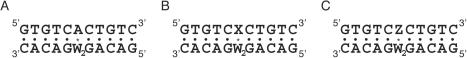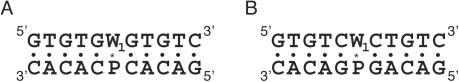Abstract
On the basis of non-covalent bond interactions in nucleic acids, we synthesized the deoxyadenosine derivatives tethering a phenyl group (X) and a naphthyl group (Z) by an amide linker, which mimic a Watson–Crick base pair. Circular dichroism spectra indicated that the duplexes containing X and Z formed a similar conformation regardless of the opposite nucleotide species (A, G, C, T and an abasic site analogue F), which was not observed for the natural duplexes. The values among the natural duplexes containing the A/A, A/G, A/C, A/T and A/F pairs differed by 5.2 kcal mol−1 while that among the duplexes containing X or Z in place of the adenine differed by only 1.9 or 2.8 kcal mol−1, respectively. Fluorescence quenching experiments confirmed that 2-amino purine opposite X adopted an unstacked conformation. The structural and thermodynamic analyses suggest that the aromatic hydrocarbon group of X and Z intercalates into a double helix, resulting in the opposite nucleotide base flipping into an unstacked position regardless of the nucleotide species. This observation implies that modifications at the aromatic hydrocarbon group and the amide linker may expand the application of the base pair-mimic nucleosides for molecular biology and biotechnology.
INTRODUCTION
Hydrogen bonds play a central role in the formation of Watson–Crick A/T and G/C base pairs. In addition to the interstrand hydrogen bonds, base stacking is important for the integrity and stabilization of the double helix structure of DNA and RNA. The stabilization energy provided by the base stacking is estimated to be 1.8–0.5 kcal mol−1 (1 kcal = 4.18 kJ) in , which is comparable with the energy provided by hydrogen bond formation (1–3). On the basis of the non-covalent interactions in nucleic acids, artificial nucleosides have been examined from the standpoints of evaluation of the base-pairing energy (1,3), recognition of a target DNA sequence and dNTPs by polymerases (4–6), expansion of the genetic alphabet (7,8) and development of functional molecules (9–16), where planar aromatic molecules are often examined because compounds having a contiguous π-system have a high potential for stacking on a nucleotide base and can intercalate between two adjacent base pairs in a duplex.
We have synthesized the deoxyadenosine derivatives tethering a phenyl group [N6-(N′-phenylcarbamoyl)-2′-deoxyadenosine, X] and a naphthyl group [N6-(N′-naphthylcarbamoyl)-2′-deoxyadenosine, Z] by an amide linker (Scheme 1), where the aromatic hydrocarbon group is expected to stack with a double helix when it orients in an appropriate position. Our previous work demonstrated that the single deoxyadenosine derivatives at the 5′ dangling ends stabilized the self-complementary DNA duplexes of 5′-ATGCGCAT-3′ and 5′-TGCGCA-3′ more than a single adenine, and the stabilization energy was as much as or greater than the formation of the Watson–Crick A/T base pair (17). The strong stabilization suggests that the deoxyadenosine derivatives stack on the adjacent terminal base pairs efficiently, probably by adopting a Watson–Crick base pair-mimic geometry.
Scheme 1.
Chemical structures of the deoxyadenosine derivatives used in this study.
In this study, the non-self complementary DNA duplexes containing X and Z in the middle of a sequence were investigated in order to estimate the stacking interaction by the deoxyadenosine derivatives in a duplex. We measured circular dichroism (CD) spectra, ultraviolet (UV) melting curves and fluorescence emission spectra of the DNA duplexes. It was demonstrated that the duplexes containing X or Z adopted a similar conformation regardless of the opposite nucleotide species, and it was suggested that X and Z could force the target base to flip into an unstacked position by intercalating their aromatic hydrocarbon group in a duplex. The lack of the pairing selectivity of the deoxyadenosine derivatives would have an advantage for the site-selective base flipping in a target sequence.
MATERIALS AND METHODS
Syntheses of the deoxyadenosine derivatives
Synthesis of the phosphoramidite derivatives of X and Z was started with 2′-deoxyadenosine (the details are described in the Supplementary Data). All chemicals were purchased from Aldrich and Wako Chemicals Co., Ltd, and the solvents were freshly distilled just before use. 1H NMR spectra were measured at 400 MHz with a Varian INOVA 400 NMR spectrometer, and electro spray ionization mass spectra were measured with a Finnigan Mat LCQ mass spectrometer. Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra were measured with a PE Biosystems Voyager DE MALDI-TOF mass spectrometer using 3-hydroxypicolinic acid as a matrix.
Synthesis of the DNA oligomers
Oligonucleotides were synthesized on a solid support using phosphoramidite chemistry by automated DNA synthesis (Applied Biosystems Model 391) and purified with reversed-phase high-performance liquid chromatography on a C18 column (TOSOH) after the removal of the protection groups, followed by desalting with a C18 cartridge column (18–20). All oligonucleotides were confirmed by MALDI-TOF mass spectra (PE Biosystems Voyager).
CD measurements
CD spectra of the DNA duplexes were obtained with a JASCO J-820 spectropolarimeter equipped with a temperature controller. All spectra were measured at 20 µM in a buffer containing 1 M NaCl, 10 mM Na2HPO4 (pH 7.0) and 1 mM Na2EDTA at 4°C. The DNA samples were heated to 90°C and cooled at a rate of 2°C min−1 before use.
Measurements and analysis of the UV melting curves
The UV absorbance was measured with a Shimadzu 1650 or 1700 spectrophotometer equipped with a temperature controller. The melting curves were monitored at 260 nm or 325 nm in a buffer containing 1 M NaCl, 10 mM Na2HPO4 (pH 7.0) and 1 mM Na2EDTA. The extinction coefficients for the single-strand oligonucleotides were calculated based on the nearest-neighbor approximation (21), and those containing the deoxyadenosine derivatives were assumed to be the same as those of the deoxyadenosine.
The thermodynamic parameters (ΔH0, ΔS0 and ΔG0 at 37°C) for duplex formations were calculated from the Tm (melting temperature) obtained at different total strand concentrations (Cts) using the versus log(Ct/4) plot and the fit of the melting curves with a theoretical equation, as reported previously (22,23).
Fluorescence measurements
Fluorescence emission spectra of the 2-amino purine-containing DNA strands were measured with a JASCO FP-6500 spectrofluorometer equipped with a temperature controller. Since 2-amino purine and the naphthyl group of Z have overlapping absorption and emission bands, reliable emission spectrum of 2-amino purine cannot be obtained for the DNAs containing Z. The emission spectra were measured at 20 µM duplex or 10 µM single-strand DNA in a buffer containing 1 M NaCl, 10 mM Na2HPO4 (pH 7.0) and 1 mM Na2EDTA at 4°C. The DNA samples were heated to 90°C before use. The emission spectra were obtained with an excitation wavelength at 305 nm, and the band path for the excitation and emission was set at 5 nm.
RESULTS
CD spectra of the DNA duplexes containing the deoxyadenosine derivatives
Scheme 1 indicates the structures of the deoxyadenosine derivatives of X (phenylurea derivative) and Z (naphthylurea derivative). The artificial nucleotides tether a phenyl group or a naphthyl group with the N6 of deoxyadenosine by an amide linker, where the aromatic hydrocarbon group is expected to stack with a double helix when it orients in an appropriate position. The DNA duplexes of 5′-GTGTCW1CTGTC-3′/5′-GACAGW2GACAC-3′ form identical Watson–Crick base pairs except the W1/W2 pair, where W1 is A (A-series), X (X-series) or Z (Z-series), and W2 is A, G, C, T or F (tetrahydrofuran abasic site analogue) (Figure 1). When W1 is A and W2 is T, the duplex has the Watson–Crick base pairs exclusively. In contrast, the duplexes formed by the DNA strands containing A as W1 and A, G, C or F as W2 have a single mismatched pair of A/A, A/G, A/C or A/F, respectively. When W1 is X or Z, the duplex contains the X/W2 or Z/W2 pair, respectively.
Figure 1.
The sequences of (A) the A-series, (B) the X-series and (C) the Z-series DNA duplexes forming the A/W2, X/W2 or Z/W2 pair, respectively, where W2 is A, G, C, T or F.
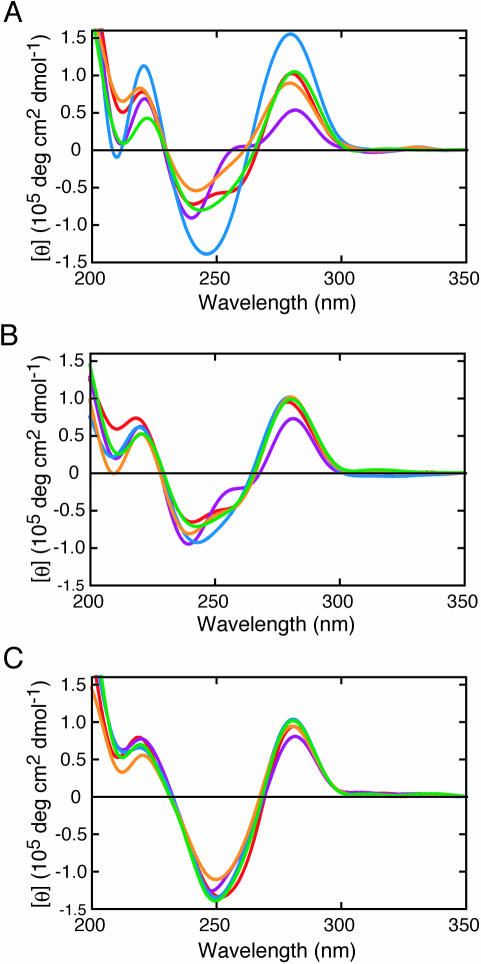
Figure 2 compares the CD spectra of the duplexes of 5′-GTGTCACTGTC-3′/5′-GACAGW2GACAC-3′ (A-series), 5′-GTGTCXCTGTC-3′/5′-GACAGW2GACAC-3′ (X-series) and 5′-GTGTCZCTGTC-3′/5′-GACAGW2GACAC-3′ (Z-series). All the duplexes showed a typical B-form signal with positive peaks around 280 and 220 nm and negative peaks around 245 and 210 nm. However, the intensity of the peaks and the overall shape of the spectra of the A-series duplexes differed substantially depending on the W2 nucleotide (Figure 2A), indicating that the A/A, A/G, A/C, A/T and A/F pairs adopt a different conformation, as has been reported for the duplexes containing a single mismatch (24–29). In contrast, differences in the CD spectra of the X-series duplexes (Figure 2B) were less than those of the A-series duplexes. Figure 2C also indicates nearly identical spectra for all the Z-series duplexes. These observations are suggestive of similar duplex structures regardless of W2 nucleotide in the X/W2 or Z/W2 pair.
Figure 2.
CD spectra of the (A) A-series, (B) X-series and (C) Z-series duplexes [W2 = A (red), G (purple), C (blue), T (green) or F (orange)]. All measurements were done in the 1 M NaCl-phosphate buffer at 4°C.
Thermodynamic parameters for duplex formations containing the deoxyadenosine derivatives
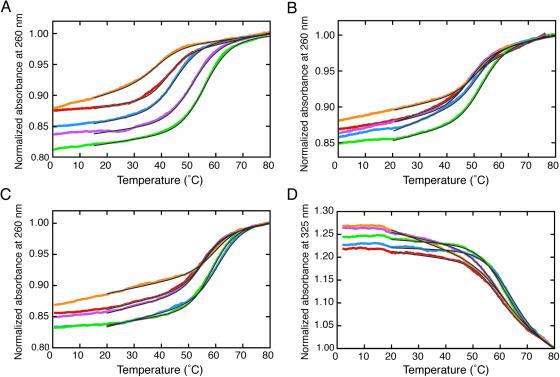
Figure 3A–C shows the typical melting curves of the duplexes monitored at 260 nm. The UV melting curves are consistent that all the duplexes denatured in a two-state transition. The Tm values of the A-series duplexes were in the range from 55.4 to 38.5°C (ΔTm = Tm,max − Tm,min = 16.9°C), while those of the X-series and Z-series duplexes were from 52.2 to 46.6°C (ΔTm = 5.6°C) and from 58.7 to 53.2°C (ΔTm = 5.5°C), respectively, consistent with the fact that the duplexes containing X or Z adopt a similar structure. The melting curves of the Z-series duplexes monitored at 325 nm are also shown in Figure 3D. The absorption at 325 nm by the naphthyl group decreased above 50°C, consistent with the Tm determined from the absorption change monitored at 260 nm. The finding suggests the naphthyl group of Z stacking on nucleotide bases in a duplex and melting simultaneously with the dissociation of nucleotide bases. In addition, the facts that the extinction coefficients of the Z-series duplexes are close (8.2–7.0 × 104 cm−1 M−1 at 325 nm, determined from the absorption at 0°C) and the hypochromicities at 325 nm owing to heat denaturation are similar imply that the naphthyl group assumes a similar environment in a duplex regardless of the type of W2 nucleotide.
Figure 3.
Melting curves of the (A) A-series, (B) X-series and (C) Z-series duplexes monitored at 260 nm [W2 = A (red), G (purple), C (blue), T (green) or F (orange)]. All measurements were carried out at ∼40 µM in the 1 M NaCl-phosphate buffer. The Tm values are as follows: A/A (42.5°C), A/G (51.3°C), A/C (44.5°C), A/T (55.4°C), A/F (38.5°C), X/A (46.6°C), X/G (48.0°C), X/C (49.3°C), X/T (52.2°C), X/F (50.8°C), Z/A (53.4°C), Z/G (53.2°C), Z/C (58.5°C), Z/T (57.4°C) and Z/F (58.7°C). (D) Melting curves of the Z-series duplexes monitored at 325 nm. The fits to a two-state model are indicated in a black line.
Table 1 compares the thermodynamic parameters for the duplex formations obtained from the UV melting curves. The of the duplexes differed depending on the W1/W2 pair. In the A-series DNAs, the fully matched duplex (W1 = A and W2 = T) presented the greatest stability (−12.6 kcal mol−1 in ), and the duplexes containing the A/A, A/G and A/C mismatches showed a lower stability (−8.3 to −10.2 kcal mol−1). The ΔH0 and ΔS0 values of these duplexes also differed considerably (−62.2 to −89.0 kcal mol−1 and −174 to −248 cal mol−1 K−1, respectively), suggesting different interactions near the A/W2 pair. In addition, the duplex containing the abasic site analogue showed −7.4 kcal mol−1 in , which was the lowest stability owing to the lack of the nucleotide base (30,31).
Table 1.
Thermodynamic parameters for the DNA duplex formations with different W1/W2 pairsa
| Sequence | ΔH0 (kcal mol−1) | ΔS0 (cal mol−1K−1) | (kcal mol−1) | Tm (°C) |
|---|---|---|---|---|
| A-Series | ||||
| 5′-GTGTCACTGTC-3′ | −62.2 ± 2.2 | −174 ± 5 | −8.3 ± 0.1 | 45.9 |
| 3′-CACAGAGACAG-5′ | ||||
| 5′-GTGTCACTGTC-3′ | −69.7 ± 1.4 | −192 ± 4 | −10.2 ± 0.3 | 54.7 |
| 3′-CACAGGGACAG-5′ | ||||
| 5′-GTGTCACTGTC-3′ | −72.4 ± 1.9 | −203 ± 3 | −8.9 ± 0.2 | 48.8 |
| 3′-CACAGCGACAG-5′ | ||||
| 5′-GTGTCACTGTC-3′ | −89.0 ± 1.4 | −248 ± 4 | −12.6 ± 0.1 | 57.4 |
| 3′-CACAGTGACAG-5′ | ||||
| 5′-GTGTCACTGTC-3′ | −69.5 ± 2.0 | −199 ± 5 | −7.4 ± 0.2 | 42.9 |
| 3′-CACAGFGACAG-5′ | ||||
| X-series | ||||
| 5′-GTGTCXCTGTC-3′ | −57.5 ± 2.1 | −157 ± 5 | −8.8 ± 0.1 | 50.4 |
| 3′-CACAGAGACAG-5′ | ||||
| 5′-GTGTCXCTGTC-3′ | −56.6 ± 2.5 | −153 ± 7 | −9.2 ± 0.4 | 52.6 |
| 3′-CACAGGGACAG-5′ | ||||
| 5′-GTGTCXCTGTC-3′ | −73.1 ± 2.4 | −202 ± 9 | −10.7 ± 0.5 | 55.2 |
| 3′-CACAGCGACAG-5′ | ||||
| 5′-GTGTCXCTGTC-3′ | −72.8 ± 3.0 | −200 ± 8 | −10.5 ± 0.3 | 55.8 |
| 3′-CACAGTGACAG-5′ | ||||
| 5′-GTGTCXCTGTC-3′ | −73.4 ± 2.2 | −203 ± 5 | −10.4 ± 0.4 | 54.9 |
| 3′-CACAGFGACAG-5′ | ||||
| Z-series | ||||
| 5′-GTGTCZCTGTC-3′ | −67.8 ± 1.8 | −184 ± 5 | −10.5 ± 0.1 | 57.3 |
| 3′-CACAGAGACAG-5′ | ||||
| 5′-GTGTCZCTGTC-3′ | −63.5 ± 1.5 | −172 ± 5 | −10.0 ± 0.2 | 54.8 |
| 3′-CACAGGGACAG-5′ | ||||
| 5′-GTGTCZCTGTC-3′ | −84.7 ± 2.1 | −232 ± 6 | −12.8 ± 0.2 | 60.9 |
| 3′-CACAGCGACAG-5′ | ||||
| 5′-GTGTCZCTGTC-3′ | −80.2 ± 1.7 | −219 ± 7 | −12.3 ± 0.6 | 60.9 |
| 3′-CACAGTGACAG-5′ | ||||
| 5′-GTGTCZCTGTC-3′ | −76.5 ± 2.1 | −209 ± 5 | −11.5 ± 0.2 | 60.2 |
| 3′-CACAGFGACAG-5′ | ||||
aAll experiments were carried out in a buffer containing 1 M NaCl, 10 mM Na2HPO4 and 1 mM Na2EDTA at pH 7.0. Underline indicates the nucleotides forming the W1/W2 pair. Tm was calculated at the total strand concentration of 100 µM.
It is remarkable that the values among the X-series duplexes differed by only 1.9 kcal mol−1 (−8.8 to −10.7 kcal mol−1) while those among the A-series duplexes differed by 5.2 kcal mol−1 (−7.4 to −12.6 kcal mol−1). The differences in the ΔH0 and ΔS0 values among the X-series duplexes (16.8 kcal mol−1 and 50 cal mol−1 K−1, respectively) are also considerably smaller than those among the A-series duplexes (26.8 kcal mol−1 and 74 cal mol−1 K−1, respectively). It is also striking that the thermodynamic parameters of the duplexes containing X opposite the purine base are quite similar (−57.5 and −56.6 kcal mol−1 in ΔH0, −157 and −153 cal mol−1 K−1 in ΔS0, and −8.8 and −9.2 kcal mol−1 in ), and those duplexes containing X opposite the pyrimidine base are identical (−73.1 and −72.8 kcal mol−1 in ΔH0, −202 and −200 cal mol−1 K−1 in ΔS0 and −10.7 and −10.5 kcal mol−1 in ). The same trends were also found in the parameters of the Z-series duplexes.
Comparison of the thermodynamic parameters of the duplexes having different adjacent base pairs
Table 1 demonstrates the results of the duplexes containing X next to two adjacent C/G base pairs (CXC/GW2G, as a trinucleotide expression). In order to investigate influences of the adjacent base pairs on the duplex stability, different sequence sets were further examined. Table 2 summarizes the thermodynamic parameters of the duplexes containing the X/A pair next to two adjacent A/T, G/C or T/A base pairs (AXA/TAT, GXG/CAC or TXT/AAA, respectively) and the corresponding natural duplexes. As demonstrated for CAC/GW2G and CXC/GW2G in Table 1, the displacement of A at W1 by X increased the duplex stability. The increases in for AXA/TAT and GXG/CAC were greater (1.7 and 1.5 kcal mol−1, respectively) than those for CXC/GAG and TXT/AAA (0.5 and 1.1 kcal mol−1, respectively). It is therefore likely that the stabilization energy provided by the displacement of adenine by X is context-dependent, affected by the two adjacent base pairs.
Table 2.
Thermodynamic parameters for the DNA duplex formations with different adjacent base pairsa
| Sequence | ΔH0 (kcal mol−1) | ΔS0 (cal mol−1K−1) | (kcal mol−1) | (kcal mol−1)b | Tm (°C) |
|---|---|---|---|---|---|
| 5′-GTGTAAATGTC-3′ | −55.9 ± 1.9 | −160 ± 4 | −6.6 ± 0.1 | 0 | 35.5 |
| 3′-CACATATACAG-5′ | |||||
| 5′-GTGTAXATGTC-3′ | −61.1 ± 1.6 | −170 ± 4 | −8.3 ± 0.1 | −1.7 | 46.9 |
| 3′-CACATATACAG-5′ | |||||
| 5′-GTGTGAGTGTC-3′ | −73.4 ± 2.4 | −208 ± 8 | −9.1 ± 0.1 | 0 | 47.4 |
| 3′-CACACACACAG-5′ | |||||
| 5′-GTGTGXGTGTC-3′ | −69.4 ± 4.6 | −189 ± 13 | −10.6 ± 0.2 | −1.5 | 56.3 |
| 3′-CACACACACAG-5′ | |||||
| 5′-GTGTCACTGTC-3′ | −62.2 ± 2.2 | −174 ± 5 | −8.3 ± 0.1 | 0 | 45.9c |
| 3′-CACAGAGACAG-5′ | |||||
| 5′-GTGTCXCTGTC-3′ | −57.5 ± 2.1 | −157 ± 5 | −8.8 ± 0.1 | −0.5 | 50.4c |
| 3′-CACAGAGACAG-5′ | |||||
| 5′-GTGTTATTGTC-3′ | −66.0 ± 2.2 | −188 ± 8 | −6.9 ± 0.3 | 0 | 41.9 |
| 3′-CACAAAAACAG-5′ | |||||
| 5′-GTGTTXTTGTC-3′ | −62.9 ± 1.7 | −175 ± 6 | −8.0 ± 0.2 | −1.1 | 47.0 |
| 3′-CACAAAAACAG-5′ |
aAll experiments were carried out in a buffer containing 1 M NaCl, 10 mM Na2HPO4 and 1 mM Na2EDTA at pH 7.0. Underline indicates the nucleotides forming the W1/W2 pair. Tm was calculated at the total strand concentration of 100 µM.
b indicates the difference between the values of the duplexes containing X/W2 and A/W2 with the same adjacent base pairs.
cThe data are derived from Table 1.
Fluorescence emission spectra of a fluorescent base opposite the deoxyadenosine derivative
To reveal the nucleotide conformation opposite the deoxyadenosine derivative, the DNA strands containing the fluorescent base, 2-amino purine (P), were examined. The 2-amino purine base can form a base pair with thymine by forming two hydrogen bonds in the same geometry as the Watson–Crick base pairs, where the fluorescence emission is efficiently quenched by the base stacking (32).
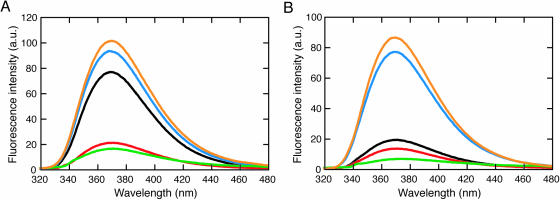
The fluorescence emission spectra were measured for the duplexes containing 2-amino purine opposite A, T and X, in addition to the 2-amino purine nucleotide forced into a single bulge and that in a single strand state (Figure 4). Figure 5A demonstrates the fluorescence emission spectra of 2-amino purine adjacent to two cytosines, 5′-GACACPCACAC-3′. The fluorescence emission around 370 nm was efficiently quenched when thymine or adenine was located opposite the 2-amino purine, resulting from base stacking. In contrast, the duplex containing the X/P pair demonstrated a significant fluorescence emission, comparable with the 2-amino purine nucleotide forced into a bulge and that in a single strand state. This observation suggests that the 2-amino purine opposite X adopts an unstacked conformation similar to the single bulge nucleotide and the nucleotide in a single-strand DNA.
Figure 4.
The duplexes containing the 2-amino purine nucleotide (P) opposite W1 (A, T, X or •), forming the A/P, T/P or X/P pair or a single P-bulge, respectively. The 2-amino purine nucleotide is adjacent to (A) two cytosines or (B) two guanines.
Figure 5.
Fluorescence emission spectra of the duplexes containing 2-amino purine adjacent to two cytosines (A) or two guanines (B), opposite A (red), T (green) or X (blue) in a duplex, in addition to a duplex forming a 2-amino purine bulge (orange) and the single-stranded DNA (black). All measurements were carried out in the 1 M NaCl-phosphate buffer at 4°C.
Figure 5B shows the fluorescence emission spectra of 2-amino purine adjacent to two guanines, 5′-GACAGPGACAC-3′. Unlike the DNA strand containing 2-amino purine adjacent to two cytosines, the fluorescence emission of the single strand was quenched, probably by partial stacking on the adjacent guanines. Nevertheless, the facts that the emission was efficiently quenched when thymine or adenine was opposite the 2-amino purine and that the duplex containing the X/P pair showed a large fluorescence emission comparable with those of the 2-amino purine nucleotide forced into a single bulge also suggest that the 2-amino purine assumes an unstacked form when locating opposite X.
DISCUSSION
Conformation of the DNA duplex containing the deoxyadenosine derivative
We have synthesized the deoxyadenosine derivatives tethering a phenyl group (X) and a naphthyl group (Z) to deoxyadenosine by means of an amide linker. We have previously reported that the single X and Z nucleoside at the 5′ ends of a self-complementary DNA duplex increased the duplex stability by 1.8–3.3 kcal mol−1 in which was equal to or more than the formation of Watson–Crick A/T base pairs (17). This observation is suggestive of a strong stacking interaction of the aromatic hydrocarbon group at a helix terminus and adoption of a geometry analogous to the Watson–Crick base pairs. Here, we investigated the structure and thermodynamic parameters of the duplexes containing X and Z in the middle of a DNA sequence in order to estimate the stacking interaction by the base pair-mimic nucleosides in a duplex.
The conformation of single mismatches in a DNA duplex has been investigated in previous studies: the geometry of the A/G mismatch in a DNA duplex is context-dependent, and it can form a base pair with two hydrogen bonds, differing from Watson–Crick base pairs, which may also interconvert to other pairing geometries (25). The A/A mismatches have a high stacking potential and can form a base pair with one hydrogen bond in anti–anti conformation by disrupting the backbone structure (24,27). The A/C mismatches are unlikely to form a stable base pair at neutral pH, although a pair may be formed with one hydrogen bond depending on the sequence (26). The abasic site perturbs the canonical helix structure and induces additional flexibility (28). Therefore, it can be concluded that most of the differences in the CD spectra in Figure 2A result from the differences in the A/W2 pair geometry. The existence of different interactions among the A-series duplexes is also suggested by the thermodynamic parameters in Table 1 that demonstrate different ΔH0, ΔS0 and values.
Contrary to the observations for the A-series duplexes, the CD spectra in Figure 2B and C suggests that all the X-series or Z-series duplexes adopt a similar geometry at the W1/W2 pair. The CD spectrum of the fully matched duplex (W1 = A and W2 = T) was similar to those of the X-series duplexes and the Z-series duplexes. A relatively large negative peak around 250 nm of the Z-series duplexes, probably partly originating from the naphthyl group, implies that the naphthyl group adopts a stacked conformation regardless of the opposite nucleotide species. One probable explanation for the similarity in the duplex conformations containing X or Z regardless of the type of the W2 nucleotide is that the aromatic hydrocarbon group intercalates into the double helix and causes the W2 nucleotide base to flip into an unstacked position.
The fluorescence emission spectra in Figure 5 indicate that the environment of the 2-amino purine opposite X differs considerably from that opposite adenine or thymine. The fluorescence emission intensity of the X/P pair was comparable with that of the 2-amino purine nucleotide forced into a single bulge and that in a single-strand DNA. Therefore, it is likely that the X opposite 2-amino purine causes the nucleotide base to flip into an unstacked position.
In our previous study, we demonstrated that the duplexes of 5′-XTGCGCA-3′ and 5′-ZTGCGCA-3′ showed a higher stability (−10.5 and −10.9 kcal mol−1, respectively) than those of 5′-XTGCGCAT-3′ and 5′-ZTGCGCAT-3′ with terminal X/T and Z/T pairs (−9.8 and −9.9 kcal mol−1, respectively), implying that the deoxyadenosine derivatives did not form a base pair with the opposing thymine (17). We have also found that the duplexes of an RNA strand associated with the DNA strand containing X showed a similar thermal stability regardless of the ribonucleotide species (A, G, C or U) opposite X and that the phosphorodiester bond of the RNA strand on the 3′-side of the ribonucleotides opposite X was prone to be hydrolysed (33). These observations also support that X induces unstacking of the opposite base regardless of the nucleotide species.
Conformation of the duplexes from the standpoint of the energetics
As suggested by the comparison of the Tm values in Figure 3, the thermodynamic parameters in Table 1 indicate that the value of the A-series duplexes is in the range from −7.4 to −12.6 kcal mol−1 in , which is a substantially larger range than those of the X-series and the Z-series duplexes (−8.8 to −10.7 kcal mol−1 and −10.0 to −12.8 kcal mol−1, respectively). In the A-series duplexes, the duplex stability containing the A/G mismatch is greater than those containing the A/C and A/A mismatches, in agreement with the nearest-neighbor parameters for the single mismatch formations determined previously (25–27). Although incorporation of a single abasic site in the duplex has a strong destabilization effect (5.2 kcal mol−1), the stability is partially restored by the displacement of A by X (3.0 kcal mol−1) or Z (4.1 kcal mol−1) opposite the abasic site. Because the duplex containing F provides a space unoccupied by a nucleotide base, the aromatic hydrocarbon group of X and Z opposite the abasic site can intercalate between the adjacent base pairs, by adopting the amide linker geometry suitable for the aromatic hydrocarbon group stacking in the helix.
In Table 1, the values of the duplexes containing the X/C and X/T mismatches (−10.7 and −10.5 kcal mol−1) are similar to that containing the X/F pair (−10.4 kcal mol−1). Furthermore, the ΔH0 and ΔS0 values of these duplexes are identical within experimental uncertainty (−72.8 to −73.4 kcal mol−1 and −200 to −203 cal mol−1 K−1, respectively), strongly suggesting that the interactions in the duplexes containing the X/pyrimidine pair are similar to those in the duplex containing X/F. This observation also implies that the pyrimidine base unstacking involves lower energy costs and that the rigidity of the duplexes is similar. On the other hand, the thermodynamic parameters of the duplexes containing the X/purine pair are similar (−57.5 and −56.6 kcal mol−1 in ΔH0, −157 and −153 cal mol−1 in ΔS0 and −8.8 and −9.2 kcal mol−1 in ). The same tendencies are also found in the Z-series duplexes. It is also demonstrated that the stability of the duplexes containing X or Z paired with a pyrimidine base was greater than those paired with a purine base and the greater stabilization observed for the duplexes containing the deoxyadenosine/pyrimidine pair results from the ΔH0 term. This observation suggests the existence of a greater penalty for the unstacking of a purine base than a pyrimidine base in duplex formation, probably because the purine bases can stack with adjacent nucleotide bases more efficiently than the pyrimidine bases.
The penalty for the nucleotide base unstacking is also implicated in Table 2. The value in Table 2 that reflects the stabilization energy by the displacement of A at W1 by X is context-dependent, and greater stabilization is observed for AXA/TAT and GXG/CAC (−1.7 and −1.5 kcal mol−1, respectively), while those for CXC/GAG and TXT/AAA are relatively small (−0.5 and −1.1 kcal mol−1, respectively). These results indicate that the adenine base unstacking next to two adjacent purine (Pu) bases (PyXPy/PuAPu) may require greater energy than that next to two adjacent pyrimidine (Py) bases (PuXPu/PyAPy). Table 3 showing the thermodynamic parameters for the trinucleotide formation also indicates lower stabilization energy for the adenine next to two adjacent purine bases, accompanied by larger positive ΔH0 and ΔS0 values. Strong stacking of a purine base next to two adjacent purine bases and less stacking of a purine base next to two adjacent pyrimidine bases were also demonstrated in the fluorescence emission spectra in Figure 5.
Table 3.
Thermodynamic parameters for the trinucleotide formationsa
| Sequence | ΔH0 (kcal mol−1) | ΔS0 (cal mol−1K−1) | (kcal mol−1) |
|---|---|---|---|
| 5′-AXA-3′ | 3.9 | 15 | −0.5 |
| 3′-TAT-5′ | |||
| 5′-GXG-3′ | 1.0 | 9 | −1.4 |
| 3′-CAC-5′ | |||
| 5′-CXC-3′ | 10.7 | 34 | 0.2 |
| 3′-GAG-5′ | |||
| 5′-TXT-3′ | 5.9 | 20 | 0.4 |
| 3′-AAA-5′ |
Energetics for the duplex formations
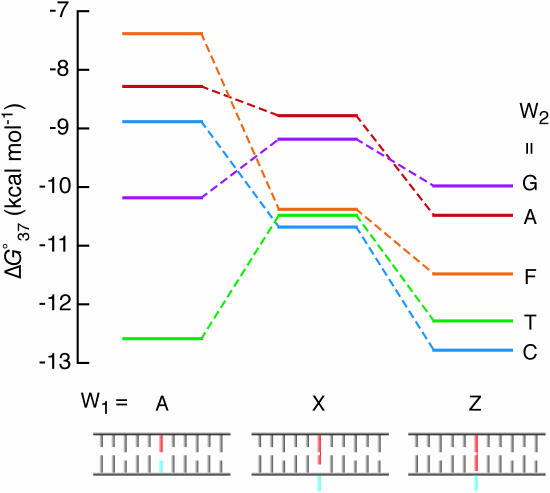
Figure 6 compares the values for the A-series, X-series and Z-series duplex formations. By the displacement of A at W1 by X, the stability of the duplexes with A, C and F at W2 increased, while those with G and T decreased. Stabilization of the duplexes containing A and C at W2 results from the ΔS0 term, consistent with the intercalation of X leading to bury its aromatic hydrocarbon group accompanied by dehydration and/or base unstacking leading to a less constrained conformation. The ΔS0-driven stabilization was also observed for the DNA duplexes containing X and Z at the 5′ dangling ends (17). In contrast, stabilization of the duplex containing the abasic site analogue is achieved by the ΔH0 term, which is in agreement with the occupancy of the abasic site by efficient stacking of the phenyl group of X. On the other hand, destabilization of the duplexes with G and T at W2 results from the loss of enthalpy energy, partly from the loss of the base pair hydrogen bonds by the replacement of A by X at W1. The displacement of X by Z stabilized all the duplexes, and the stabilization originates from the ΔH0 term, probably owing to greater stacking interaction by the naphthyl group. It is probable that Z is more efficient in the base flipping because of incorporation of the larger aromatic group into the duplex, consistent with more similarity in the CD spectra of the Z-series duplexes (Figure 2C) than those of the X-series duplexes (Figure 2B). In addition, the stacking of the aromatic group may also be partly driven by the release of water molecules leading to favorable ΔS0 contributions, as suggested for the adenosine derivatives as a 5′ dangling end (17) and this could be more significant for the larger aromatic group.
Figure 6.
Comparison of the values for the DNA duplex formations. A model of the base flipping is also indicated, containing the W1 (red) and the W2 (blue) nucleotides.
Since it is concluded that the duplexes containing X/T and Z/T adopt a conformation similar to that of other types of duplexes, it is probable that the deoxyadenosine derivatives do not form a base pair with thymine. We have also previously found for the RNA/DNA duplexes containing X that the phosphorodiester bond of an RNA strand on the 3′-side of the ribonucleotides (U as well as A, G, C) opposite X in the DNA strand was hydrolysed with similar efficiency (33). This finding implies that the stacking of the aromatic hydrocarbon group of X and Z is sufficiently strong to disrupt the base pair formation. The ability of the deoxyadenosine derivatives to stack in a duplex is surprising because only the stacking interaction can compensate for the loss of the energy for nucleotide base stacking and interstrand hydrogen bonds. However, it has been reported that a porphyrin with a copper atom binds the DNA duplex of (CGATCG)2 by intercalation between cytosine and guanosine accompanied by extrusion of the terminal cytosine from the helix (34), and that quinoline at the 5′ end of the duplex of (TGCGCA)2 disrupts the terminal T/A base pair formation and stacks on the G/C base pair next to the terminal A/T pair (35). These observations indicate the disruption of the Watson–Crick base pair by interacting with aromatic ring molecules that can efficiently stack with the nucleotide bases. Furthermore, a biascridine macrocyle (BisA) is able to recognize DNA-base mismatches and induce base flipping by the insertion of BisA into the DNA helix displacing the mispaired thymine into an extrahelical position (36). Polycyclic aromatic DNA-base surrogates, such as 1-pyrenyl, 1-napthyl, 2-naphthyl, acenaphthyl and 4-biphenyl residues, have been synthesized and found to disrupt the base stacking of the opposite strand (37,38). It has also been reported that the pyrene nucleotide in a DNA strand opposite uracil acts as a wedge to push the uracil from the base stack and restore the catalytic activity of mutant uracil-DNA glycosylases (39–41). Like the polycyclic aromatic nucleosides, the non-hydrogen-bonding Watson–Crick base pair analogues of X and Z, although these nucleosides do not have a large contiguous π-system but have an amide linker that covalently connects deoxyadenosine with a phenyl group or a naphthyl group, would be useful to flip the target base into an extrahelical position, and the lack of the pairing selectivity would have an advantage for the site-selective base flipping in a target sequence. The base flipping can be used for molecular biology and biotechnology, such as a selective cleavage of a target nucleotide and protein associations that recognize an unpaired nucleotide.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research and ‘Academic Frontier’ Project (2004–2009) from MEXT, Japan. Funding to pay the Open Access publication charges for this article was provided by MEXT, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Turner D.H., Sugimoto N., Kierzek R., Dreiker S.D. Free energy increments for hydrogen bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987;109:3783–3784. [Google Scholar]
- 2.Bommarito S.V., Peyret N., SantaLucia J., Jr Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moody E.M., Bevilacqua P.C. Folding of a stable DNA motif involves a highly cooperative network of interactions. J. Am. Chem. Soc. 2003;125:16285–16293. doi: 10.1021/ja038897y. [DOI] [PubMed] [Google Scholar]
- 4.Morales J.C., Kool E.T. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 5.Kool E.T., Morales J.C., Guckian K.M. Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed. Engl. 2000;39:990–1099. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Hendrickson C.L., Devine K.G., Benner S.A. Probing minor groove recognition contacts by DNA polymerases and reverse transcriptases using 3-deaza-2′-deoxyadenosine. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger M., Luzzi A., Henry A.A., Romesberg F.E. Stability and selectivity of unnatural DNA with five-membered-ring nucleobase analogues. J. Am. Chem. Soc. 2002;124:1222–1226. doi: 10.1021/ja012090t. [DOI] [PubMed] [Google Scholar]
- 8.Liu H., Gao J., Maynard L., Saito D., Kool E.T. Toward a new genetic system with expanded dimensions: size-expanded analogues of deoxyadenosine and thymidine. J. Am. Chem. Soc. 2004;126:1102–1009. doi: 10.1021/ja038384r. [DOI] [PubMed] [Google Scholar]
- 9.Arkin M.R., Stemp E.D., Pulver S.C., Barton J.K. Long-range oxidation of guanine by Ru(III) in duplex DNA. Chem. Biol. 1997;4:389–400. doi: 10.1016/s1074-5521(97)90129-0. [DOI] [PubMed] [Google Scholar]
- 10.Kuzuya A., Mizoguchi R., Morisawa F., Machida K., Komiyama M. Metal ion-induced site-selective RNA hydrolysis by use of acridine-bearing oligonucleotide as cofactor. J. Am. Chem. Soc. 2002;124:6887–6894. doi: 10.1021/ja025653p. [DOI] [PubMed] [Google Scholar]
- 11.Lermer L., Roupioz Y., Ting R., Perrin D.M. Toward an RNaseA mimic: a DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002;124:9960–9961. doi: 10.1021/ja0205075. [DOI] [PubMed] [Google Scholar]
- 12.Lewis F.D., Liu J., Weigel W., Rettig W., Kumikov I.V., Beratan D.N. Donor-bridge-acceptor energetics determine the distance dependence of electron tunneling in DNA. Proc. Natl Acad. Sci. USA. 2002;99:12536–12541. doi: 10.1073/pnas.192432899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K., Tengeiji A., Kato T., Toyama N., Shionoya M. A discrete self-assembled metal array in artificial DNA. Science. 2003;299:1212–1213. doi: 10.1126/science.1080587. [DOI] [PubMed] [Google Scholar]
- 14.Liu H., Gao J., Lynch S.R., Saito Y.D., Maynard L., Kool E.T. A four-base paired genetic helix with expanded size. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- 15.Amann N., Huber R., Wagenknecht H.A. Phenanthridinium as an artificial base and charge donor in DNA. Angew. Chem. Int. Ed. Engl. 2004;43:1845–1847. doi: 10.1002/anie.200353153. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto A., Ichiba T., Saito I. Pyrene-labeled oligodeoxynucleotide probe for detecting base insertion by excimer fluorescence emission. J. Am. Chem. Soc. 2004;126:8364–8365. doi: 10.1021/ja049061d. [DOI] [PubMed] [Google Scholar]
- 17.Nakano S., Uotani Y., Nakashima S., Anno Y., Fujii M., Sugimoto N. Large stabilization of a DNA duplex by the deoxyadenosine derivatives tethering an aromatic hydrocarbon group. J. Am. Chem. Soc. 2003;125:8086–8087. doi: 10.1021/ja034465b. [DOI] [PubMed] [Google Scholar]
- 18.Kierzek R., Caruthers M.H., Longfellow C.E., Swinton D., Turner D.H., Freier S.M. Polymer-supported RNA synthesis and its application to test the nearest-neighbor model for duplex stability. Biochemistry. 1986;25:7840–7846. doi: 10.1021/bi00372a009. [DOI] [PubMed] [Google Scholar]
- 19.Sugimoto N., Nakano S., Yoneyama M., Honda K. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res. 1996;24:4501–4505. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano S., Kanzaki T., Sugimoto N. Influences of ribonucleotide on a duplex conformation and its thermal stability: study with the chimeric RNA–DNA strands. J. Am. Chem. Soc. 2004;126:1088–1095. doi: 10.1021/ja037314h. [DOI] [PubMed] [Google Scholar]
- 21.Richards E.G. Use of tables in calculation of absorption, optical rotatory dispersion and circular dichroism of polyribonucleotides. In: Fasman G.D., editor. Handbook of Biochemistry and Molecular Biology, 3rd edn. Vol. 1. Cleveland, OH: CRC Press; 1975. pp. 596–603. [Google Scholar]
- 22.Marky L.A., Breslauer K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 23.Puglisi J.D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 24.Maskos K., Gunn B.M., LeBlanc D.A., Morden K.M. NMR study of G.A and A.A pairing in (dGCGAATAAGCG)2. Biochemistry. 1993;32:3583–3595. doi: 10.1021/bi00065a009. [DOI] [PubMed] [Google Scholar]
- 25.Allawi H.T., SantaLucia J., Jr Nearest neighbor thermodynamic parameters for internal G.A mismatches in DNA. Biochemistry. 1998;37:2170–2179. doi: 10.1021/bi9724873. [DOI] [PubMed] [Google Scholar]
- 26.Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry. 1998;37:9435–9444. doi: 10.1021/bi9803729. [DOI] [PubMed] [Google Scholar]
- 27.Peyret N., Seneviratne P.A., Allawi H.T., SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 28.Barsky D., Foloppe N., Ahmadia S., Wilson D.M., III., MacKerell A.D., Jr New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res. 2000;28:2613–2626. doi: 10.1093/nar/28.13.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugimoto N., Nakano M., Nakano S. Thermodynamics-structure relationship of single mismatches in RNA/DNA duplexes. Biochemistry. 2000;39:11270–11281. doi: 10.1021/bi000819p. [DOI] [PubMed] [Google Scholar]
- 30.Gelfand C.A., Plum G.E., Grollman A.P., Johnson F., Breslauer K.J. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 31.Sági J., Guliaev A.B., Singer B. 15-mer DNA duplexes containing an abasic site are thermodynamically more stable with adjacent purines than with pyrimidines. Biochemistry. 2001;40:3859–3868. doi: 10.1021/bi0024409. [DOI] [PubMed] [Google Scholar]
- 32.Pompizi I., Häberli A., Leumann C.J. Oligodeoxynucleotides containing conformationally constrained abasic sites: a UV and fluorescence spectroscopic investigation on duplex stability and structure. Nucleic Acids Res. 2000;28:2702–2708. doi: 10.1093/nar/28.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano S., Uotani Y., Uenishi K., Fujii M., Sugimoto N. Site-selective RNA cleavage by DNA bearing a base pair-mimic nucleoside. J. Am. Chem. Soc. 2005;127:518–519. doi: 10.1021/ja045445s. [DOI] [PubMed] [Google Scholar]
- 34.Lipscomb L.A., Zhou F.X., Presnell S.R., Woo R.J., Peek M.E., Plaskon R.P., Williams L.D. Structure of DNA-porphyrin complex. Biochemistry. 1996;35:2818–2823. doi: 10.1021/bi952443z. [DOI] [PubMed] [Google Scholar]
- 35.Tuma J., Connors W.H., Stitelman D.H., Richert C. On the effect of covalently appended quinolones on termini of DNA duplexes. J. Am. Chem. Soc. 2002;124:4236–4246. doi: 10.1021/ja0125117. [DOI] [PubMed] [Google Scholar]
- 36.David A., Bleimling N., Beuck C., Lehn J.-M., Weinhold E., Teulade-Fichou M.-P. DNA mismatch-specific base flipping by a bisacridine macrocycle. Chembiochem. 2003;4:1326–1331. doi: 10.1002/cbic.200300693. [DOI] [PubMed] [Google Scholar]
- 37.Singh I., Hecker W., Prasad A.K., Parmar V.S., Seitz O. Local disruption of DNA-base stacking by bulky base surrogates. Chem. Commun. 2002;7:500–501. doi: 10.1039/b110842e. [DOI] [PubMed] [Google Scholar]
- 38.Beuck C., Singh I., Bhattacharya A., Hecker W., Parmar V.S., Seitz O., Weinhold E. Polycyclic aromatic DNA-base surrogates: high-affinity binding to an adenine-specific base-flipping DNA methyltransferase. Angew. Chem. Int. Ed. Engl. 2003;42:3958–3960. doi: 10.1002/anie.200219972. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y.L., Kwon K., Stivers J.T. Turning On uracil-DNA glycosylase using a pyrene nucleotide switch. J. Biol. Chem. 2001;276:42347–42354. doi: 10.1074/jbc.M106594200. [DOI] [PubMed] [Google Scholar]
- 40.Jiang Y.L., Stivers J.T., Song F. Base-flipping mutations of uracil DNA glycosylase: substrate rescue using a pyrene nucleotide wedge. Biochemistry. 2002;41:11248–11254. doi: 10.1021/bi026227j. [DOI] [PubMed] [Google Scholar]
- 41.Kwon K., Jiang Y.L., Stivers J.T. Rational engineering of a DNA glycosylase specific for an unnatural cytosine:pyrene base pair. Chem. Biol. 2003;10:351–359. doi: 10.1016/s1074-5521(03)00077-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.