs were used during the UV crosslinking. The results show that a 100-fold molar excess of unlabeled ARS (Figure 1A, lane 7) or poly(A)50 RNA (Figure 1A, lane 8) competed efficiently with the [32P]-labeled ARS RNA for binding to these polypeptides. Whereas a 100-fold molar excess of an unrelated RNA of size similar to the ARS RNA derived from the transcription of an empty pGEM-T plasmid vector (Promega) was unable to compete with the [32P]-labeled ARS RNA for binding to all four polypeptides (Figure 1A, lane 6). To further examine whether the ARS binding polypeptides are also present in different cell types, UV crosslinking experiments were performed using mouse NIH3T3 and C2 cells, and human HEK293 cells. The results (Figure 1A, lanes 2–4) show that similar polypeptides could be seen in all four crosslinking experiments.
Figure 1.
Formation of ARS RNA–protein complex. (A) RNA–protein crosslinking by UV. The in vitro synthesized [32P]-labeled ARS RNA (≈3 ng, 3 × 105 c.p.m.) was incubated with different cell extract (≈60 µg protein) in the flat cap of a 0.2 ml PCR tube. Following the UV treatment, the samples were treated with RNaseA/RNase T1, analyzed by 10% SDS–PAGE and autoradiographed as described under experimental procedures. Cell extracts from mouse NIH3T3 fibroblasts (lane 2) and C2 myoblasts (lane 3), and human HEK293 (lane 4) and HeLa cells (lane 5) were used for these studies. One sample containing HeLa cell extract (lane 1) was analyzed without UV treatment as a control. Approximately 300 ng of the non-radioactive pGEM-T (lane 6), ARS (lane 7) or poly(A)50 RNA (lane 8) was used for competition studies. (B) Analysis of RNP complex by REMSA. The in vitro synthesized [32P]-labeled ARS RNA (≈1 ng, 1 × 105 c.p.m.) was incubated with different cell extracts (20 µg) as indicated above each lane. The unbound RNA was digested with RNaseT1 and subjected to electrophoresis in a 2% agarose as described in the experimental procedures. Lane 1, radioactive ARS RNA incubated without the cell extract; lanes 2–5, radioactive ARS RNA incubated with NIH3T3, C2, HEK293 and HeLa cell extracts, respectively. Lanes 6–8, competition with 100 ng of non-radioactive pGEM-T, ARS, or poly(A)50 RNA, respectively. Lane 9, cell extract was pre-incubated with the PABP antibody and precleared with protein A-sepharose beads before being used for REMSA. Approximately 20 µg of PABP deficient cell extract was used for REMSA. Lane 10, cell extract was similarly treated with the GFP antibody (BD Biosciences) before being used for REMSA. Lane 11, radiolabeled ARS RNA (≈ 1 ng, 1 × 105 c.p.m.) incubated with ≈2 ng of purified 6×His-PABP. (C) Analysis of RNP complexes by SDS–PAGE. RNP complex formation was initiated as described above and the samples were irradiated by UV before being resolved in a 2% agarose gel. The RNP bands (as shown in Figure 1B) were excised from the gel, treated with RNaseA/RNase T1, and analyzed by 10% SDS–PAGE as described in the Materials and Methods. Lane 1, ARS RNA and HeLa extracts treated with UV and analyzed before gel purification. Lane 2, polypeptides from the gel purified slower migrating ARC. Lane 3, polypeptides from the faster migrating minor complex.
To further examine the ARS RNP complex, we used RNA gel shift analyses. The results show that one major (ARC) and one minor RNP complex are formed with all four cell extracts tested (Figure 1B, lanes 2–5). The electrophoretic mobility shift of the [32P]ARS RNA was inhibited by unlabeled ARS (Figure 1B, lane 7) or poly(A)50 RNA (Figure 1B, lane 8) while the unrelated pGEM-T empty vector derived RNA failed to compete (Figure 1B, lane 6). The ARC migrated considerably slower in the gel compared with the RNP complex formed by incubating ARS-RNA with purified PABP (Figure 1B, lane 11), which suggests that the ARC is a heteromeric complex. Further analyses using HeLa cell extracts show that formation of the ARC can be inhibited by depleting PABP from the extract with PABP antibody (Figure 1B, lane 9) but not by a GFP antibody (Figure 1B, lane 10). These results suggest that PABP plays an essential role in the formation of the heteromeric ARS-RNA–protein complex.
As more than one polypeptide may be present in the RNP complex formed between the cellular proteins and the ARS RNA, we analyzed the polypeptide complements of this complex. The UV irradiated RNP complexes were resolved and extracted from an agarose gel and the presence of polypeptides bound to the ARS RNA was analyzed by SDS–PAGE (Figure 1C). The results show that three polypeptides of Mr 105, 72 and 63 kDa were present in the slower migrating major complex (Figure 1C, lane 2). The faster migrating minor complex consisted of two of these three polypeptides which included the 72 and 63 kDa polypeptides (Figure 1C, lane 3). These results suggest that a multimeric complex consisting of three different polypeptides can be formed with the ARS. It is not certain whether the minor complex was the dissociation product of the larger complex or represents an intermediate complex. Therefore, we investigated this possibility by analyzing the formation of these complexes at different incubation times. The results (not shown here) did not show any changes in the relative ratios between the two complexes. Therefore, the minor complex is likely a dissociation product of the ARC, which implies that the 105 kDa polypeptide was less tightly bound to the ARS RNA than the 72 and the 63 kDa polypeptides.
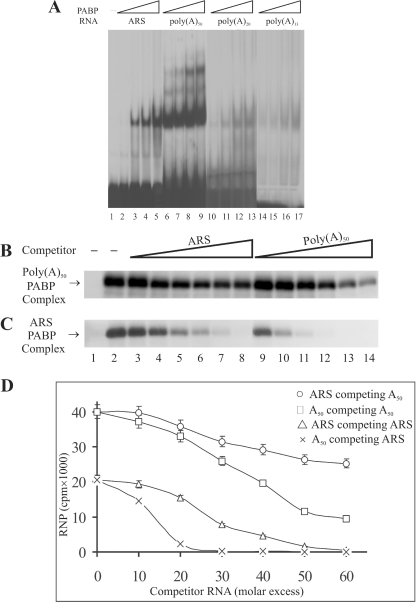
The ability of ARS to form homo-oligimeric complexes with purified PABP was further investigated by gel shift assays. The results show that in the presence of a limiting amount of PABP, ARS formed a monomeric complex (Figure 2A, lanes 2–5) whereas the poly(A)50 RNA, which has approximately the same number of adenines as the ARS, under similar conditions formed predominantly a monomeric complex, but also formed significant levels of both dimeric and trimeric complexes (Figure 2A, lanes 6–9). Also, as judged by the amount of radioactivity present in the RNP complex, PABP bound to poly(A)50, almost two times more efficiently than what was observed with the ARS RNA at all protein concentrations tested. RNAs with shorter stretches of adenine residues including poly(A)20 and poly(A)13 did not bind to PABP efficiently and could not form detectable levels of dimeric and trimeric complexes (Figure 2A, lanes 10–13 and lanes 14–17, respectively). In a previous study formation of both dimeric and trimeric complexes with the ARS was reported (15). However, in these studies PABP was present in several fold molar excess to that of the ARS RNA; under these conditions the formation of oligomeric RNP complex through PABP–PABP interaction may be facilitated. Nevertheless, this group also reported formation of a predominantly monomeric ARS RNA–PABP complex [Ref. (15), Figure 4B, lanes with 3.8 ng PABP] at a lower PABP concentration. Results of our studies, therefore, suggest that the slower migrating RNP complex of the ARS seen with the total HeLa cell extract consists of more than one polypeptide. This complex may more efficiently repress mRNA translation than PABP alone which predominantly forms a monomeric complex with the ARS RNA.
Figure 2.
Comparison of the ability of the ARS and poly(A) RNA to bind PABP. (A) Gel-shift assays of binding of PABP to the ARS and poly(A) RNAs of various length. REMSA was performed using 0.2, 0.4, 0.8 and 1.6 ng of purified 6× His-PABP and ≈1.5 ng of [32P]-labeled ARS (lanes 2–5), poly(A)50 (lanes 6–9), poly(A)20 (lanes 10–13) and poly(A)13 (lanes 14–17) RNAs. Samples were analyzed on 5% PAGE under non-denaturing conditions. (B and C) UV crosslinking assays of binding of PABP to ARS and poly(A)50 RNAs. The in vitro synthesized [32P]-labeled ARS and poly(A)50 RNAs (≈3 ng, 3 × 105) were incubated with the purified PABP (≈2 ng) for 5 min at room temperature. Unlabeled ARS (lanes 3–8) or poly(A)50 (lanes 9–14) competitor RNAs were added (10-fold molar excess increment) and incubated further at the room temperature for 3 min. Following the UV treatment, the samples were treated with RNase A/RNase T1, fractionated on a 10% SDS–PAGE and visualized by autoradiography. Lane 1, samples without UV treatment; lane 2, samples without unlabeled competitor RNA. (D) The RNP bands in (A and B) were excised by superimposing the radiograph and the level of radioactivity was measured by scintillation counter. The average level of radioactivity of the RNP complex in each band from three separate competition experiments was plotted against the molar concentration of the competitor RNA.
Figure 4.
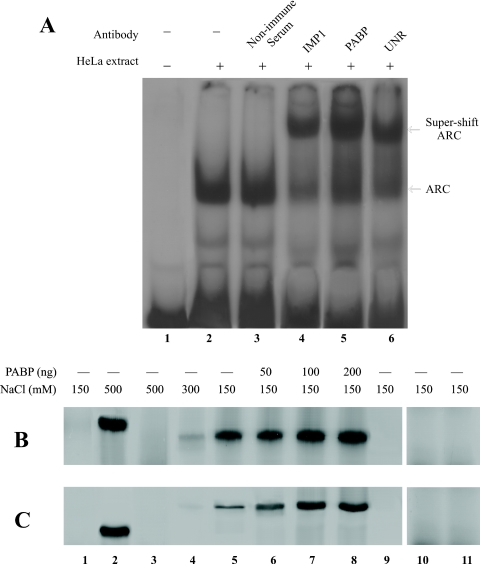
Interaction of IMP1 and UNR with RNA. (A) Presence of IMP1, PABP and UNR in the ARS RNA–protein complex. REMSA was performed using [32P]-labeled ARS RNA and HeLa cell extract as described. Cell extract (≈20 µg) was incubated with ≈1 µg of either non-immunized serum (lane 3), IMP1 (lane 4), PABP (lane 5) or UNR (lane 6) antibody for 10 min on ice prior to the addition of the labeled ARS RNA (≈1 ng, 1 × 105 c.p.m.). Samples were subjected to 5% PAGE under non-denaturing conditions. (B) Binding of IMP1 to the ARS RNA. [35S]methionine labeled 6× His-tagged IMP1, PABP, UNR and β-galactosidase were expressed in E.coli and purified by Ni-NTA agarose as described in Materials and Methods. The purified polypeptides were incubated with ARS or poly(A)50–agarose beads and the bound proteins were eluted by boiling the beads in protein sample loading buffer and analyzed by 10% SDS–PAGE. In some binding reactions the indicated amount of purified non-radiolabeled PABP was added. Lane 1, [35S]methionine labeled total cell extract from non-transformed E.coli DH5α; Lane 2, [35S]methionine labeled PABP; Lanes 3–5, [35S]methionine labeled IMP1 at different NaCl concentration; Lanes 6–8, [35S]methionine labeled IMP1 and indicated amount of unlabeled PABP. Lane 9, [35S]methionine labeled Luciferase. Lane 10, [35S]methionine labeled His-tagged β-galactosidase. Lane 11, binding of IMP1 to poly(A)50–agarose. (C) Binding of UNR to the ARS RNA. Experiment was performed under similar conditions as in (B). Lanes 1, 2, 9 and 10 are same as in (A); Lanes 3–5, [35S]methionine labeled UNR at different NaCl concentration; Lane 6–8, [35S]methionine labeled UNR and indicated amount of unlabeled PABP. Lane 11, binding of UNR to poly(A)50–agarose.
In addition to the gel shift assays, we compared the ability of the ARS and poly(A)50 RNA to bind to PABP by UV crosslinking studies. The results show that under similar conditions poly(A)50 (Figure 2B) binds to PABP almost two times more efficiently than the ARS (Figure 2C). This was also evident in competition studies, as ∼2-fold more unlabeled ARS (Figure 2C, lanes 3–8 and triangles in Figure 2D) was required than that of the unlabeled poly(A)50 RNA (Figure 2C, lanes 9–14 and legend crosses in Figure 2D) to attain a similar level of inhibition of the binding of radioactively labeled ARS RNA to PABP. Similar competition studies of binding of radioactively labeled poly(A)50 to PABP also show that the poly(A)50 binds more tightly to PABP than the ARS, as more unlabeled poly(A)50 RNA (Figure 2D, legend circles or squares, respectively) was required for this competition, than what was necessary to obtain a similar level of competition with the radio labeled ARS RNA (Figure 2D, legend crosses or triangles, respectively). Also, in these binding studies using the labeled poly(A)50, the unlabeled ARS competed less efficiently than the unlabeled poly(A)50.
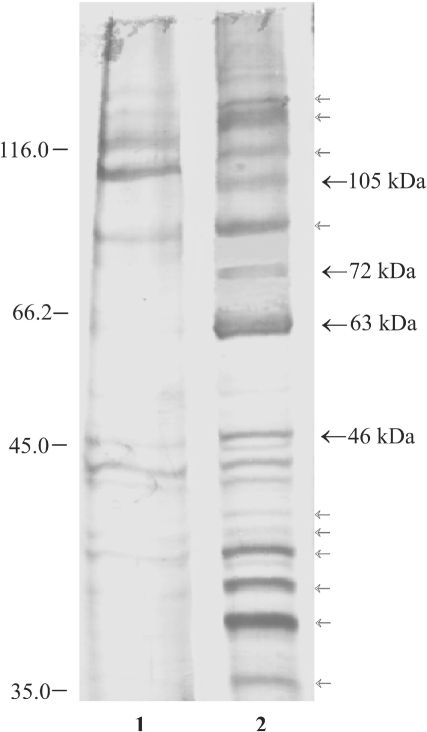
Identification of polypeptides of the ARC
To identify the polypeptides of the ARC, we purified the ARS binding polypeptides by RNA-affinity chromatography (Figure 3). The results show that several polypeptides bind tightly to the ARS RNA (Figure 3, lane 2). Control experiments using the pGEM-T vector derived RNA ligand in affinity chromatography show that several of these polypeptides are probably non-specific RNA-binding proteins (Figure 3, lane 1). The polypeptides that are unique to the ARS RNA affinity chromatography are marked by arrows. A total of 14 ARS RNA specific polypeptides including the 105, 72, 63 and 46 kDa polypeptides that were seen in the gel purified ARC or UV crosslinking studies can be seen in the affinity purified material. As 10 of these polypeptides were not present in the gel purified complexes, it is likely that they were not authentic partners of the ARS-RNA. However, it is possible that these polypeptides bind to the ARS RNA through protein–protein interactions. Since visualization of the polypeptides in the gel purified ARC or in the UV crosslinking samples depends on the presence of small fragments of covalently linked [32P]-labeled RNA, this method may exclude polypeptides that are present in the RNP complex through their direct interaction with RNA-binding proteins. The affinity purified 72 kDa polypeptide was less abundant in the 2 M NaCl eluted fraction than other polypeptides. This was primarily due to the tight binding of the 72 kDa (PABP) to the ARS RNA. This polypeptide was present in relatively large quantities in the SDS eluted fraction (result not shown).
Figure 3.
Affinity chromatography of ARS RNA-binding proteins. In vitro synthesized ARS RNA was covalently linked to agarose beads and incubated with HeLa cell extract. The bound polypeptides were eluted, resolved on 10% SDS–PAGE and visualized by silver staining. Lane 1, pGEM-T and lane 2, ARS RNA bound protein fractions. The polypeptides specific for the ARS RNA affinity chromatography are shown by arrows. The polypeptides with bold arrows were seen in the UV crosslinking experiments.
Among the 14 polypeptides binding specifically to the ARS RNA containing matrix we analyzed the 105, 72, 63 and 46 kDa polypeptides by mass spectrometry of tryptic digests since these were the only polypeptides that showed binding to the ARS RNA in UV crosslinking studies. The results of LC/MS/MS analyses are summarized in Table 4. As expected, the 72 kDa polypeptide was identified as PABP. The 63 kDa polypeptide is a known RNA-binding protein IMP1 and the 105 kDa polypeptide was identified as UNR, a known PABP interacting protein. The 46 kDa polypeptide was identified as β-actin. As β-actin and 11 other polypeptides were not present in the gel purified RNP complex, we considered them not to be the parts of a stable core complex and did not pursue investigating these polypeptides further.
Table 4.
Identity of polypeptides binding to the ARS-RNA by LC/MS/MS
| MW (kDa) | Peptide sequence | Peptide ID | Homologs | References |
|---|---|---|---|---|
| 105 | ATNIEVLSNTFQFTNEAR DQFGFINYEVGDSKK LLGYVATLK IKVDFVIPK EAFGFIER | UNR | KIAA0885, D3Jfr1, NRAS, mKIAA0885, D1S155E | (46,47) |
| 72 | SLGYAYVNFQQPADAER EAAQKAVNSATGVPTV ALYDTFSAFGNILSCK GFGFVCFSSPEEATK GYGFVHFETQEAAER | PABP | PABP1, PAB1, PABPC1 | (47) |
| 65 | VNELQNLTAAEVVVPR ISYSGQFLVK RLEIEHSVPK QQQVDIPLR | IMP1 | MGC68429, Zipcode-binding protien | (37–40,48) |
To determine whether PABP, IMP and UNR polypeptides are present in the ARS-RNA protein complex, we tested the ability of PABP, IMP1 and UNR antibodies to bind to the ARC. The results of REMSA super-shift analyses show that both IMP1 and UNR antibodies were able to super-shift the ARC (Figure 4A, lanes 4 and 6). Both antibodies behaved similarly to that of the PABP antibody (Figure 4A, lane 5). The non-immunized serum was ineffective in the super-shift assay (Figure 4A, lane 3).
We further tested the ability of IMP1 and UNR to directly interact with the ARS RNA in in vitro binding assays. The 6× His-tagged PABP, IMP1 and UNR were labeled with [35S] during expression in E.coli and purified by Ni-NTA affinity chromatography. The ability of purified radiolabeled proteins to bind ARS-RNA conjugated agarose beads was tested as described previously (11). The results show that PABP binds to the ARS-agarose beads efficiently and this binding was resistant to 0.5 M NaCl treatment (Figure 4B and C, lane 2). IMP1 alone was also able to bind to ARS-agarose beads almost as efficiently as PABP at physiological salt concentration (Figure 4B, lane 5), but this binding was sensitive to 0.5 M NaCl, as very little binding of IMP1 to ARS-agarose beads at this salt concentration was observed (Figure 4B, lane 3). UNR alone on the other hand showed a considerable lower binding ability to the ARS-agarose beads than that of either PABP or IMP1 at physiological salt concentration (Figure 4C, lane 5). As controls, we used [35S]-labeled total cell extract from non-transformed E.coli (Figure 4B and C, lane 1), in vitro translated [35S]methionine labeled luciferase (Figure 4B and B, lane 9) and purified [35S]-labeled 6× His-tagged β-galactosidase (Figure 4B and C, lane 10). No detectable binding of these polypeptides to the ARS-agarose beads was observed, which suggests that non-specific binding of polypeptides to the RNA was minimal under our experimental conditions. Therefore, the results of these binding assays suggest that both IMP1 and UNR could bind independently to the ARS RNA, albeit, with lower affinity than PABP. Similar pull-down studies with poly(A)50–agarose beads did not show detectable binding to IMP1 (Figure 4B, lane 11) and UNR (Figure 4C, lane 11). Thus, a stretch of 50 adenine residues alone is not sufficient for binding IMP1 and UNR, the pyrimidine bases that separate the A-rich regions of the ARS must be present to form the correct binding site.
There is an apparent contradiction between the inability of IMP1 and UNR to bind to the poly(A)50–agarose beads (Figure 4B and C, lane 11) and the results of UV crosslinking studies (Figure 1A) which showed that poly(A)50 RNA competed with the binding of both 63 (IMP1) and 105 (UNR) kDa polypeptides to the labeled ARS RNA. A possible interpretation of this difference is that both IMP1 and UNR can make contact with the poly(A)50 in presence of PABP in the total cell extract, and therefore, can compete with the ARS when present in several fold molar excess. However, this interaction is weak in absence of PABP; therefore, both proteins were removed from the poly(A)50–agarose beads during the washing procedure. The inability of UNR to bind poly(A) RNA has also been reported earlier (8).
We also tested whether PABP facilitates the binding of IMP1 and UNR to the ARS RNA. The results show that the presence of unlabeled PABP during the binding reaction stimulated binding of both IMP1 and UNR to the ARS-agarose beads in a dose-dependent manner (Figure 4B and C, lanes 6–8). Maximum stimulation of binding to ARS-agarose was observed when ∼4-fold molar excess of PABP was used (Figure 4B and C, lane 7). In the presence of PABP the binding ability of IMP1 and UNR to ARS RNA was similar since PABP had a greater stimulatory effect on the binding of UNR to the ARS RNA than that of the IMP1. PABP also stimulated binding of IMP1 and UNR to the ARS RNA at 0.5 M NaCl concentration (data not shown). Results of both gel supershift assays and in vitro RNA–protein binding studies, therefore, suggest that IMP1 and UNR are legitimate partners of the ARC, and PABP stabilizes their interaction with the ARS.
Formation of ARC and translational repression
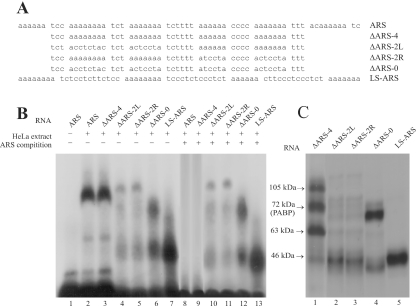
To obtain insights into the role of ARC formation in repressing PABP mRNA translation, the effect of various mutations in the ARS region on the RNP complex formation and its ability to repress mRNA translation was examined. Altogether five mutant RNAs were used in our studies (Figure 5A). The authentic ARS region of human PABP mRNA contains six oligo(A) rich regions which are separated by 3–6 pyrimidine bases. The first mutant RNA tested for RNP formation by REMSA had four internal oligo(A) rich regions, one oligo(A) containing region from both the 5′ and the 3′ ends were deleted. This shorter ARS RNA (ΔARS-4) formed the same RNP complex (Figure 5B, lane 3) as the wild-type ARS region (Figure 5B, lane 2). Two additional mutant RNAs which had only the two oligo(A) regions at either the 5′ end (ΔARS-2L) or the 3′ end (ΔARS-2R) were unable to form the RNP complex similar to the wild-type RNA (Figure 5B, lanes 4 and 5, respectively). For both mutant RNAs, a low level of a slower migrating complex than the ARC was observed. This complex most likely contained different polypeptides than those found in the ARC since its formation was not competed by unlabeled wild-type ARS RNA (Figure 5B, lanes 10 and 11, respectively). Deletion of all six oligo(A)s from the ARS RNA and replacing them with mostly pyrimidine bases (ΔARS-0) also prevented ARC formation (Figure 5B: lane 6). The ΔARS-0 RNA formed two faster migrating complexes than the ARC. As formation of these two complexes was also not inhibited by unlabeled ARS RNA (Figure 5B, lane 12), it is unlikely that they shared any polypeptides with the ARC. We also tested another mutant ARS RNA which had all six oligo(A) regions but the spacings between them were uniformly increased to 13 nt. This mutant ARS RNA (LS-ARS) also failed to form the ARC, instead it formed a significant level of a much faster migrating complex (Figure 5B: lane 7). Again formation of this faster migrating complex was not inhibited by unlabeled ARS RNA (Figure 5B: lane 13), therefore, suggesting that this complex did not share any polypeptides with the ARC. The results of RNP formation by different mutant RNAs suggest that binding of IMP1, PABP and UNR to the ARS to form the ARC requires the presence of at least four stretches of oligo(A)s and also an appropriate length of spacer sequences between them, which is probably between 3 and 6 nt in length. The spacer length of 13 nt used in our studies was not suitable for ARC formation.
Figure 5.
The ability of different mutant ARS RNAs to form the ARC. (A) Wild-type and various mutants ARS RNA sequences. (B) [32P]-labeled wild-type or mutant ARS RNAs (A) were prepared as described and used in REMSA. For competition, ≈20-fold molar excess of unlabeled ARS RNA was added to the cell extract prior to the addition of the radiolabeled RNA (≈1 ng). The samples were treated with RNase T1 and analyzed by electrophoresis on a 2% agarose gel. (C) Mutant ARS-protein crosslinking by UV. Approximately 3 ng of [32P]-labeled mutant ARS RNA (A) was incubated with the HeLa cell extract (≈60 µg) in the flat cap of a 0.2 ml PCR tube. Following the UV treatment, the samples were treated with RNaseT1/RNaseA, analyzed by 10% SDS–PAGE and autoradiographed as described under Materials and Methods.
We also used these mutant ARS RNAs in UV crosslinking assays to detect the nature of bound polypeptides (Figure 5C). The ΔARS-4 showed binding to 105, 72, 63 and 46 kDa polypeptides (Figure 5C: lane 1) as seen previously with the wild-type ARS RNA (Figure 1A). However, mutant ARS RNAs with fewer oligo(A) stretches and increased spacer lengths between the oligo(A)s did not bind to these polypeptides (Figure 5C, lanes 2–5). As the ability of mutated ARS RNA to bind 63 (IMP1), 72 (PABP) and 105 (UNR) kDa polypeptides were affected simultaneously it is likely that all three polypeptides bind to the ARS as a multimeric protein complex. These results also suggest that the IMP1 and UNR binding site are present within the 47 nt-long region of the ΔARS-4 RNA. Since poly(A)50 can not bind IMP1 and UNR (Figure 4B and C, lane 11), the conserved pyrimidine stretches between the oliog(A)s of the ΔARS-4 RNA are necessary to form the IMP1 and UNR binding site.
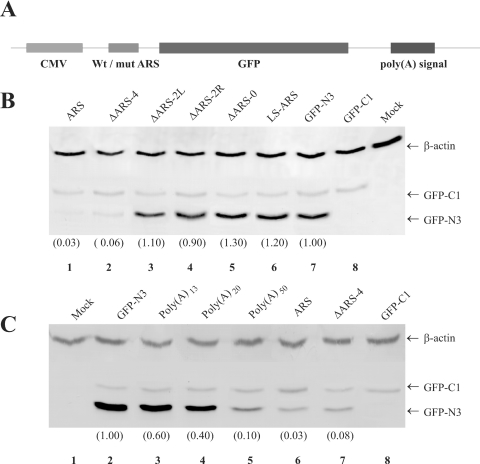
The physiological significance of RNP formation was examined by using expression of the normal or the mutant ARS containing GFP reporter gene. In all cases the mutant or the normal ARS sequences (Figure 5A) were placed at the 5′-UTR of the GFP reporter gene derived from the pEGFP-N3 expression vector (Figure 6A). Analyses of the GFP polypeptide levels in transiently transfected HeLa cells by western blotting show that the ARS (Figure 6B, lane 1) or the ΔARS-4 (Figure 6B, lane 2) containing constructs did not express high levels of GFP. In contrast, replacing the wild-type ARS by one of the mutant ARS represented either by ΔARS-2L, ΔARS-2R, ΔARS-0 or LS-ARS in our constructs restored high levels of GFP expression. The GFP expression level in ΔARS-2L (Figure 6B, lane 3) and ΔARS-2R (Figure 6, lane 4) containing transcripts were similar to what was observed in cells transfected with the parent GFP-N3 construct lacking the ARS (Figure 6B: lane 7). The GFP level in cells transfected with ΔARS-0 and LS-ARS (Figure 6B, lanes 5 and 6, respectively) was a little higher than that of the control cells (GFP-N3, Figure 6B, lane 7). This increase was probably due to insertion of pyrimidine nucleotides in the ARS. As transfection efficiency controls we co-transfected cells with a GFP construct that lacked the ARS and produced a slightly longer protein owing to translation of the sequences within the multiple cloning site (pEGFP-C1, Clontech). Almost equal levels of this longer GFP-C1 were produced in cells transfected with the different reporter constructs. The GFP-N3 levels in all experiments were corrected for differences in loading and transfection efficiencies using β-actin and GFP-C1 levels, respectively.
Figure 6.
GFP expression from wild-type and mutant ARS containing reporter genes. Cells were transfected with various GFP reporter constructs (pEGFP-N3) containing either poly(A) of different length, wild-type or mutant ARS regions in the 5′-UTR. To evaluate the transfection efficiency cells were also co-transfected with an unmodified pEGFP-C1 plasmid, which encodes a larger GFP than the pEGFP-N3. The cellular levels of GFP-N3, GFP-C1 (transfection control) and β-actin (loading control) polypeptides were measured by western blotting. The results presented here are representative of five separate transfection experiments. (A) pEGFP-N3 plasmid constructs containing wild-type or mutant ARS region. (B and C) Western blots, the numbers at the bottom within parentheses show the relative levels of GFP-N3 after correcting for the variations in the loading and transfection efficiency.
Additional transfection studies using constructs expressing the reporter GFP mRNA with either a poly(A)13, poly(A)20 or a poly(A)50 containing region at its 5′-UTR were performed to correlate the translational repression by the ARS and poly(A) tracts of different length. The results (Figure 6C) show that the GFP level in cells expressing poly(A)13 (Figure 6C, lane 3) and poly(A)20 (Figure 6C, lane 4) containing mRNA was almost 50% less than the control GFP-N3 mRNA expressing cells (Figure 6C, lane 2). Almost 90% reduction of GFP level was observed in cells expressing poly(A)50 containing reporter mRNA (Figure 6C, lane 5). The presence of full-length ARS at the 5′-UTR was, however, significantly more effective than the poly(A)50 in preventing GFP expression (Figure 6C, lane 6).
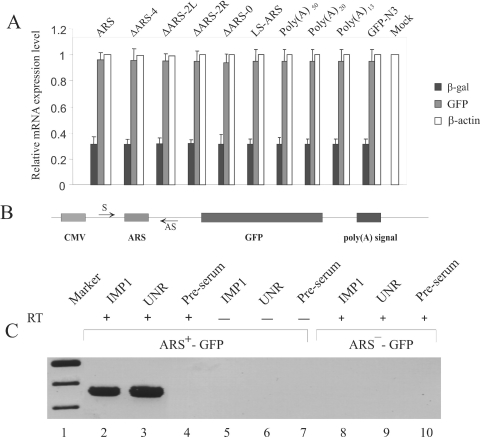
We then determined whether the reporter mRNA levels in cells transfected with different ARS containing constructs were similar. In earlier studies, we showed that presence of the ARS in the 5′-UTR of a reporter β-gal mRNA repressed its translation (24). In these studies we observed that the same level of the reporter mRNA can be detected by real-time RT–PCR (Figure 7A). As internal controls to monitor the transfection efficiencies the cells were co-transfected with a pCMV-SPORT-β-gal plasmid (Invitrogen). The GFP mRNA levels were normalized by the level of the β-gal mRNA. Since earlier studies (24,29) using the sucrose density gradient profile of the ARS containing reporter mRNA had established that the translation of the reporter mRNA is repressed by the ARS, we did not perform similar sucrose gradient analyses of the mutant ARS containing transcripts. The results of analysis of the mRNA levels show that similar levels of GFP mRNA were present in cells transfected with different constructs.
Figure 7.
Analysis of the reporter mRNA. (A) Measurement of mRNA levels by real-time RT–PCR. HeLa cells were transfected with various GFP reporter constructs to express GFP mRNA containing either poly(A) of different length, wild-type or mutant ARS elements in their 5′-UTR. Cells were co-transfected with pCMV-SPORT-β-gal vector as a control for the transfection efficiency between experiments. Total cellular RNA from the transfected cells was analyzed by real-time RT–PCR using gene specific primers (Table 3) as described in Materials and Methods. Two separate analyses for each of the four independent transfection experiments were performed and averages of eight measurements are presented here. The β-actin mRNA level was measured as an internal loading control. PCR of RNA from the ARS-pEGFP-N3 transfected cells was carried out without performing the reverse transcription step as a negative control. (B) In vivo RNA–protein crosslinking and immunoprecipitation. In vivo crosslinked RNPs were immunoprecipitated using IMP1 and UNR antibodies. The presence of GFP reporter mRNA in the immunoprecipitae was analyzed by RT–PCR. Samples without the reverse transcription step (RT-) were also used in PCRs to monitor the absence of any contaminating plasmid DNA. Lane 1, DNA marker; lanes 2–4, RT–PCR analysis of ARS+-GFP samples immunoprecipitated using IMP1, UNR, and rabbit pre-serum, respectively. Lanes 5–7, analysis of ARS+-GFP samples immunoprecipitated using IMP1, UNR and rabbit pre-serum without RT reaction, respectively. Lanes 8–10, RT–PCR analysis of ARS−-GFP samples immunoprecipitated using IMP1, UNR and rabbit pre-serum, respectively.
Taken together the results of GFP protein level and mRNA level measurements strongly suggest that while the ARS or ΔARS-4 containing mRNAs were translationally repressed, the mRNAs containing the mutated ΔARS-0, LS-ARS, ΔARS-2L and ΔARS-2R were not repressed. Thus, there is good correlation between the inability of the mutant ARS to form the ARC and repress mRNA translation. Also the result of GFP polypeptide and mRNA levels in cells expressing poly(A)13, poly(A)20 and poly(A)50 containing mRNAs suggest a direct relationship of the length of the poly(A) tract with the formation of multimeric complexes, and ability to repress translation when placed at the 5′-UTR of a reporter mRNA.
Ribonucleoprotein immunoprecipitation (RIP) assays (36) were performed to determine if IMP1 and UNR bind to the ARS region of the reporter GFP mRNA in vivo. HeLa cells were transfected with either the wild-type or the ARS containing pEGFP-N3 reporter construct (Figure 7B), and 24 h after transfection, the RNA–protein interaction was stabilized in situ by formaldehyde treatment. The RNP complex was immunoprecipitated using specific antibodies and the presence of GFP reporter mRNA in the sample was analyzed using RT–PCR. The results of RIP assay (Figure 7C) show that the ARS containing GFP reporter mRNA was specifically precipitated by the IMP1 (Figure 7C, lane 2) and UNR (Figure 7C, lane 3) antibodies, whereas the same antibodies failed to precipitate the wild-type GFP reporter mRNA that did not contain ARS (Figure 7C, lanes 8 and 9, respectively). The PCR analyses of samples without reverse transcription (Figure 7C, lanes 5 and 6) or RT–PCR analysis of samples treated with RNase (results not shown) revealed no amplification of the target RNA sequence. Also, the presence of GFP reporter mRNA was not detected in samples immunoprecipitated using rabbit pre-serum (Figure 7C, Lanes 4, 7 and 10). Thus, the results show that IMP1 and UNR can bind specifically to the ARS and not to any other region in the GFP mRNA in vivo.
DISCUSSION
PABP mRNA translation is autoregulated by an A-rich cis element at its 5′-UTR. We have shown earlier that the ARS binds to PABP and can repress translation of a reporter mRNA (24,27). Although presence of a short, 10–60 nt long poly(A) sequence at the 5′-UTR of a reporter mRNA can also inhibit translation (15,37), the precise mechanism how ARS and poly(A) sequence inhibits translation could differ. Result of our studies suggests that, indeed, this is the case. We showed that the ARS is almost three times more effective than the poly(A)50 in repressing mRNA translation (Figure 6C). This difference is probably due to the greater ability of the ARS to form a trimeric RNP complex with IMP1, PABP and UNR than what was observed for the poly(A)50. We have shown that the poly(A)50 can not bind to IMP1 and UNR, and forms largely a monomeric and a small amount of dimeric and trimeric complexes with PABP at a limiting concentration of this polypeptide. A previous study (37) has shown that a stretch of as few as 10 adenines at the 5′-UTR of an mRNA can repress translation as efficiently as the ARS in H293 cells. The reasons for this difference in results between the two studies are not known. However, the use of different cell-lines and promoters for reporter mRNA expression in these two studies could be a contributing factor. Furthermore, an earlier report suggested that H293 cells are refractory to the ARS mediated repression of PABP mRNA translation and a region near the initiation codon of PABP mRNA have the ability to override ARS mediated repression (23). In this context it is noteworthy, that in UV crosslinking studies significantly more ARS bound 63 kDa polypeptide (IMP1) was observed with HeLa cell extract than what were seen with H293, 3T3 and C2 cell extracts (Figure 1A). Therefore, the greater availability of IMP1 to bind to the ARS in HeLa cells could enhance the ability of ARS to repress translation in these cells.
We have shown earlier that the ARS prevents mRNA translation by stalling the 40S ribosomal subunit at a site adjacent to the ARS (29). It is likely that the critical factor in determining the ability of ARS to repress translation is the formation of a multisubunit RNP complex, sufficiently large, to prevent the scanning 40S ribosomal subunits from reaching the initiation codon. It is possible that at a sufficiently high cellular PABP level a homotrimeric complex between PABP and the ARS could form (15). However, this process would allow more PABP synthesis before the repression mechanism is triggered, than, if a multimeric complex with additional proteins is formed.
Survey of the 5′-UTR region of PABP mRNA from different species revealed that not only the presence of the A-rich region is highly conserved but also the sequence of the pyrimidine rich spacers between the oligo(A) regions is conserved between several species (Table 5). Therefore, it is not surprising that the conserved pyrimidine rich regions of the ARS are also required for binding to additional polypeptides to repress PABP mRNA translation. In these studies we have shown that two additional polypeptides of 63 and 105 kDa can be crosslinked to the ARS RNA in vitro by UV. These two polypeptides were purified by affinity chromatography and identified by mass spectrometry as IMP1 and UNR, respectively. We have further shown that both IMP1 and UNR are present in the ARS RNA–protein complex by gel-supershift assays and we have coined the term ARC for this RNA–protein complex. Both IMP1 and UNR can directly bind to the ARS RNA but PABP has a stimulatory effect on their binding to the ARS. This explains why depletion of PABP from the HeLa cell extract almost abolished binding of these two polypeptides to the ARS. The precise RNA sequence within the ARS recognized by IMP1 and UNR is not known. However, our results suggest that both polypeptides make contact with adenine nucleotides since both polypeptides were detected by UV crosslinking using [32P]ATP labeled RNA. We had also used [32P]CTP or [32P]UTP to label the ARS RNA and used it for UV crosslinking studies. Both labeled RNAs showed crosslinking to 72, 63 and 105 kDa polypeptides (results not shown). Therefore, it is likely that these polypeptides make contact with a large region of the ARS. A 47 nt-long domain of the ARS has been identified as the IMP1, PABP and UNR binding region. Presence of the pyrimidine stretches within this region is necessary for binding to IMP1 and UNR. Stimulation of the interactions of ARS with IMP1 and UNR by PABP also suggest a co-operative nature of binding of these polypeptides to the ARS and formation of a multicomponent RNP complex. To validate the results of in vitro studies, we showed here that the ARS containing reporter GFP mRNA, but not the wild-type GFP mRNA without the ARS, can be immunoprecipitated with both the IMP1 and UNR antibodies.
Table 5.
ARS sequence homology among different species
| Organisms | ARS sequence |
|---|---|
| Homo Sapiens | aaaaaaa tcc aaaaaaaa tct aaaaaaa tctttt aaaaaa cccc aaaaaaa tttac aaaaaa |
| Pleioblastus Pygmaeus | aaaaaa tcc aaaaaaaa tct aaaaaaa tctttt aaaaaa cccc aaaaaaa tttac aaaaaaa |
| Mus Musculus | aaaaaa tcc aaaaaaaa tct aaaaaat cctttg aaaaaa accc aaaaaaa tttac aaaaaaa |
| Rattus Norvegicus | aaaaaa tcc aaaaaaaa tct aaaaaat cctttg aaaaaa accc aaaaaaa tttac aaaaaa |
| Xenopus Laevis | aaaaa tcc aaaaaaaa tct aaaaaat cctttg aaaaaa ccca aaaaaaa tttac aaaaaaa |
Although presence of a long (50 or more) stretch of adenines at the 5′-UTR of a reporter mRNA can repress translation (15), this is not biologically relevant as a similar poly(A) sequence has not been found in any PABP mRNA. The translational control element present in human and other mammalian PABP mRNA consists of several stretches of oligo(A)6–8 separated by conserved pyrimidine bases. Therefore, it is important to determine how the naturally occurring translational control element of PABP mRNA functions. The evidences presented in this manuscript suggest that the ARS forms a hetromeric complex to repress translation. The need for the formation of a multimeric RNP complex at the ARS for suppressing PABP mRNA translation is further supported by two observations, (i) that the ARS RNA is weaker than a similar size poly(A) RNA, in its interaction with PABP (Figure 2) and (ii) the peptide binding C-terminal PABC domain is required for the ARS mediated translational control (15). Often, control of mRNA translation is not an all or none phenomenon. The degree to which the translation of a specific mRNA is regulated depends on several factors including the availability of translational repressors and enhancers, the mRNA and the precise nature of its cis element.
IMP1 belongs to the VICKZ family of zipcode binding proteins and is similar to chicken ZBPI (38,39) and Xenopus, VgIRBP/vera (40). These groups of proteins are composed of two RNA recognition motifs and four hn-RNP homology domains. IMP1 is known to bind to the 5′-UTR of IGF-II mRNA to repress its translation (40). Like its ortholog chicken ZBPI which is involved in the localization of β-actin mRNA to the leading edge of fibroblast cells, IMP1 is also implicated in the subcytoplasmic localization of mRNAs (41,42). As a modular protein IMP1 binds to various RNA sequences generally rich in A, U and C residues. Chicken ZBPI can be crosslinked to RNA containing ACACCC sequences (39) but its Xenopus ortholog, vera, binds to UUCAC and UUUCUA sequences of vg1 mRNA (40). The human IMP1 appears to recognize the UUCACGUUCAC sequence in the 5′-UTR of IGF-II mRNA (40). Thus, the IMP1 binding domain of the ARS is different from any previously characterized IMP1 binding sequence.
The third member of the ARC described in our studies is a known PABP interacting protein UNR, which is also a RNA-binding protein with five cold-shock domains (43–45). The main cellular function of UNR includes internal initiation of uncapped mRNA translation (46) and translation dependent destabilization of mRNA (43). Recent studies have shown that UNR is involved in the decay of c-fos mRNA through the major protein-coding determinant of instability (mCRD). UNR binds to both mCRD and PABP to form a platform for the formation of a deadenylation/decay mRNA protein complex (43). We showed that UNR also binds to the same 47 nt-long region of the ARS as IMP1 and PABP does. The known UNR binding sequence of mCRD (43) and internal ribosome entry site (46) are longer and more complex than the UNR binding region of the ARS and bear no homology with this region. Previously we showed that the ARS containing repressed and ARS minus reporter mRNAs have similar half lives in transfected cells (24). Therefore, it is unlikely that UNR is involved in destabilizing the ARS containing repressed mRNA. Whether UNR is involved in any internal initiation of PABP mRNA, which may impair proper initiation, could be the subject for further investigation. As a PABP interacting protein UNR could form a larger RNP complex with the ARS than what could be achieved by PABP alone. This larger RNP complex should be better able to prevent movement of the scanning 40S subunits.
Analysis of various mutations of the ARS have shown that binding of all three polypeptides, IMP1, PABP and UNR, are similarly affected by these mutations. The presence of a four oligo(A) rich stretches was shown to be sufficient for both ARC formation and repression of mRNA translation. In addition, the length of the spacer sequences between the oligo(A)s is important for the function of ARS. In these studies we have not tested whether the presence of T and Cs in the spacer is also important. In this report we have mostly focused on demonstrating the importance of ARC formation for repression of mRNA translation. The three polypeptides present in the ARC are RNA-binding polypeptides, which can also interact with other polypeptides (17,44,45). Whether a larger RNP is formed with the ARS through protein–protein interactions with the core RNP complex needs to be further investigated. The presence of several additional ARS specific polypeptides in the affinity purified sample (Figure 3) supports the formation of a large multimeric complex with the ARS through both RNA–protein and protein–protein interactions.
Acknowledgments
Human PABP and UNR antibody was generously provided by Drs G. Dreyfuss and A. Shyu, respectively. Human PABP, IMP1 cDNA and pET28b-β-galactosidase clones were generously provided by Drs R. Pictet, J. Christiansen and L. Holland, respectively. We thank Dr Richard Mosser for a critical reading of the manuscript. This work was supported by a grant from the CIHR. Funding to pay the Open Access publication charges for this article was provided by the grant from the CIHR (Canada).
Conflict of interest statement. None declared.
REFERENCES
- 1.Amrani N., Minet M., Le Gouar M., Lacroute F., Wyers F. Yeast Pab1 interacts with Rna15 and participates in the control of the poly(a) tail length in vitro. Mol. Cell. Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le H., Tanguay R.L., Balasta M.L., Wei C.C., Browning K.S., Metz A.M., Goss D.J., Gallie D.R. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J. Biol. Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 3.Tarun S.Z., Jr, Sachs A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson B., Berkova N., Couturier A., Chabelskaya S., Philippe M., Zhouravleva G. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell. 2002;94:205–216. doi: 10.1016/s0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 5.Craig A.W., Harhighat A., Yu A.T., Sonenberg N. Interaction of polyadenylate-binding protein with eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 6.Khaleghpour K., Svitkin Y.V., Craig A.W., DeMaria C.T., Deo R.C., Burley S.K., Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 7.D'Orso I., Frasch A.C. TcUBP-1, an mRNA destabilizing factor from Trypanosomes, homodimerizes and interacts with novel AU-rich element and poly(A)-binding proteins forming a ribonucleoprotein complex. J. Biol. Chem. 2002;277:50520–50528. doi: 10.1074/jbc.M209092200. [DOI] [PubMed] [Google Scholar]
- 8.Chang T.C., Yamashita A., Chen C.A., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z., Day N., Trifillis P., Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorgoni B., Gary N.K. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental prespective. Brief. Funct. Genomics Proteomics. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 11.Burd C.G., Matunis E.L., Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nietfeld W., Mentzel H., Pieler T. The Xenopus laevis poly(A) binding protein is composed of multiple functionally independent RNA binding domains. EMBO J. 1990;9:3699–3705. doi: 10.1002/j.1460-2075.1990.tb07582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn U., Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein–protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 14.Kozlov G., Trempe J.F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl Acad. Sci. USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melo E.D., Dhalia R., Martins de sa C., Standart N., de Melo Neto O.P. Identification of a C-terminal poly(A)-binding protein (PABP)–PABP interaction domain. J. Biol. Chem. 2003;278:46357–46368. doi: 10.1074/jbc.M307624200. [DOI] [PubMed] [Google Scholar]
- 16.Sachs A,B., Davis R.W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobson A. Poly(A) metabolism and translation: the closed loop model. In: Hershey J.W., Methews M.B., Sonenberg N., editors. Translation Control. NY: Cold Spring Harbor Press; 1996. pp. 451–480. [Google Scholar]
- 19.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 20.Wei C.C., Balasta M.L., Ren J., Goss D.J. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998;37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- 21.Kozak M. How strong is the case for regulation of the initiation step of translation by elements at the 3′ end of eukaryotic mRNAs? Gene. 2004;343:41–54. doi: 10.1016/j.gene.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Kahvejian A., Svitki Y.V., Sukarieh R., M'Boutchou M., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornstein E., Git A., Braunstein I., Avni D., Meyuhas O. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J. Biol. Chem. 1999;274:1708–1714. doi: 10.1074/jbc.274.3.1708. [DOI] [PubMed] [Google Scholar]
- 24.Wu J., Bag J. Negative control of the poly(A) binding protein mRNA translation is mediated by the adenine-rich region of its 5′-UTR. J. Biol. Chem. 1998;273:34535–34542. doi: 10.1074/jbc.273.51.34535. [DOI] [PubMed] [Google Scholar]
- 25.Shama S., Meyuhas O. The translational cis-regulatory element of mammalian ribosomal protein mRNAs is recognized by the plant translational apparatus. Eur. J. Biochem. 1996;236:383–388. doi: 10.1111/j.1432-1033.1996.00383.x. [DOI] [PubMed] [Google Scholar]
- 26.Avni D., Shama S., Loreni F., Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell. Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bag J., Wu J. Translational control of poly(A)-binding protein expression. Eur. J. Biochem. 1996;237B:143–152. doi: 10.1111/j.1432-1033.1996.0143n.x. [DOI] [PubMed] [Google Scholar]
- 28.de Melo Neto O.P., Standart N., Martins de Sa C. Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 1995;23:2198–2205. doi: 10.1093/nar/23.12.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bag J. Feedback inhibition of poly(A)-binding protein mRNA translation: A possible mechanism of translation arrest by stalled 40S ribosomal subunits. J. Biol. Chem. 2001;276:47352–47360. doi: 10.1074/jbc.M107676200. [DOI] [PubMed] [Google Scholar]
- 30.Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K., editors. Current Protocols in Molecular Biology (Online Version) NY: John Wiley & Sons, Inc.; 2004. [Google Scholar]
- 31.Wildeman A.G., Nazar R.N. Studies on the secondary structure of 5.8S rRNA from a thermophile, Thermomyces lanuginosus. J. Biol. Chem. 1981;256:5675–5682. [PubMed] [Google Scholar]
- 32.Thomson A.M., Rogers J.T., Walker C.E., Staton J.M., Leedman P.J. Optimized RNA gel-shift and UV cross-linking assays for characterization of cytoplasmic RNA–protein interactions. Biotechniques. 1999;27:1032–1042. doi: 10.2144/99275rr03. [DOI] [PubMed] [Google Scholar]
- 33.Caputi M., Zahler A.M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 34.Podtelejnikov A.V., Mann M. Identification of yeast proteins by mass spectrometry. Methods Enzymol. 2002;351:296–321. doi: 10.1016/s0076-6879(02)51854-1. [DOI] [PubMed] [Google Scholar]
- 35.Heid C.A., Stevens J., Livak K.J., Williams P. Real time quantitative PCR. Genome Res. 1996;10:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 36.Niranjanakumari S., Lasda E., Brazas R., Garcia-Blanco M.A. Reversible cross-linking combined with immunoprecipitation to study RNA–protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 37.Melo E.O., de Melo Neto O.P., Martins de Sa C. Adenosine-rich elements present in the 5′-untranslated region of PABP mRNA can selectively reduce the abundance and translation of CAT mRNAs in vivo. FEBS Lett. 2003;546:329–334. doi: 10.1016/s0014-5793(03)00620-3. [DOI] [PubMed] [Google Scholar]
- 38.Eom T., Antar L.N., Singer R.H., Bassell G.J. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross A.F., Oleynikov Y., Kislauskis E.H., Taneja K.L., Singer R.H. Characterization of a beta-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshler J.O., Highett M.I., Abramson T., Schnapp B.J. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A.H., Wewer U.M., Nielsen F.C. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielson F.C., Nielson J., Kristensen M.A., Koch G., Christiansen J. Cytoplasmic trafficking of IGF-II mRNA-binding protein by conserved KH domains. J. Cell Sci. 2002;115:2087–2097. doi: 10.1242/jcs.115.10.2087. [DOI] [PubMed] [Google Scholar]
- 43.Chang T.C., Yamashita A., Chen C.Y., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Havin L., Git A., Elisha Z., Oberman F., Yaniv K., Schwartz S.P., Standart N., Yisraeli J.K. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.TriQueneaux G., Velten M., Franzon P., Dautry F., Jaquemin-Sablon H. RNA binding specificity of Unr, a protein with five cold shock domains. Nucleic Acids Res. 1999;27:1926–1934. doi: 10.1093/nar/27.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt S.L., Hsuan J.J., Totty N., Jackson R.J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeffers M., Paciucci R., Pellicer A. Characterization of unr; a gene closely linked to N-ras. Nucleic Acids Res. 1990;18:4891–4899. [PMC free article] [PubMed] [Google Scholar]
- 48.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15 000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]