Abstract
Multiple sequence alignments play a central role in the annotation of novel genomes. Given the biological and computational complexity of this task, the automatic generation of high-quality alignments remains challenging. Since multiple alignments are usually employed at the very start of data analysis pipelines, it is crucial to ensure high alignment quality. We describe a simple, yet elegant, solution to assess the biological accuracy of alignments automatically. Our approach is based on the comparison of several alignments of the same sequences. We introduce two functions to compare alignments: the average overlap score and the multiple overlap score. The former identifies difficult alignment cases by expressing the similarity among several alignments, while the latter estimates the biological correctness of individual alignments. We implemented both functions in the MUMSA program and demonstrate the overall robustness and accuracy of both functions on three large benchmark sets.
INTRODUCTION
The alignment of multiple protein sequences is central in the annotation of genomes (1). Given the pivotal role of multiple sequence alignments (MSAs), the field has received a lot of attention in the recent years. Several new and accurate alignment methods have been introduced (2–5) and even relatively minor algorithmic details attract attention (6). A critical, yet largely unsolved, problem in the field is how to assess the quality of alignments.
When benchmarking alignment programs, reference alignments are used as the gold standard against which test alignments are compared. The accuracy of an individual method is then described by the average score over all test cases in a benchmark test set. The hope is that the test sets are comprehensive and that high accuracies are transferable to real cases.
A problem here is the use of an overall average to describe the accuracy of alignment programs. High scores often reflect the inherent easiness of the benchmark set rather than potentially beneficial properties of an alignment program. Furthermore, in difficult alignment cases all programs typically fail to reflect the biological relations between the sequences, and reporting whether method A fails less than method B is inconsequential. Hence, Notredame (7) suggested that it is more important to assess the quality of individual cases than improving average accuracies.
So why is it so difficult to produce multiple sequence alignments in the first place? The problem can be split up into the following areas: the choice of sequence, the objective function used and the heuristics (7). All alignment programs to date require homologous sequences as input. If this central requirement is not met, alignments become meaningless. This may happen for example when using Blast (8,9) or FASTA (10,11) to gather input sequences as they can potentially collect non-homologous sequences if only part of the sequences match.
Second, the majority of alignment programs available today require that the sequences are related to each other in a linear fashion. Proteins related to each other through a process other than short insertions, deletions and substitutions simply do not fit the models used within alignment programs. Good examples are multi-domain proteins containing repeated domains and, less frequently, domains in different orders. Notable exceptions are the alignment methods Poa (12,13) and ABA (14) which model domain recombinations and shuffled or repeated domains, respectively.
Third, the choice of objective function and heuristics is critically important in obtaining high quality alignments. Most of the work in this field is focused on this area, exemplified by the large number of alignment programs that emerged recently. We will not give a comprehensive review of the methods here, but we will focus on the main problem they all share: the choice of parameters. It is clear that the choice of alignment parameters, especially gap penalties (15), has a substantial effect on the alignment quality (16,17). Furthermore, the choice of gap penalties can have a bigger effect than the choice of alignment program (18). Clearly, default parameter settings only give the best results in few cases, and hence it is preferable to have a method insensitive to parameters (19).
Finally, there are cases in which sequences cannot be aligned unambiguously (20). In such cases, a biologist has to decide which alignment is the most appropriate for the purpose at hand. For example, a structure-based alignment might be used for the identification of a residue critical to function, while an evolutionary-based alignment may be more appropriate for phylogenetic reconstruction.
Clearly, it is unlikely that all of the before mentioned sources of problems will be eliminated in the near future; hence, the automatic alignment of multiple sequences will remain a challenging area of research. So how can a molecular biologist decide on which alignment program in combination with which parameters will give the most biologically correct alignment in a specific case?
Our solution is to use several alignment programs and cross-examine the resulting alignments. The underlying idea of comparing alignments of the same sequences in the context of accuracy assessment is not new (21–23) and the concept of consistency (24) has been used to increase the accuracy of multiple sequence alignment algorithms (2,3,25,26). In addition, the ComAlign algorithm (27) combines several multiple sequence alignments into one improved alignment. Here, we distinguish between two types of consistency: intra-consistency being the consistency between pairwise alignments within a single multiple alignment and inter-consistency being the consistency, or better similarity, between alternate pairwise alignments of the same sequences. The former is applied in consistency-based alignment algorithms while the latter is primarily used to compare of alignments. The T-Coffee program is unique in that it uses both types of consistency: inter-consistency to create a weighted library of pairwise alignments and intra-consistency to extend this library. Given such an extended library, T-Coffee can also assess the support for a given multiple alignment (28).
In this paper, we use an approach that employs solely inter-consistency for the purpose of assessing alignment quality. As such, our approach is conceptually similar to the evaluation part of T-coffee. Essentially, we search for regions which are identically aligned in many alignments, assuming that these are more reliable than regions differently aligned in many alignments. By doing so our method can establish the difficulty of each alignment case and assess the biological correctness or quality of individual alignments. For clarification, we define an alignment case as a set of sequences, for example globins, to be aligned. The important point here is that our method works independently of the presence of reference alignments or the presence of secondary information that can be utilized to determine the quality (29). Hence, our method can be applied to any alignment that may arise in molecularbiology.
We envision our method to be used in two main applications: first, as a quality filter in large scale automatic or semi-automatic genome annotation pipelines and second as a tool to facilitate the critical assessment of individual alignments by human experts. Indirectly, we also hope that our method will stimulate the development of alignment methods by focusing the attention of alignment program developers (ourselves included) to real biological problems in this field.
MATERIALS AND METHODS
Algorithm overview
Our approach is based on the comparison of multiple sequence alignments. To represent each alignment, we use the concept of pairs-of-aligned residues. For example, one such pair might consist of the following statement: residue 3 in sequence 1 aligned to residue 5 in sequence 7. All such pairs are extracted from all input alignments m. Effectively, each alignment is atomized into the set of their smallest components that still allows reassembly of the original alignment. The intersection between two such sets, or input alignments, represents the regions which are aligned identically. In practice, this is usually a biologically conserved block, while the remainder of the sequences are less conserved regions. The overlap score O (22), reflecting the similarity between two alignments Qa and Qb, is defined as the ratio between the cardinality of the intersection of two sets of aligned residues and the average cardinality of each set:
| 1 |
We define the difficulty of an alignment case by the average overlap score between all input alignments:
| 2 |
Basically, this is a crude measure of how dispersed alignments are in the space of all solutions. In simple alignment cases, alignment programs produce similar alignments and the average overlap will approach 1, while in difficult cases the score approaches 0.
To calculate the accuracy of individual alignments we assign scores to each pair of aligned residues reflecting their proliferation in all alignments: Let n(σ) be the number of m − 1 alignments that contain σ. A pair occurring in all alignments is thus given the highest score (m − 1) while a pair occurring in a single alignment is given the lowest score of zero. These scores are then summed for alignment Qa to determine its multiple overlap score (MOS):
| 3 |
The numerator sums up the scores of each pair of aligned residues occurring in alignment Qa. The denominator reflects the maximum possible score, i.e. if all pairs of aligned residues in alignment Qa occur in all m alignments. Basically, aligned residues that are found in many alignments are more reliable, and the alignment with the highest number of such pairs is assumed to be the most biologically correct one.
Implementation
We implemented both score functions in the MUMSA program written in C. By default, the program produces a summary of the results and two graphs generated by the R package (30) (if available). The first is a histogram of similarities between the alignments while the second is a tree showing the relations between the alignment programs. Computationally, the program is very fast and its running time negligible compared with the running time of the alignment programs used to generate the input alignments. The program is freely available under the GNU license upon request from the author. An on-line server is available at http://msa.cgb.ki.se where users can submit their alignments to be analyzed.
Testing methodology
To demonstrate the predictive power of our method we used three alignment benchmark sets, Balibase (31,32), Prefab (5) and SABmark (33) in combination with seven commonly used multiple alignment programs (Table 1). We ran all programs with default parameters on all test sets and assessed the ‘true’ alignment accuracy by comparison to the reference alignments. In addition, we ran Muscle in three different ways and Mafft in four different ways, for a total of 12 automatically generated alignments for each test case in the databases.
Table 1.
Alignment methods and parameters used in this study
| Method | Description/Options |
|---|---|
| Poa version 2 (12,13) | Local unprogressive mode using blosum80.mat |
| ClustalW version 1.83 (36) | Default parameters |
| Muscle version 3.52 (5) | One iteration: -stable -maxiters 1 |
| Two iterations: -stable -maxiters 2 | |
| Default: -stable | |
| Probcons version 1.09 (2) | Default parameters |
| Dialign version 2.2 (21,23) | Default parameters |
| Mafft version 5.63 (3,37) | -Localpair |
| -Localpair -maxiterate 100 | |
| -Globalpair | |
| -Globalpair -maxiterate 100 | |
| Kalign version 1.03 (manuscript submitted) | Default parameters |
In total, we generated 30 408 alignments (12 for each of the 218, 1682, 425 and 209 test cases in Balibase, Prefab, SABmark ‘superfamily’ and SABmark ‘twilight’, respectively) for which we could determine their real accuracy through comparisons to reference alignments.
Benchmark sets
We used the Balibase 3.0, Prefab 4.0 and SABmark 1.65 alignment benchmark set for the validation of our method.
Balibase contains 218 alignment cases, each of which contains two reference alignments: an alignment of only partial, or truncated, sequences and an alignment of the same full-length sequences. Here, we choose to use only the full-length alignments as we feel that this agrees more with real applications. The Prefab 4.0 benchmark test set contains two sets of benchmark cases: a standard set similar to the one found in previous versions and a weighted set. The latter contains over-represented sub-families of sequences and is designed to test programs for their ability to weight sequences differently. Since the benchmarking of the alignment programs themselves is not the focus here, we limited our analysis on the main test set containing 1686 test cases. Similarly, we limited our analysis to the standard SABmark test sets (Superfamily, 425 cases and Twilight, 209 cases) and omitted the false-positive test sets, designed to test the ability of programs to detect non-homologs.
Alignment programs
For this study, we chose to generate alignments using a multitude of alignment programs. It has been shown on the Balibase benchmark set that methods with a very low average accuracy outperform the best methods in many individual cases (5). In practice, it is therefore always recommended to use as many different methods. We therefore did not restrict our analysis to only a few of the best alignment methods but aimed to use as many methods as possible. Moreover, we did not make any attempts to optimize the set of programs used here to inflate the accuracy of MUMSA and based our choice of programs solely on practical issues, such as computational aspects and past experience. A detailed list of the programs we used and their respective options are summarized in Table 1. We ran all programs with default parameters on all test sets. In addition, we ran Muscle in three different ways and Mafft in four different ways, for a total of 12 automatically generated alignments for each test case in the databases. The motivation for running these two programs several times was the fact that alignments generated prior to the respective iterative refinement are often quite different from the final alignment.
We attempted to use T-Coffee (25), but found it to be too memory demanding.
Other quality assessment programs
We compared our method against norMD (34) and al2co (35). Both of these programs analyze individual alignments column by column and have both been suggested for the analysis of alignment quality. In addition, we calculated the average sequence identity for each alignment as a measure of quality. The assumption is that alignments with a high average identity are more accurate than alignments of the same sequences with low identity.
Quality assessment
For our analysis, the assessment of the real accuracy using reference alignments is of paramount importance since we use the resulting values as a gold standard against which we compare the performance of our method. Each of the benchmark test set used here comes with its own program to calculate the accuracy of test alignments compared with the respective reference alignments. In the case of Balibase, we used the bali_score program to assess the accuracy of each test case. For each of the reference alignments, core blocks, or reliably aligned regions, are defined. The accuracy of alignments can be defined based on core blocks or on the whole alignment. We chose to use the core block definition to achieve the most reliable accuracy values. Additionally, the accuracy of test alignments can be reflected using two scores: the column score (CS) and the sum-of-pairs score (SP). The column score looks for identical columns in reference and test alignments and can become 0 if only one sequence is misaligned. We chose to use the SP score for our analysis here.
The Prefab database consists of pairwise structural alignments to which a number of homologs have been added to make up multiple sequence alignment cases. Programs generate alignments from all sequences but the accuracy (here Q score) of each individual case is then only defined based on the pairwise alignment of the two original sequences. This represents a problem since the accuracies estimated by MUMSA, norMD and al2co are calculated from whole alignments. To make our results comparable, we chose to remove all added sequences from the completed alignments and calculated the average overlap, MOS and norMD scores based on the alignment of the original two sequences. This procedure is analogous to the one used to calculate the Q scores. In 25 out of the 20 184 alignments (12 alignments for each of 1682 test cases), or 0.1%, our program failed to produce meaningful scores because in these cases no residues were aligned anywhere within the pairwise alignment. Such cases are artifacts of the testing procedure, i.e. extracting pairwise alignments from multiple alignments, and similar cases do not occur in real application. The al2co program produced no meaningful results on pairwise alignments and was omitted from this test.
The SABmark benchmark is supplied with scripts that automatically assess the accuracy of individual alignments. A minor complication is that the accuracies (here fD scores) for each case are expressed as a list of scores for each of the pairwise alignments contained within the resulting multiple sequence alignments. For example, an alignment of 10 sequences will have 45 pairwise fD scores. To obtain the accuracy of the whole alignment we took the average of all the pairwise scores. Occasionally, in cases when no residues were aligned in the reference alignments, no meaningful fD score is reported. In such cases, we omitted the score from the calculation of the average (Ivo Van Walle, personal communication).
RESULTS
Benchmark difficulty
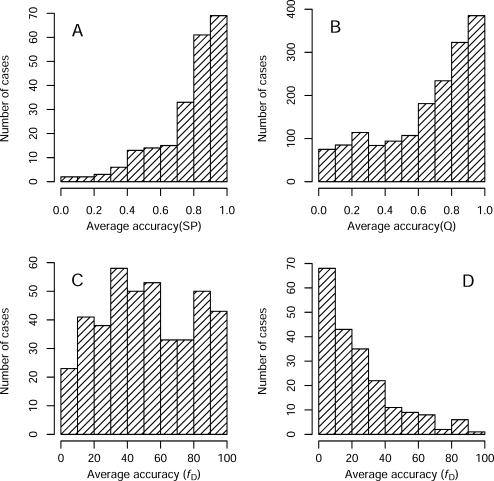
First, we wanted to look at how challenging each of the alignment benchmark test sets are. For this purpose, we calculated the average accuracy of the 12 alignment methods for each test case. The accuracy was measured by the method provided with each benchmark, which all range from 0 to 100%. We asserted that the average accuracy of all the alignment programs was low in difficult alignment cases and high in easy cases. By enumerating the number of difficult and easy cases we can establish the overall difficulty of each benchmark (Figure 1). This strategy is more direct and practical than attempting to differentiate among alignments based on features such as large internal deletions or the presence of remote homologs. The histograms for Balibase and Prefab (Figure 1A and B) reveal that in the majority of cases all alignment programs produce correct alignments and hence we conclude that these benchmark sets are comparatively simple. The super-family subset of SABmark contains a roughly equal number of difficult and easy cases. The ‘twilight’ subset of SABmark is the most challenging test set and most alignment programs produce very low scoring, or incorrect, alignments.
Figure 1.
Histograms of the distribution of difficult/easy alignment cases in Balibase (A), Prefab (B), SABmark superfamily (C) and the SABmark twilight (D) benchmark test sets. The accuracy of each alignment was calculated by comparison to reference alignments using the sum-of-pairs (SP), Q and fD scores, respectively (see Materials and Methods). The SABmark twilight set consists of predominantly difficult cases while Balibase and Prefab sets contains mainly easy cases. The superfamily subset of SABmark is made up of an equal number of difficult and easy alignment cases.
Determining alignment difficulty
The first use of MUMSA is to predict the difficulty of an alignment case. We consider the average accuracy (same as above) a good measurement for the difficulty of an alignment. Simply put, a case in which all methods fail to reproduce a reference alignment is probably more difficult than a case in which all methods produce similarly correct alignments. We ran MUMSA on all 12 alignments for each benchmark case and also calculated the average overlap score (AOS) between them. Our hypothesis states that in difficult cases alignment programs will generate more dissimilar alignments than in trivial cases. On all four test sets we found a clear correlation between the average accuracy scores and the average overlap scores (Figure 2), indicating the good predictive power of this method. The correlation coefficient was lowest in the Balibase test set. The reason is that the accuracy was assessed based only on core blocks, or reliable regions within alignments, while MUMSA operates on whole alignments (see Materials and Methods). Basically, we added noise from ambiguous regions which were excluded when the real accuracy was calculated. Despite of this the correlation coefficient was high (0.81), indicating that several alignments of the same case differ primarily in unconserved rather than in conserved regions. An important question is how high the average overlap score should be to be of practical use in real genome annotation applications. Since alignments usually represent the initial step in annotation pipelines, only high quality alignments should be accepted. At a cutoff of 0.8 average overlap, MUMSA detects between 96 and 100% of all alignment cases in which the accuracy is below 80%.
Figure 2.
Scatter-plots of estimated case difficulty using the average overlap score versus real difficulty: Balibase (A), Prefab (B), SABmark superfamily (C) and SABmark twilight (D). The Pearson correlation coefficient (r) is high for all test sets.
Determining alignment quality
The second objective of MUMSA is to assess the quality of automatically generated alignments. Using the same datasets, we ran MUMSA on all 12 alignments for each test case and compared the MOSs with the real accuracy scores for each alignment. For comparison, we also evaluated alignments using al2co, the norMD objective function and the average sequence identity. To present the results, we calculated the correlation coefficients between the estimated and real accuracy scores for each benchmark set (Table 2). For visual inspection, we included corresponding scatter plots for norMD and MUMSA in the Supplementary Data. On all test sets the scores produced by our method are better correlated to the real alignment accuracies than scores produced by other methods. For all methods the correlation coefficient is the lowest in the case of Balibase. Again, this is due to the fact that the real accuracy here is estimated from core regions while both methods take whole alignments as input. An unexpected result was the comparatively high correlation coefficient for the SABmark twilight test set. Given the overall difficulty of this test set (Figure 1D), we expected the accuracy of our method to deteriorate just as the accuracy of the alignment programs does. However, the correlation coefficient remains high (0.78, only 8% points less than the SABmark superfamily subset) and we conclude that our estimation of alignment quality is largely independent of the difficulty of each alignment case. Moreover, our method is equally accurate at predicting the quality of easy and difficult alignments. Since we hope our method will be used as a quality control measure this inherent robustness is a very important feature.
Table 2.
Pearson correlation coefficients between real alignment accuracy and predicted accuracy by MOS (bold), norMD, al2co and the average sequence identity
| Balibase | Prefab | SABmark sup | SABmark twi | |
|---|---|---|---|---|
| MOS | 0.76 | 0.87 | 0.86 | 0.78 |
| NorMD | 0.50 | 0.56 | 0.66 | 0.54 |
| Average sequence ID | 0.61 | 0.51 | 0.65 | 0.44 |
| Al2co 1_1 | 0.07 | — | 0.32 | 0.32 |
| Al2co 1_2 | 0.04 | — | 0.31 | 0.32 |
| Al2co 1_3 | −0.05 | — | 0.29 | 0.32 |
| Al2co 2_1 | 0.23 | — | 0.37 | 0.34 |
| Al2co 2_2 | 0.18 | — | 0.37 | 0.35 |
| Al2co 2_3 | 0.01 | — | 0.33 | 0.34 |
| Al2co 3_1 | 0.28 | — | 0.39 | 0.35 |
| Al2co 3_2 | 0.22 | — | 0.38 | 0.35 |
| Al2co 3_3 | 0.06 | — | 0.35 | 0.34 |
For al2co, the first number refers to the way conservation was calculated: 1, entropy-based measure; 2, variance-based measure; 3, sum-of-pairs measure. The second number refers to the weighting strategy used: 1, Unweighted amino acid frequency; 2, Henikoff weighting scheme; 3, estimated independent counts.
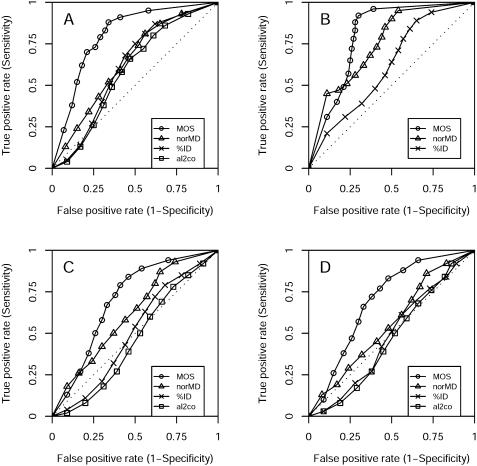
Another interesting question is how well the ranking according to the MOS corresponds to the real ranking of the test alignments. For example, is the alignment with the highest MOS really the most accurate alignment? To make this assessment, we ranked the 12 alignments for each test case in the databases according to their accuracy as well as according to the MOS and performed a standard sensitivity/specificity analysis (Figure 3). In comparison with the other approaches, our predictor is clearly superior, accepting fewer false-positive predictions at comparable levels of true positives. The AUC (area under ROC curve) values, reflecting the likelihood of making correct predictions, are consistently higher for our method than for norMD, al2co and average identity (Table 3). The performance of all methods drops noticeably on both SABmark test sets, but the same value was obtained for both, underscoring MUMSA's robustness on difficult cases. Intuitively, the lower performance is expected as these test sets are the most challenging ones used here (Figure 1). Basically, it is more difficult for our method to determine the correct ranks among several equally incorrect alignments than among a mixture of moderately correct alignments. Nevertheless, the accuracy of our predictor remains high even in the most difficult alignment cases.
Figure 3.
ROC curves demonstrating the agreement between real and predicted rank of several alignments of the same sequences: Balibase (A), Prefab (B), SABmark superfamily (C) and SABmark twilight (D). For al2co, we only show the best curve among all 9 combination of methods (Table 3, italic). For the Prefab set no meaningful results could be obtained using al2co. The rankings based on our MOSs are more accurate than the rankings according to norMD, al2co and sequence identity scores, accepting fewer false positives at comparable levels of of true positives. The predictions of all scores are less accurate on the SABmark sets than on Balibase and Prefab.
Table 3.
The AUC (area under the ROC curve) values for each benchmark set and method
| Balibase | Prefab | SABmark sup | SABmark twi | |
|---|---|---|---|---|
| MOS | 0.80 | 0.80 | 0.70 | 0.70 |
| NorMD | 0.64 | 0.75 | 0.62 | 0.56 |
| Average sequence ID | 0.62 | 0.62 | 0.51 | 0.48 |
| Al2co 1_1 | 0.55 | — | 0.45 | 0.44 |
| Al2co 1_2 | 0.56 | — | 0.44 | 0.44 |
| Al2co 1_3 | 0.55 | — | 0.44 | 0.44 |
| Al2co 2_1 | 0.56 | — | 0.45 | 0.46 |
| Al2co 2_2 | 0.58 | — | 0.46 | 0.46 |
| Al2co 2_3 | 0.57 | — | 0.45 | 0.46 |
| Al2co 3_1 | 0.57 | — | 0.46 | 0.47 |
| Al2co 3_2 | 0.59 | — | 0.46 | 0.47 |
| Al2co 3_3 | 0.59 | — | 0.46 | 0.46 |
On all benchmark sets out method (bold) is superior to norMD, al2co and the average sequence identity. See Table 2 for description of the al2co modes. All methods are less accurate at predicting the correct rank of alignments on the SABmark benchmark sets.
In conclusion, the MOS values for individual alignments are well correlated to the real accuracies in the overwhelming majority of cases. When multiple alignments of the same sequences are present, the MOS is sufficiently accurate to differentiate between correct and incorrect alignments.
Additionally, the ranking of alignments according to the MOS usually reflects the true ranking of the alignments, but becomes slightly flawed when the alignment cases themselves are difficult. In practice, however, the alignments in such cases are equally incorrect, and hence the correct ranking becomes less important.
The comparison with norMD clearly indicates the superiority of our method. However, the norMD objective function has the notable advantage of being able to be applied to single alignments, while our method requires several alignments—we used 12 here as input. In a sense, our method has several times the input and taking this into account, the norMD score performs considerably well and is more practical. Nevertheless, when aiming for optimal assessment of alignment accuracy, the advantage of our strategy, based on the comparison of several multiple alignments, is apparent.
How important is the selection of input alignments?
For this study, we used a selection of alignment methods to generate the alignments to be analyzed by MUMSA. Intuitively, we used as many different alignment methods as possible for this study. A valid question is whether comparably good results can be obtained using fewer input alignments or whether a certain selection of methods can further improve MUMSAs performance. It is beyond this paper to answer these questions satisfactorily. Preliminary results suggest that the accuracy of MUMSA is slightly decreased when fewer input alignments are used (Supplementary Data). Although it is possible to optimize the choice of input alignments, the accuracy estimates presented here are much better than any previously published. More importantly, they are good enough to be used in real applications. We therefore recommend to use as many different input alignments as practically possible.
DISCUSSION
The alignment of multiple sequences remains a challenging problem today. Here, we do not discuss possible strategies to improve alignment quality, but instead focus on the equally important task of assessing the biological correctness of automatically generated alignments. Of course, both problems are related: being able to diagnose problematic alignments is the first step in improving quality, and a function used to assess the quality of completed alignments can potentially be used in reverse as an objective function in an alignment algorithm.
To achieve the goal of assessing alignment quality, we introduced the concept of comparing several multiple sequence alignments of the same sequences. It is generally recommended to always run several multiple sequence alignment programs and compare their results to find the most suited alignment. In previous studies, we noticed that alignments, whether created by the same program with different parameters or by different programs altogether, often agreed in the way conserved blocks are aligned. Therefore, we developed a strategy to identify identically aligned residues across several alignments in order to determine the alignment sharing the most aligned residues with other alignments.
Conversely, requiring several multiple sequence alignments is also a drawback of our method. It is practical or computationally feasible to perform many alignments, especially when the volume of data is high? To answer this question, we have to look at the computational properties of current alignment programs. In our experience, high quality alignment programs take at least one order of magnitude longer to align sequences than faster but less accurate alignment programs. So by the time one high quality alignment is produced, several other alignments can be produced alongside by faster methods. The extra time penalty required to use our method is therefore negligible in practice when compared with only using one slow and accurate alignment program.
A key point of this work is the ability of our method to identify difficult or even un-alignable sets of sequences. In large scale automatic annotation projects, the quality of alignments is of paramount importance. By being able to diagnose difficult alignments early, our method guarantees that only high-quality alignments enter annotation pipelines. At the same time, our method helps to focus the attention of human experts to the more difficult, and usually more interesting cases. The workload of annotators should also be reduced, since the alignment of most protein families is relatively simple. Here, our method correctly identifies the case as an easy one, making manual inspection or refinement largely unnecessary. Our extensive benchmark tests revealed the high accuracy of our method. The overall message is clear: in easy cases alignment programs produce similar alignments while in difficult cases the alignments tend to vary to a much greater extent.
The second use of our method is to assess the accuracy of individual alignments. We demonstrated that the MOSs correlates well with the real accuracy values determined by comparisons to reference alignments. Our method was also accurate in determining the correct rank among several alignments. A major remaining question is how accurate alignments should be when used for sequence annotation. Picking the appropriate cutoff clearly depends on the exact purpose of the alignments. For example, the performance of profile Hidden Markov models is less dependent on alignment quality than is the accuracy of phylogenetic reconstruction (18,36). In our experience, a cutoff of 0.8 MOS is practical for trusting the quality of an alignment. Conversely, alignments scoring 0.5 MOS or less should be considered incorrect and need to be manually refined. For example, an MOS of 0.5 on a Balibase alignment implies that the alignment method failed to recognize a biologically conserved core block.
Since we envision our method to be used as an quality assessment tool, its own accuracy is vital. Therefore, we conducted extensive tests and found the performance of our method to be robust on four distinct benchmark sets. We carried out 30 408 alignments covering everything from easy alignment cases to difficult cases in which all current alignment methods fail. There was no dramatic drop of performance observed on any particular test set, reflecting the robustness of our method. In the case of Balibase, we deliberately made it more difficult for our method by including unconserved regions into our accuracy assessment, but this had no noticeable effects.
The concept of comparing several multiple alignments is clearly a valuable tool for assessing alignment accuracy.
SUPPLEMENTARY DATA
Supplementary data are available at NAR online.
Supplementary Material
Acknowledgments
The authors would like to thank Carsten Daub for many useful discussions, Markus Wistrand for the critical assessment of the manuscript and Ivo VanWalle and Robert Edgar for help with the SABmark and Prefab test set respectively. Funding to pay the Open Access publication charges for this article was provided by Swedish Graduate School for Functional Genomics and Bioinformatics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lecompte O., Thompson J.D., Plewniak F., Thierry J., Poch O. Multiple alignment of complete sequences (MACS) in the postgenomic era. Gene. 2001;270:17–30. doi: 10.1016/s0378-1119(01)00461-9. [DOI] [PubMed] [Google Scholar]
- 2.Do C.B., Mahabhashyam M.S.P., Brudno M., Batzoglou S. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katoh K., Kuma K.-i., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Walle I., Lasters I., Wyns L. Align-m—a new algorithm for multiple alignment of highly divergent sequences. Bioinformatics. 2004;20:1428–1435. doi: 10.1093/bioinformatics/bth116. [DOI] [PubMed] [Google Scholar]
- 5.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace I.M., O'Sullivan O., Higgins D.G. Evaluation of iterative alignment algorithms for multiple alignment. Bioinformatics. 2005;21:1408–1414. doi: 10.1093/bioinformatics/bti159. [DOI] [PubMed] [Google Scholar]
- 7.Notredame C. Recent progress in multiple sequence alignment: a survey. Pharmacogenomics. 2002;3:131–144. doi: 10.1517/14622416.3.1.131. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearson W.R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 12.Lee C., Grasso C., Sharlow M.F. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 13.Grasso C., Lee C. Combining partial order alignment and progressive multiple sequence alignment increases alignment speed and scalability to very large alignment problems. Bioinformatics. 2004;20:1546–1556. doi: 10.1093/bioinformatics/bth126. [DOI] [PubMed] [Google Scholar]
- 14.Raphael B., Zhi D., Tang H., Pevzner P. A novel method for multiple alignment of sequences with repeated and shuffled elements. Genome Res. 2004;14:2336–2346. doi: 10.1101/gr.2657504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh O. Significant improvement in accuracy of multiple protein sequence alignments by iterative refinement as assessed by reference to structural alignments. J. Mol. Biol. 1996;264:823–838. doi: 10.1006/jmbi.1996.0679. [DOI] [PubMed] [Google Scholar]
- 16.Vogt G., Etzold T., Argos P. An assessment of amino acid exchange matrices in aligning protein sequences: the twilight zone revisited. J. Mol. Biol. 1995;249:816–831. doi: 10.1006/jmbi.1995.0340. [DOI] [PubMed] [Google Scholar]
- 17.Vingron M., Waterman M.S. Sequence alignment and penalty choice. Review of concepts, case studies and implications. J. Mol. Biol. 1994;235:1–12. doi: 10.1016/s0022-2836(05)80006-3. [DOI] [PubMed] [Google Scholar]
- 18.Morrison D.A., Ellis J.T. Effects of nucleotide sequence alignment on phylogeny estimation: a case study of 18S rDNAs of apicomplexa. Mol. Biol. Evol. 1997;14:428–441. doi: 10.1093/oxfordjournals.molbev.a025779. [DOI] [PubMed] [Google Scholar]
- 19.Hickson R.E., Simon C., Perrey S.W. The performance of several multiple-sequence alignment programs in relation to secondary-structure features for an rRNA sequence. Mol. Biol. Evol. 2000;17:530–539. doi: 10.1093/oxfordjournals.molbev.a026333. [DOI] [PubMed] [Google Scholar]
- 20.Durbin R., Eddy S., Krogh A., Mitchison G. Biological Sequence Analysis. Cambridge University Press; 1998. pp. 134–159. [Google Scholar]
- 21.Thompson J.D., Plewniak F., Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lassmann T., Sonnhammer E.L.L. Quality assessment of multiple alignment programs. FEBS Lett. 2002;529:126–130. doi: 10.1016/s0014-5793(02)03189-7. [DOI] [PubMed] [Google Scholar]
- 23.Morgenstern B., Goel S., Sczyrba A., Dress A. AltAVisT: comparing alternative multiple sequence alignments. Bioinformatics. 2003;19:425–426. doi: 10.1093/bioinformatics/btf882. [DOI] [PubMed] [Google Scholar]
- 24.Morgenstern B., Dress A., Werner T. Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc. Natl Acad. Sci. USA. 1996;93:12098–12103. doi: 10.1073/pnas.93.22.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 26.Morgenstern B. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 27.Bucka-Lassen K., Caprani O., Hein J. Combining many multiple alignments in one improved alignment. Bioinformatics. 1999;15:122–130. doi: 10.1093/bioinformatics/15.2.122. [DOI] [PubMed] [Google Scholar]
- 28.Poirot O., O'Toole E., Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Sullivan O., Zehnder M., Higgins D., Bucher P., Grosdidier A., Notredame C. APDB: a novel measure for benchmarking sequence alignment methods without reference alignments. Bioinformatics. 2003;19(Suppl. 1):215–221. doi: 10.1093/bioinformatics/btg1029. [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing Vienna, Austria. 2005. ISBN 3-900051-07-0.
- 31.Thompson J.D., Plewniak F., Poch O. BAliBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics. 1999;15:87–88. doi: 10.1093/bioinformatics/15.1.87. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J.D., Koehl P., Ripp R., Poch O. BaliBASE 3.0: Latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 33.Van Walle I., Lasters I., Wyns L. SABmark—a benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J.D., Plewniak F., Ripp R., Thierry J.C., Poch O. Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol. 2001;314:937–951. doi: 10.1006/jmbi.2001.5187. [DOI] [PubMed] [Google Scholar]
- 35.Pei J., Grishin N.V. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17:700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths-Jones S., Bateman A. The use of structure information to increase alignment accuracy does not aid homologue detection with profile HMMs. Bioinformatics. 2002;18:1243–1249. doi: 10.1093/bioinformatics/18.9.1243. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katoh K., Misawa K., Kuma K.-i., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.