Abstract
PPARs (peroxisome-proliferator-activated receptors) α, β/δ and γ are a group of transcription factors that are involved in numerous processes, including lipid metabolism and adipogenesis. By comparing liver mRNAs of wild-type and PPARα-null mice using microarrays, a novel putative target gene of PPARα, G0S2 (G0/G1 switch gene 2), was identified. Hepatic expression of G0S2 was up-regulated by fasting and by the PPARα agonist Wy14643 in a PPARα-dependent manner. Surprisingly, the G0S2 mRNA level was highest in brown and white adipose tissue and was greatly up-regulated during mouse 3T3-L1 and human SGBS (Simpson–Golabi–Behmel syndrome) adipogenesis. Transactivation, gel shift and chromatin immunoprecipitation assays indicated that G0S2 is a direct PPARγ and probable PPARα target gene with a functional PPRE (PPAR-responsive element) in its promoter. Up-regulation of G0S2 mRNA seemed to be specific for adipogenesis, and was not observed during osteogenesis or myogenesis. In 3T3-L1 fibroblasts, expression of G0S2 was associated with growth arrest, which is required for 3T3-L1 adipogenesis. Together, these data indicate that G0S2 is a novel target gene of PPARs that may be involved in adipocyte differentiation.
Keywords: adipogenesis, G0/G1 switch gene 2 (G0S2), growth arrest, peroxisome-proliferator-activated receptor (PPAR)
Abbreviations: aP2, adipocyte fatty-acid-binding protein; Avg Diff, average difference; BAT, brown adipose tissue; BMP-2, bone morphogenetic protein-2; BODIPY®, 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene; ChIP, chromatin immunoprecipitation; CYP4A10, cytochrome P450, family 4, subfamily a, polypeptide 10; DMEM, Dulbecco's modified Eagle's medium; DsRed, Discosoma sp. red fluorescent protein; ER, endoplasmic reticulum; GFP, green fluorescent protein; G0S2, G0/G1 switch gene 2; GPDH, glycerol 3-phosphate dehydrogenase; h, human; HEK-293 cells, human embryonic kidney-293 cells; m, mouse; PPAR, peroxisome-proliferator-activated receptor; PPRE, PPAR-responsive element; Q-PCR, real-time quantitative PCR; RT, reverse transcriptase; RXR, retinoid X receptor; SEAP, secreted alkaline phosphatase; SGBS, Simpson–Golabi–Behmel syndrome; WAT, white adipose tissue
INTRODUCTION
PPARs (peroxisome-proliferator-activated receptors) represent a group of nuclear receptors that play pivotal roles in the regulation of energy metabolism [1]. These receptors function as ligand-activated transcription factors by binding to the promoters of target genes and inducing transcription upon activation by small lipophilic compounds. Three different PPARs can be distinguished: PPARα, PPARβ/δ and PPARγ. All three receptors are activated by (mainly polyunsaturated) fatty acids and various fatty-acid-derived compounds, such as eicosanoids.
PPARγ, which is most highly expressed in adipose tissue, is known as the master regulator of adipogenesis. Numerous studies, both in vivo and in vitro, have pointed to PPARγ as the transcription factor that drives adipocyte differentiation [2–5]. The role of PPARγ in adipogenesis is diverse, and concerns the regulation of cell-cycle withdrawal, as well as induction of fat-specific target genes that are involved in adipocyte metabolism. Indeed, PPARγ stimulates the expression of numerous genes that are involved in lipogenesis, including those for aP2 (adipocyte fatty-acid-binding protein), lipoprotein lipase and CD36/fatty acid translocase. Previous microarray studies have yielded a comprehensive picture of the likely target genes of PPARγ in adipose tissue and indicate a general role for PPARγ in the regulation of lipid metabolism [6], which is underlined by the therapeutic utilization of the PPARγ ligands thiazolidinediones in obesity-linked Type II diabetes.
Expression of PPARβ/δ is ubiquitous, which has been a major impediment in elucidating its assorted functions. The most compelling recent studies indicate that PPARβ/δ stimulates fatty acid oxidation in both adipose tissue and skeletal muscle [7,8], regulates hepatic VLDL (very-low-density lipoprotein) production and catabolism [9], and is involved in wound healing by governing keratinocyte differentiation [10]. Furthermore, PPARβ/δ has been connected with colon carcinogenesis, although conflicting results have been reported [11,12].
The last PPAR isotype, PPARα, has mostly been studied in the context of liver metabolism and is known to be a central regulator of hepatic fatty acid catabolism [13]. Evidence is accumulating that PPARα also governs several aspects of glucose metabolism [14]. Furthermore, it potently represses the hepatic inflammatory response by down-regulating the expression of numerous genes [15,16]. Indeed, up-regulation of various acute-phase proteins during hepatic inflammation may be linked directly to down-regulation of hepatic PPARα mRNA under these conditions [17]. Importantly, PPARα is the molecular target for the hypolipidaemic fibrates, a group of drugs that are prescribed for their ability to lower plasma triacylglycerols and elevate plasma HDL (high-density lipoprotein) levels.
Although much is already known about PPARs, significant gaps remain in our knowledge, particularly with respect to the set of genes that are regulated by PPARs in various organs. In the present study, we applied microarray technology to find putative targets of PPARα by comparing liver mRNA from PPARα-knockout mice and wild-type mice. One of the putative target genes identified, called G0S2 (G0/G1 switch gene 2), was subjected to detailed follow-up investigation. The collective data indicate that G0S2 is a direct PPAR target gene, with a functional PPRE (PPAR-responsive element) in its promoter, and may be involved in adipocyte differentiation.
EXPERIMENTAL
Materials
Wy14643 was obtained from ChemSyn laboratories. Rosiglitazone was from Alexis. Recombinant human insulin (Actrapid) was from Novo Nordisk. Recombinant human BMP-2 (bone morphogenetic protein-2) was from R&D systems. BODIPY® (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene) 493/503 was from Molecular Probes. SYBR Green was from Eurogentec. DMEM (Dulbecco's modified Eagle's medium), foetal calf serum, calf serum and penicillin/streptomycin/fungizone were from Cambrex Bioscience. The 3T3-L1 cell line was purchased from ECACC (European Collection of Cell Culture). HEK-293 (human embryonic kidney-293) cells were from BD Biosciences. All other chemicals were from Sigma.
Animals
Male pure-bred Sv129 and PPARα-null mice (2–3-month-old) on a Sv129 background were used. Fed mice were killed at the end of the dark cycle. Fasting was started at the onset of the light cycle for 6, 12 or 24 h (n=5 per group). For the feeding experiment with Wy14643, 3–5-month-old male wild-type and PPARα-null mice were fed with 0.1% Wy14643 for 5 days by mixing it in their food. Alternatively, they received a single dose of Wy14643 (400 μl of 10 mg/ml Wy14643 dissolved in 0.5% carboxymethylcellulose) and were killed 6 h later (n=5 per group). Blood was collected via orbital puncture. Livers were dissected and directly frozen in liquid nitrogen.
The animal experiments were approved by the animal experimentation committee of the Etat de Vaud (Switzerland) or Wageningen University.
Affymetrix microarray
Total RNA was prepared from mouse livers using TRIzol® reagent (Invitrogen). For each microarray experiment, 10 μg of total liver RNA pooled from four mice was used for cRNA synthesis. RNA was pooled because pilot experiments with Affymetrix chips at Pfizer had indicated that the inter-animal variability in gene expression (determined by performing eight separate hybridizations of eight different mice of the same strain), as well as variability between repeated hybridizations of the same pooled RNA sample, were statistically insignificant. Hybridization, washing and scanning of Affymetrix Genechip Mu6500 probe arrays was according to standard Affymetrix protocols. Fluorimetric data were processed by Affymetrix GeneChip3.1 software, and the gene chips were globally scaled to all the probe sets with an identical target intensity value. Affymetrix software measures the expression level of a gene as an average difference value (Avg Diff) by comparing the intensity of hybridization of 20 sets of perfect match oligonucleotide probes to 20 sets of mismatch probes. Only genes with an Avg Diff above the threshold of 100 and with a difference in Avg Diff values between wild-type and PPARα-null mutant mice at least 2-fold were considered.
RT (reverse transcriptase)-PCR
Total RNA was extracted from cells or tissue with TRIzol® reagent following the supplier's protocol. Total RNA (3–5 μg) was treated with amplification grade DNAse I (Invitrogen), then reverse-transcribed with oligo(dT) using Superscript II RT RNase H− (Invitrogen) following the supplier's recommendation. cDNA was PCR-amplified with Platinum Taq DNA polymerase (Invitrogen). Primer sequences used in the PCRs were chosen based on the sequences available in GenBank®. Primer sequences to amplify mG0S2 cDNA were 5′-TGCTGCCTCTCTTCCCACTGC-3′ (forward) and 5′-GTAGGGTCAGTTCTGGATTCGGTG-3′ (reverse). Other sequences are available from S.K. on request.
Q-PCR (real-time quantitative PCR)
Primers were designed to generate a PCR-amplification product of 100–200 bp. Only primer pairs yielding unique amplification products without primer dimer formation were subsequently used for Q-PCR assays. The primer pairs listed in Table 1 were used.
Table 1. Primer pairs used in Q-PCR.
| Primer name | Sequence |
|---|---|
| hG0S2 (forward) | 5′-CGCCGTGCCACTAAGGTC-3′ |
| hG0S2 (reverse) | 5′-GCACACAGTCTCCATCAGGC-3′ |
| m/rG0S2 (forward) | 5′-AGTGCTGCCTCTCTTCCCAC-3′ |
| m/rG0S2 (reverse) | 5′-TTTCCATCTGAGCTCTGGGC-3′ |
| mPPARγ (forward) | 5′-CACAATGCCATCAGGTTTGG-3′ |
| mPPARγ (reverse) | 5′-GCTGGTCGATATCACTGGAGATC-3′ |
| mA-FABP (aP2) (forward) | 5′-AAGAAGTGGGAGTGGGCTTT-3′ |
| mA-FABP (aP2) (reverse) | 5′-AATCCCCATTTACGCTGATG-3′ |
| mOsteocalcin (forward) | 5′-GCAGCTTGGTGCACACCTAG-3′ |
| mOsteocalcin (reverse) | 5′-GGAGCTGCTGTGACATCCAT-3′ |
| mGPDH (forward) | 5′-GCCTTCGCCAAGCTCTTCTG-3′ |
| mGPDH (reverse) | 5′-TAGCAGGTCGTGATGAGGTCTG-3′ |
| mβ-Actin (forward) | 5′-GATCTGGCACCACACCTTCT-3′ |
| mβ-Actin (reverse) | 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
| m36B4 (forward) | 5′-AGCGCGTCCTGGCATTGTGTGG-3′ |
| m36B4 (reverse) | 5′-GGGCAGCAGTGGTGGCAGCAGC-3′ |
| r36B4 (forward) | 5′-CGGGAAGGCTGTGGTGCTGATG-3′ |
| r36B4 (reverse) | 5′-TCGGTGAGGTCCTCCTTGGTGAAC-3′ |
PCR was carried out using Platinum Taq polymerase and SYBR green on an iCycler PCR machine (Bio-Rad) according to the instructions from the manufacturer.
Primary hepatocytes
Rat and mouse hepatocytes were isolated by two-step collagenase perfusion as described previously [18]. Viability was determined by Trypan Blue exclusion, and was at least 75%. Hepatocytes were suspended in William's E medium (Cambrex) supplemented with 10% (v/v) foetal calf serum, 20 m-units/ml insulin, 50 nM dexamethasone, 100 units of penicillin, 100 μg of streptomycin, 0.25 μg/ml fungizone and 50 μg/ml gentamycin. The next day, cells were incubated in fresh medium in the presence or absence of Wy14643 (25 μM) for 24 h.
3T3-L1 and SGBS (Simpson–Golabi–Behmel syndrome) adipogenesis assay
3T3-L1 fibroblasts were amplified in DMEM plus 10% (v/v) calf serum. At 2 days after reaching confluence (=day 0), the medium was changed to DMEM plus 10% (v/v) foetal calf serum and the following compounds were added: isobutyl-methylxanthine (0.5 mM), dexamethasone (1 μM) and insulin (5 μg/ml). On day 3, the medium was changed to DMEM plus 10% (v/v) foetal calf serum and insulin (5 μg/ml). The medium was subsequently changed every 3 days, and, from day 9 onwards, no further insulin was added.
The culture of the SGBS cells as well as their induction into mature human adipocytes were performed exactly as described previously [19].
Western blot
The combined human/mouse polyclonal antibody used was directed against epitopes TVLGGRALSNRQHAS and EATLCSRALSLRQHAS of the human and mouse G0S2 proteins respectively. The peptide affinity-purified antibodies were generated in rabbit and ordered via Eurogentec's customized antibody production service. Western blot was carried out as described previously [20].
Transactivation assay
HepG2 cells were co-transfected by calcium phosphate precipitation with an mPPARα (m is mouse), mPPARβ/δ or mPPARγ1 expression vector and pGL3 reporter vector containing different size fragments of the hG0S2 (h is human) promoter. A β-galactosidase reporter vector was co-transfected to normalize for differences in transfection efficiency. After transfection, cells were incubated in the presence or absence of Wy14643 (50 μM), L165041 (5 μM) or rosiglitazone (5 μM) respectively for 24 h before lysis. A Promega luciferase assay and a standard β-galactosidase assay using 2-nitrophenyl-β-D-galactopyranoside as a substrate were used to measure the relative promoter activities.
To disable the G0S2 PPRE within the hG0S2 promoter, two separate partially overlapping PCR fragments were generated using the wild-type hG0S2 promoter as a template. The mutant sequence was verified by automated sequencing.
A 200 nt fragment surrounding the putative PPRE within the mG0S2 promoter was PCR-amplified from mouse genomic DNA (strain C57/B6) and subcloned into the KpnI and BglII sites of pTAL-SEAP (BD Biosciences). This reporter vector was transfected into HepG2 cells by calcium phosphate precipitation together with an expression vector for mPPARα, mPPARβ/δ or mPPARγ1 in the presence or absence of their respective ligands. A β-galactosidase reporter was co-transfected to normalize for differences in transfection efficiency. SEAP (secreted alkaline phosphatase) activity was measured in the medium 24 h post-transfection via the chemiluminescent SEAP reporter assay (Roche).
Gel shift assay
hRXRα (retinoid X receptor), hPPARα and hPPARγ proteins were generated from pSG5 expression vectors using the TNT (transcription and translation)-coupled in vitro system (Promega). The following oligonucleotides were annealed: G0S2-PPRE, 5′-CTGGCCAGAAAATTGCAAAGGTCACTGA-3′ and 5′-CTGGTCAGTGACCTTTGCAATTTTCTGG-3′; G0S2-PPREmut, 5′-CTGGCCAGAAAATTGCTAAGGACACTGA-3′ and 5′-CTGGTCAGTGTCCTTAGCAATTTTCTGG-3′; for specific competition malic enzyme PPRE, 5′-TCGCTTTCTGGGTCAAAGTTGATCCA-3′ and 5′-CTGGTGGATCAACTTTGACCCAGAAAG-3′; and for non-specific competition Ets, 5′-TGGAATGTACCGGAAATAACACCA-3′ and 5′-TGGTGTTATTTCCGGTACATTCCA-3′. Oligonucleotides were annealed and labelled by Klenow filling (Roche) using Redivue [α-32P]dCTP (3000 Ci/mmol) (Amersham Biosciences). In vitro translated proteins (0.5–0.8 μl per reaction) were pre-incubated for 15 min on ice in 1× binding buffer [80 mM KCl, 1 mM dithiothreitol, 10 mM Tris/HCl (pH 7.4), 10% (v/v) glycerol plus protease inhibitors] in the presence of 2 μg of poly(dI-dC)·(dI-dC), 5 μg of sonicated salmon sperm DNA and competitor oligonucleotides in a final volume of 20 μl. Then 1 ng (1 ng/μl) of radiolabelled oligonucleotide was added, and incubation proceeded for another 10 min at room temperature (25 °C). Complexes were separated on a 4% polyacrylamide gel (acrylamide/bisacrylamide, 37.5:1) equilibrated in 0.5× TBE (Tris/borate/EDTA) at 25 mA.
In vivo ChIP (chromatin immunoprecipitation)
Pure-bred wild-type or PPARα-null mice on a Sv129 background were used. Mice were fed by gavage with either Wy14643 (50 mg/kg per day) or vehicle (0.5% carboxymethylcellulose) for 5 days. Alternatively, mice were fasted or not for 24 h. After the indicated treatment, mice were killed by cervical dislocation. The liver was rapidly perfused with pre-warmed (37 °C) PBS for 5 min followed by 0.2% collagenase for 10 min. The liver was diced and forced through a stainless steel sieve, and the hepatocytes were collected directly into DMEM containing 1% (w/v) formaldehyde. After incubation at 37 °C for 15 min, the hepatocytes were pelleted, and ChIP was carried out using PPARα-specific antibodies as described previously [10].
3T3-L1 cells were differentiated as described above. After cell lysis and sonication, the supernatant was diluted 20-fold in re-ChIP dilution buffer (1 mM EDTA, 20 mM Tris/HCl, pH 8.1, 50 mM NaCl and 1% Triton X-100) before incubation with antibodies against mouse PPARγ or PPARβ/δ. The remainder of the assay was carried out as described previously [10]. PCR was performed using primers flanking the putative PPRE in the mG0S2 promoter (amplified product −1937 to −1357) and a control sequence (amplified product −3555 to −3107).
C2C12 osteo- and myo-genesis
C2C12 mesenchymal progenitor cells were differentiated into myoblasts by letting the cells grow to confluence. C2C12 cells were differentiated into osteoblasts by the addition of BMP-2 (500 ng/ml).
Cell-cycle synchronization
3T3-L1 cells were seeded at low confluence in DMEM plus 10% (v/v) foetal calf serum. After 9 h, the medium was replaced by DMEM plus 0.2% (v/v) foetal calf serum for 33 h. After that, foetal calf serum was re-added to the cells at 10%, and cells were taken at regular intervals for RNA preparation.
Cellular localization studies
The mG0S2 open reading frame was cloned into the EcoRI and BamHI sites of pEGFP-N2 and pDsRed1-N1 (BD Biosciences). The ER (endoplasmic reticulum) localization vector pDsRed2-ER (BD Biosciences) was used as a control vector for the ER. The mG0S2-containing pEGFP-N2 vector was co-transfected with pDsRed2-ER into HEK-293 cells. The mG0S2-containing pDsRed1-N1 vector was transfected into 3T3-L1 cells. Transfections were performed by calcium phosphate precipitation on 60% confluent cell cultures. For 3T3-L1 cells, after 8 h, medium was replaced with medium containing 0.1% (v/v) Tween 80. After 48 h, cells were washed with PBS and fixed in 3.5% (w/v) formaldehyde in PBS. BODIPY® 493/503 (saturated solution in 100% ethanol) was added to the fixing solution at 1:100, after which the cells were examined by fluorescence spectroscopy. Fluorescence microscopy was carried out 48 h post-transfection using a LSM510 confocal laser-scanning microscope (Zeiss).
RESULTS
Microarray studies identify the G0S2 gene as a potential PPAR target gene
Microarray studies permit the expression monitoring of thousands of genes. To identify new putative PPAR target genes, mRNA from livers of wild-type and PPARα-null mice at different stages of fasting was compared using Affymetrix murine 6500 oligonucleotide microarrays.
Out of a total of 6519 genes present on the array, mRNA levels of 50 genes were at least 2-fold lower in the livers of 24-h-fasted PPARα-null mice compared with 24-h-fasted wild-type mice. In fed mice, the number of genes fulfilling the same criteria was much lower (11 genes) (Table 2), indicating that deletion of the PPARα gene has much more severe consequences in the fasted state than in the fed state. Interestingly, there was very little overlap between the two sets of genes.
Table 2. Genes differentially expressed between wild-type and PPARα-null mice in fasted (top list) and fed (bottom list) state according to microarray analysis.
Fold D., fold difference; FXR, farnesoid X receptor; RIP14, receptor-interacting protein 14; ss, single-stranded.
| Accession no. | Gene | Fold D. | Avg Dif | Function |
|---|---|---|---|---|
| X69296 | Fatty acid ω hydroxylase (CYP4A10) | 56.5 | 120.4 | Fatty acid oxidation/ketogenesis |
| X95280 | G0S2 | 9.2 | 152.6 | Unknown |
| M13366 | NAD-linked GPDH | 7.5 | 167.2 | Glycerol metabolism |
| U14390 | Aldehyde dehydrogenase 3 | 7.4 | 371.9 | Biotransformation |
| U12791 | HMG-CoA synthase (hydroxymethylglutaryl-CoA synthase) | 7.3 | 1490.1 | Fatty acid oxidation/ketogenesis |
| Z14050 | Dodecenoyl-CoA δ isomerase | 5.5 | 423.5 | Fatty acid oxidation/ketogenesis |
| X89998 | 17β-Hydroxysteroid dehydrogenase type IV | 5.2 | 494.7 | Steroid metabolism |
| L11163 | Short-chain-acyl-CoA dehydrogenase | 5 | 208.9 | Fatty acid oxidation/ketogenesis |
| U01163 | Carnitine palmitoyltransferase II | 4.1 | 505.3 | Fatty acid oxidation/ketogenesis |
| U60987 | FAD-linked GPDH | 4.1 | 172.8 | Glycerol metabolism |
| L40406 | Heat-shock protein 105 kDa β | 4 | 150.8 | Heat-shock protein |
| X74938 | HNF3γ (hepatic nuclear factor 3γ) | 3.5 | 103.2 | Transcription factor |
| J03398 | ABC-B4 (ATP-binding cassette B4) (mdr2) | 3.4 | 642.2 | Transporter |
| M22432 | Protein synthesis elongation factor Tu | 3.3 | 186.9 | Protein synthesis |
| L05439 | Insulin-like growth factor-binding protein 2 | 3.2 | 2083.7 | Plasma-binding protein |
| X85983 | Carnitine acetyltransferase | 3.2 | 424.5 | Fatty acid oxidation/ketogenesis |
| U41497 | Very-long-chain-acyl-CoA dehydrogenase | 3.1 | 659.1 | Fatty acid oxidation/ketogenesis |
| U58883 | CAP (c-Cbl-associated protein) | 3 | 158.8 | Insulin signalling |
| X78709 | NRF1 (nuclear respiratory factor 1) | 2.9 | 144.8 | Transcription factor |
| U09010 | Mannose-binding protein A (Mbl1) | 2.9 | 577.2 | Immune function? |
| X98848 | 6-Phosphofructo-2-kinase/fructose 2,6-bisphosphate | 2.8 | 115.2 | Glucose metabolism |
| U33557 | Folylpolyglutamate synthetase | 2.6 | 275 | Folate metabolism |
| J02652 | Malate NADP-oxidoreductase (malic enzyme) | 2.6 | 158.2 | Fatty acid synthesis |
| U44389 | NAD-dependent 15-hydroxyprostaglandin dehydrogenase | 2.5 | 244.7 | Eicosanoid metabolism |
| AA120387 | Mitochondrial Lon protease holomogue 1 precursor | 2.5 | 535.4 | Unknown |
| AA016431 | E-FABP (epidermal fatty-acid-binding protein) | 2.5 | 157.2 | Fatty acid binding |
| U02098 | Pur-α | 2.5 | 122.2 | ssDNA-binding protein |
| U09416 | Bile acid receptor (BAR) (FXR, RIP14) | 2.5 | 505.7 | Transcription factor |
| Y00309 | Lactate dehydrogenase A | 2.4 | 4506.4 | Glucose metabolism |
| U15977 | Long-chain-acyl-CoA synthetase | 2.4 | 1214.7 | Fatty acid activation |
| U15977 | Long-chain-acyl-CoA dehydrogenase | 2.4 | 301 | Fatty acid oxidation/ketogenesis |
| Z19581 | Siah-2 protein | 2.4 | 107 | Unknown |
| U41751 | EI24 (etoposide-induced protein 2.4) | 2.3 | 276.5 | Cell growth/apoptosis |
| J05186 | ERp72 (endoplasmic reticulum protein 72 kDa) | 2.3 | 286.5 | Heat-shock protein |
| U48420 | Gluthatione transferase type 2θ class | 2.3 | 809.8 | Biotransformation |
| U57368 | DBI-1 (epidermal growth factor repeat transmembrane protein) | 2.3 | 102.1 | Mitogen signalling? |
| X70887 | p59 immunophilin | 2.3 | 259.9 | Binds heat-shock proteins |
| D49744 | Farnesyltransferase α subunit | 2.2 | 339.9 | Post-translational processing |
| M65255 | Hydrophilic protein KE2 | 2.2 | 130.1 | Unknown |
| X674689 | LRP1 (LDL receptor-related protein 1) | 2.2 | 402.1 | Cholesterol metabolism |
| D29639 | 3-Hydroxyacyl-CoA dehydrogenase | 2.2 | 2020.7 | Fatty acid oxidation/ketogenesis |
| AA028398 | Tubulin β | 2.2 | 107.8 | Cytoskeleton |
| X51971 | Carbonic anhydrase V | 2.1 | 186.7 | Ureagenesis/gluconeogenesis |
| U76832 | Plasma membrane protein syntaxin | 2.1 | 148.3 | Endo-/exo-cytosis |
| U49878 | HMG-CoA lyase (hydroxymethylglutaryl-CoA lyase) | 2.1 | 1933.9 | Fatty acid oxidation/ketogenesis |
| U48403 | Glycerol kinase | 2.1 | 164 | Glycerol metabolism |
| AA068057 | Ras-related protein RAB21 | 2 | 199.8 | Vesicular transport |
| U06837 | β-Hexosaminidase | 2 | 114.6 | Lysosomal enzyme |
| W30496 | Galactokinase | 2 | 139.7 | Galactose metabolism |
| U07159 | Medium-chain-acyl-CoA dehydrogenase | 2 | 2037.1 | Fatty acid oxidation/ketogenesis |
| M13522 | Serum amyloid A isoform 2 | 15.2 | 829.2 | Acute-phase protein |
| M13521 | Serum amyloid A isoform 1 | 11.9 | 1194.9 | Acute-phase protein |
| V00835 | Metallothionein-I | 5.9 | 130.8 | Acute-phase protein |
| M21285 | Stearoyl-CoA desaturase 1 | 3.2 | 2099.9 | Fatty acid desaturation |
| U73037 | Interferon regulatory factor 7 | 2.9 | 101.5 | Transcription factor |
| Z14050 | Dodecenoyl-CoA δ-isomerase | 2.8 | 331.7 | Fatty acid oxidation |
| X64070 | Mannose phosphate receptor 46 | 2.3 | 109.3 | Lysosomal transport |
| M96823 | Nucleobindin (Calnuc) | 2.2 | 296.9 | Ca2+ binding protein |
| U01163 | Carnitine palmitoyl-transferase II | 2.1 | 536.7 | Fatty acid oxidation |
| X07295 | Mitochondrial malate dehydrogenase | 2 | 295.5 | TCA cycle |
| M28666 | Porphobilinogen deaminase | 2 | 663.6 | Haem synthesis |
Many of the genes that were down-regulated in the PPARα-null mice compared with the wild-type mice after 24 h of fasting are classical PPARα target genes involved in fatty acid oxidation and ketogenesis, including CYP4A10 (cytochrome P450, family 4, subfamily a, polypeptide 10), HMG-CoA synthase (hydroxymethylglutaryl-CoA synthase), very-long-chain-acyl-CoA dehydrogenase and many others. However, there were also a significant number of differentially expressed genes that thus far have not been associated with PPARα and may represent novel PPARα target genes. Of these genes, G0S2, which encodes a small protein of unknown function, showed the largest decrease in mRNA levels in PPARα-null mice second to CYP4A10.
The G0S2 gene was first identified approx. 10 years ago in a screen to find genes that are differentially expressed during the lectin-induced switch of lymphocytes from G0 to the G1 phase of the cell cycle. It was found that G0S2 expression increased transiently within 1–2 h of addition of lectin or cycloheximide to blood mononuclear cells [21]. Additional information about the potential function of this gene is lacking. The G0S2 gene encodes a protein of 103 amino acids with 78% identity between mouse and human and contains one predicted transmembrane domain. Remarkably, G0S2 protein seems to be unique: no homologous protein could be found in lower organisms (including Caenorhabditis elegans and Drosophila), and it does not seem to contain any domain shared by other proteins.
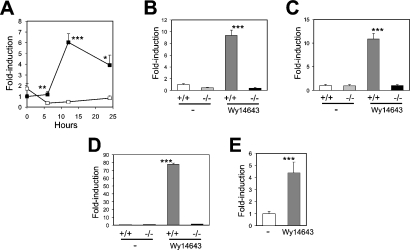
Q-PCR analysis showed that hepatic expression of G0S2 was highly increased during fasting, reaching a peak after 12 h (Figure 1A). This fasting-induced increase in expression was absent in PPARα-null mice. Administration of the synthetic PPARα agonist Wy14643 increased G0S2 mRNA in mouse liver (Figure 1B and 1C) and primary hepatocytes (Figure 1D) of wild-type, but not PPARα-null, mice. Furthermore, addition of Wy14643 increased G0S2 mRNA expression in primary rat hepatocytes (Figure 1E). These results suggest that G0S2 may be a direct target gene of PPARα.
Figure 1. G0S2 is a PPARα-regulated gene in mouse.
(A) Hepatic expression of G0S2 after 0, 6, 12 or 24 h of fasting in wild-type (■) and PPARα-null mice (□). (B) Hepatic G0S2 expression in wild-type (+/+) and PPARα-null (−/−) mice 6 h after oral gavage of 4 mg of Wy14643. (C) Hepatic G0S2 expression in wild-type (+/+) and PPARα-null (−/−) mice after 5 days of feeding with Wy14643 (0.1%). (D) G0S2 expression in primary hepatocytes of wild-type (+/+) and PPARα-null (−/−) mice incubated for 24 h in the presence or absence of Wy14643 (25 μM). (E) G0S2 expression in primary rat hepatocytes incubated for 24 h in the presence or absence of Wy14643 (25 μM). G0S2 expression was determined by Q-PCR. Results are means±S.E.M. Differences were evaluated by student's t test (*P<0.05, **P<0.01, ***P<0.001).
G0S2 is connected with adipocyte differentiation
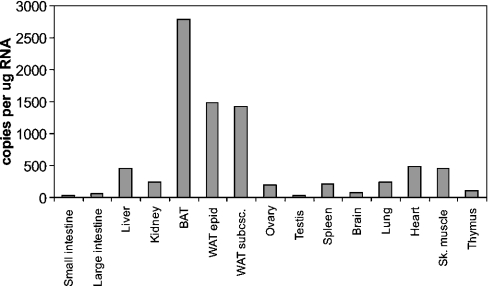
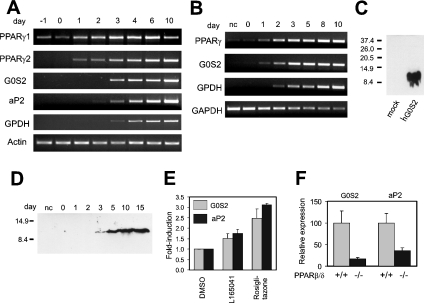
Although G0S2 was identified in liver, it may be expressed elsewhere as well. Indeed, Q-PCR showed that G0S2 mRNA levels were highest in BAT (brown adipose tissue) and WAT (white adipose tissue), followed by muscle, heart and liver (Figure 2). In contrast, expression was very low in testes, small and large intestine, and thymus. While PPARα is highly expressed in BAT and liver, it is virtually absent from WAT. In contrast, PPARγ is highly expressed in WAT, where it plays an important role in adipocyte differentiation. The high expression of G0S2 in WAT suggests that it could be a target of PPARγ. To find out whether this is true, the 3T3-L1 adipogenesis system was used. Expression of G0S2 rose dramatically during 3T3-L1 adipocyte differentiation, shortly after PPARγ1 and 2 (Figure 3A). Quantification of the changes in expression by Q-PCR indicated that G0S2 mRNA levels went up approx. 250-fold from day 0 to day 10 (see Figure 7). To find out whether G0S2 is similarly up-regulated during human adipogenesis, expression was monitored during human SGBS adipocyte differentiation. Similarly to that in 3T3-L1 cells, G0S2 was dramatically increased during SGBS adipogenesis (over 300-fold according to Q-PCR; see Figure 7), again shortly after PPARγ and jointly with the adipogenic marker GPDH (Figure 3B). According to Western blot using an anti-G0S2 antibody (Figure 3C), in parallel with the mRNA data with a delay of 1–2 days, a clear increase in G0S2 protein was observed, indicating that changes at the mRNA level were translated at the protein level (Figure 3D). Taken together, these data demonstrate that G0S2 expression is highly up-regulated during mouse and human adipocyte differentiation, together with PPARγ targets and late adipogenesis marker genes aP2 and GPDH, suggesting that G0S2 may be regulated directly by PPARγ.
Figure 2. G0S2 is expressed mainly in adipose tissue.
Total RNA was prepared from tissues of one adult male mouse (NMRI strain) and G0S2 expression was determined by Q-PCR. Ovary was sampled from a female mouse of the same age and strain. WAT epid, epididymal WAT; WAT subsc., subscapular WAT; Sk. muscle, skeletal muscle.
Figure 3. G0S2 mRNA expression is induced during 3T3-L1 and SGBS adipogenesis.
Post-confluent 3T3-L1 (A) or SGBS (B) fibroblasts were induced to differentiate into adipocytes. Expression of G0S2 and several adipogenic genes was determined at regular intervals by RT-PCR. (C) HEK-293 cells were transfected with empty vector (lane 1) or vector expressing hG0S2 (lane 2). Molecular-mass sizes are given in kDa. (D) Lysates from SGBS cells at different stages of differentiation were analysed for hG0S2 protein by Western blotting (15 μg of protein/lane) using a polyclonal anti-G0S2 antibody. Molecular-mass sizes are given in kDa. (E) Differentiated 3T3-L1 cells at day 10 were incubated with L165041 (2.5 μM) or rosiglitazone (1 μM) for 40 h, and the effect on G0S2 and aP2 expression was determined by Q-PCR. Results are means±S.E.M. (F) G0S2 and aP2 mRNA were measured by Q-PCR in WAT of wild-type (+/+) and PPARβ/δ-null (−/−) mice. Results are means±S.E.M.
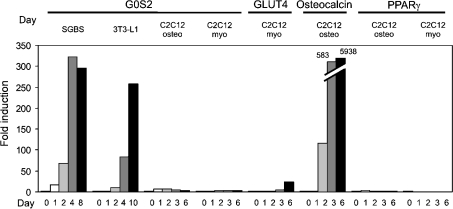
Figure 7. G0S2 is not a general marker of cell differentiation.
C2C12 cells were differentiated into osteoblasts (osteo) or myoblasts (myo) by growing them to confluence in the presence or absence of BMP-2 respectively. Expression of G0S2, PPARγ, the myogenic marker GLUT4 and the osteogenic marker osteocalcin was determined by Q-PCR. Expression of G0S2 during SGBS and 3T3-L1 adipogenesis is shown for comparison.
To substantiate further this notion, differentiated 3T3-L1 cells were treated with the synthetic PPARγ agonist rosiglitazone. Rosiglitazone at 1 μM caused an increase in G0S2 expression of approx. 2.5-fold, while the PPARβ/δ agonist L165041, at a concentration at which it specifically activates PPARβ/δ [22], increased G0S2 mRNA approx. 1.5-fold (Figure 3E). Similar changes in gene expression were observed for aP2, a well known PPARγ target gene. Regulation by PPARβ/δ was confirmed by the significantly decreased expression of G0S2 in WAT of homozygous PPARβ/δ-null mice (Figure 3F). Together, these data suggest that G0S2 may be a direct target gene of PPARγ and possibly PPARβ/δ.
A PPRE is present within the G0S2 promoter
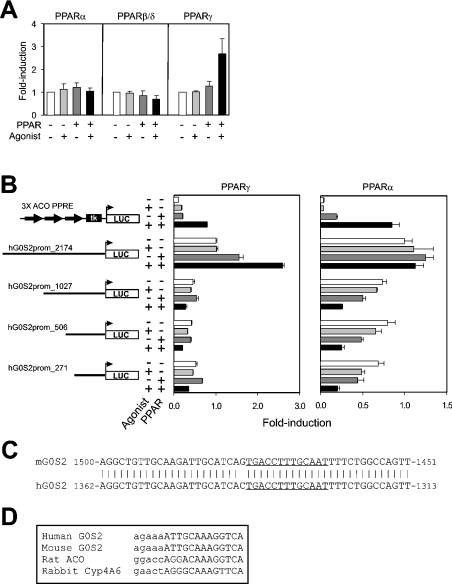
To determine what genomic region is responsible for PPAR-induced up-regulation of G0S2 expression, 2.2 kb of hG0S2 promoter sequence immediately upstream of the transcription start site was cloned in front of a luciferase reporter, and transactivation studies were carried out in HepG2 cells. Whereas PPARγ markedly increased reporter activity (Figure 4A), the other receptors showed little to no effect. This response to PPARγ and its ligand was abolished completely upon deletion of the promoter to 1.0, 0.5 or 0.27 kb (Figure 4B), indicating that the PPRE was located in the region between −2.2 and −1 kb. Interestingly, after deleting the promoter to 1.0, 0.5 or 0.27 kb, PPARα and Wy14643 decreased reporter activity (Figure 4B), suggesting that regulation of G0S2 promoter activity by PPARα is more complex.
Figure 4. G0S2 promoter is regulated by PPARs.
(A) hG0S2 reporter construct containing 2174 bp of immediate upstream promoter region was transfected into HepG2 cells together with a PPAR expression vector. Transfected cells were incubated for 24 h in the presence or absence of ligand. Normalized luciferase activity in the absence of PPAR and ligand was set at 1. Results are means±S.E.M. for at least three independent experiments. (B) hG0S2 reporter constructs containing 2174, 1027, 506 or 271 bp of immediate upstream promoter region were transfected into HepG2 cells together with an mPPARγ1 or mPPARα expression vector. Transfected cells were incubated for 24 h in the presence or absence of ligand. A luciferase reporter containing three copies of the acyl-CoA oxidase PPRE was used as a positive control. Normalized luciferase activity of the 2174 bp reporter in the absence of PPAR and ligand was set at 1. Results are means±S.E.M. (C) Alignment of a putative regulatory region within mG0S2 and hG0S2 promoter about 1.5 kb upstream of transcription start site. The putative PPRE is underlined. (D) Alignment of a putative PPRE present within G0S2 promoter with established PPREs. Lower-case letters indicate the DNA base-pair sequence preceding the PPRE, which is represented by upper-case letters.
In the region −2.2 to −1 kb, a 45 bp sequence was identified that was extremely well conserved between the mouse and human G0S2 promoter, suggesting that it is important for regulation (Figure 4C). Close inspection of this sequence revealed the presence of a putative PPRE that is highly homologous with existing PPREs (Figure 4D).
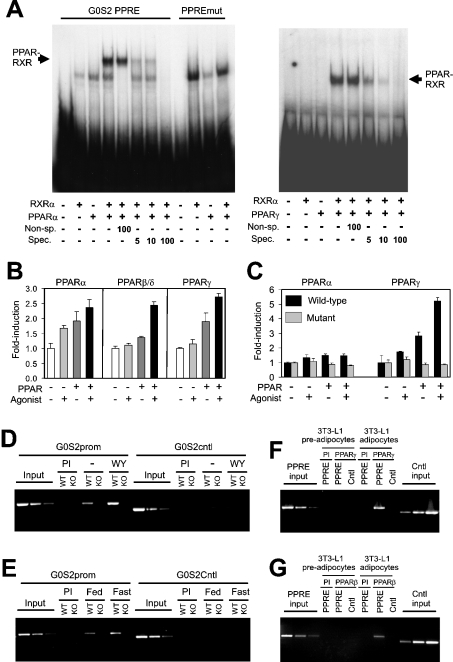
To determine whether this PPRE binds PPAR in vitro, we performed a gel shift assay. In the presence of PPARα or RXRα only, a single complex was observed, which originated from the reticulocyte lysate (Figure 5A). An additional, more intense, slower moving complex was observed only in the presence of both receptors, indicating that it represents a PPAR–RXR heterodimer. The complex disappeared in the presence of an excess of unlabelled specific oligonucleotide, but not non-specific oligonucleotide. The PPAR–RXR heterodimer did not form on an oligonucleotide that contained two substitutions within the G0S2 PPRE. Very similar results were observed for PPARγ (Figure 5A, right-hand panel) and PPARβ/δ (results not shown). These results indicate that all three PPARs are able to bind to the G0S2 PPRE in vitro.
Figure 5. G0S2 is a direct PPAR target gene.
(A) Binding of the PPAR–RXR heterodimer to the putative G0S2 PPRE as determined by gel shift assay. A double-stranded oligonucleotide containing the G0S2 PPRE was incubated with in vitro transcribed/translated hRXRα and hPPARα (left-hand panel) or hPPARγ (right-hand panel), and binding complexes were separated by electrophoresis. Fold-excess of specific (Spec.: malic enzyme PPRE) or non-specific (Non-sp.: Ets oligonucleotide) unlabelled probe is indicated. (B) HepG2 cells were transfected with a SEAP reporter vector containing a 200 bp fragment of the mG0S2 promoter and a PPAR expression vector. SEAP activity was determined in the medium 24 h post-transfection and normalized to β-galactosidase. Normalized SEAP activity in the absence of PPAR and ligand was set at 1. Results are means±S.E.M. (C) Reporter vector containing 2174 bp of hG0S2 promoter, with or without the PPRE disabled by site-directed mutagenesis, was transfected into HepG2 cells together with an expression vector for mPPARα or mPPARγ. Normalized luciferase activity in the absence of PPAR and ligand was set at 1. Results are means±S.E.M. (D–G) ChIP of G0S2 PPRE using antibodies against mPPARα, mPPARγ or mPPARβ/δ. The gene sequence spanning the putative PPRE and a random control sequence (Cntl) were analysed by PCR in the immunoprecipitated chromatin of livers of wild-type (WT) and PPARα-null (KO) mice treated or not with Wy14643 (D), livers of fed or fasted wild-type (WT) and PPARα-null (KO) mice (E), and 3T3-L1 pre-adipocytes and adipocytes (F) and (G). Pre-immune serum (PI) was used as a control.
To assess whether the G0S2 PPRE is able to mediate PPAR-dependent transactivation, a 200-nucleotide fragment surrounding the human PPRE was cloned in front of the thymidine kinase promoter followed by an SEAP reporter. In a transactivation assay, the reporter responded to PPARα, PPARβ/δ and PPARγ (Figure 5B), indicating that the PPRE identified is functional. The importance of the G0S2 PPRE for PPAR-dependent promoter activation was shown by the failure of PPARγ to stimulate hG0S2 promoter activity when, within the complete 2.2 kb promoter reporter construct, the PPRE was disabled (Figure 5C). Supporting the results in Figure 4(A), PPARα decreased reporter activity of this mutated promoter construct.
Finally, to investigate whether PPARα is bound to the G0S2 PPRE in mouse liver, in vivo ChIP was performed using an anti-PPARα antibody. In mice, treatment with Wy14643 enhanced binding of PPARα to the PPRE sequence in liver, which was not observed in PPARα-null mice (Figure 5D). Similarly, fasting enhanced binding of PPARα to the PPRE sequence, which was not observed in the PPARα null mice (Figure 5E). No detectable immunoprecipitation was observed with pre-immune serum and no amplification was observed for a control sequence. Furthermore, using ChIP, we observed binding of PPARγ (Figure 5F) and PPARβ/δ (Figure 5G) to the PPRE sequence in differentiated 3T3-L1 adipocytes, but not in pre-adipocytes. These results demonstrate that PPARα, PPARγ, and PPARβ/δ bind to the PPRE identified within the G0S2 promoter in vivo. Thus G0S2 can be formally classified as a direct PPAR target gene in human and mouse.
G0S2 protein can be localized to the ER
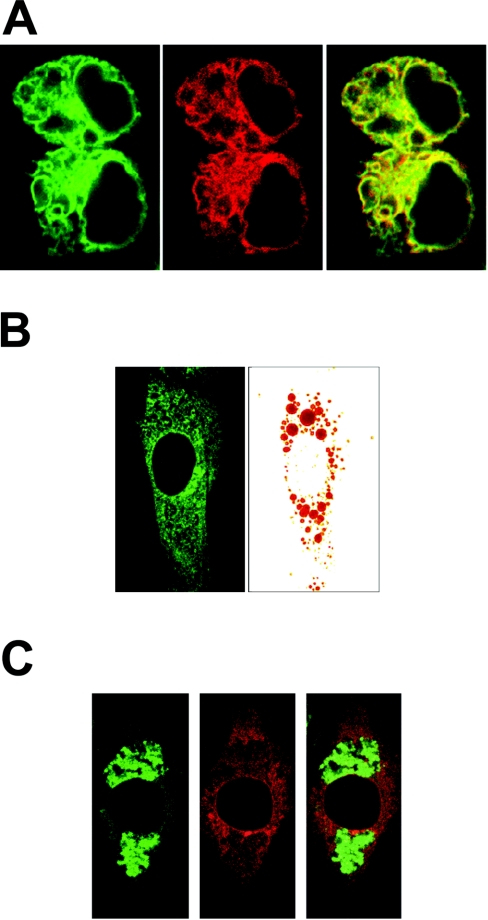
To get a better understanding of the function of G0S2, it is important to determine its intracellular localization. The presence of a single transmembrane helix indicated that G0S2 was probably anchored in a (sub)cellular membrane. To determine the precise intracellular localization of G0S2, a fusion construct was created between G0S2 and GFP (green fluorescent protein), which was transfected into HEK-293 cells. Because the Internet-based program PSORTII predicted G0S2 to be present in the ER, co-transfection was carried out with a marker vector for ER (pDsRed2-ER). Confocal fluorescence microscopy showed that GFP fluorescence was present in discrete regions within the cytoplasm, and that it perfectly overlapped with the DsRed (Discosoma sp. red fluorescent protein) fluorescence (Figure 6A), indicating that G0S2 protein is probably present in the ER.
Figure 6. G0S2 protein localizes to the ER.
HEK-293 cells were co-transfected with GFP–G0S2 fusion construct and ER localization vector pDsRed2-ER. (A) Left-hand panel: confocal image of GFP fluorescence. Middle panel: confocal image of DsRed fluorescence of the same cells as in the left-hand panel. Right-hand panel: overlay of left-hand and middle panels. (B) 3T3-L1 fibroblasts were transfected with fusion constructs of G0S2 to GFP and loaded with lipids by incubation with Tween 80 (0.1%). Lipid droplets were visualized with Oil Red O. (C) 3T3-L1 fibroblasts were transfected with fusion constructs of G0S2 to DsRed and loaded with lipids by incubation with Tween 80 (0.1%). Lipid droplets were visualized with BODIPY® 493/503.
Unfortunately, our anti-G0S2 antibody was not functional in immunohistochemistry, which precluded localization of endogenous G0S2 protein in differentiated adipocytes. To examine whether G0S2 protein might be associated with lipid droplets, which originate from the ER, and/or to study the effect of lipid droplets on the intracellular localization of G0S2, undifferentiated 3T3-L1 cells were transfected with fusion constructs of G0S2 to GFP (Figure 6B) and DsRed (Figure 6C) and were loaded with lipids by incubation with Tween 80. Lipid droplets were visualized with Oil Red O and BODIPY® 493/503, a green fluorescent dye that is compatible with DsRed. Fluorescence microscopy showed that G0S2 protein was present in distinct structures outside the nucleus, corresponding to the ER, and was not associated with lipid droplets.
G0S2 up-regulation is specifically associated with adipogenesis
Although our results indicate that G0S2 is highly up-regulated during 3T3-L1 and SGBS adipogenesis, it is unclear whether this effect is specific to adipocyte differentiation or whether it may extend to cell differentiation in general. To answer this question, G0S2 mRNA was monitored during C2C12 osteo- and myo-genesis. In this model, C2C12 cells are differentiated into myoblasts by letting them grow to post-confluence or into osteoblasts by incubation with BMP-2. In clear distinction to SGBS and 3T3-L1 adipogenesis, neither C2C12 osteogenesis nor myogenesis was associated with significantly increased G0S2 expression (Figure 7). The same was true for PPARγ. In contrast, the osteogenic marker osteocalcin showed a dramatic increase in expression during osteogenesis, while the glucose transporter GLUT4 was markedly increased during myogenesis. These results indicate that G0S2 is not involved in cell differentiation in general, but rather that GOS2 seems to be connected specifically to adipocyte differentiation.
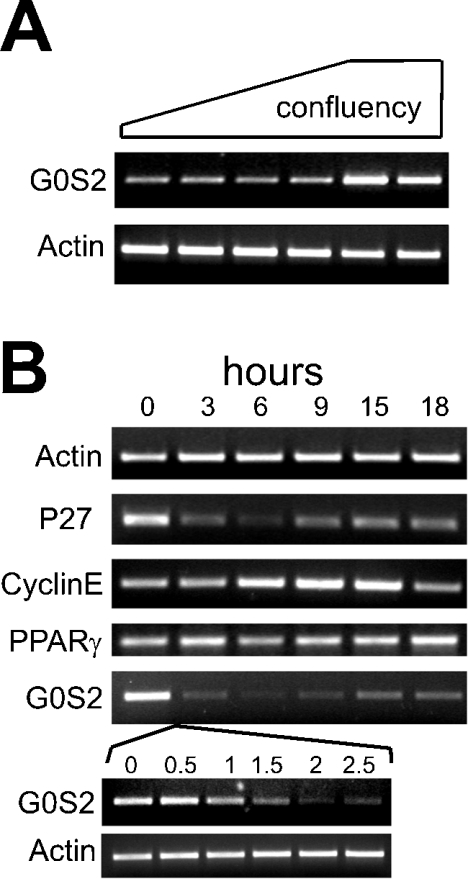
G0S2 mRNA is up-regulated during growth arrest in 3T3-L1 cells
Adipogenesis in 3T3-L1 cells is a complex process that involves numerous steps, including clonal expansion, growth arrest, and lipid synthesis and accumulation. In an effort to connect G0S2 to growth arrest in 3T3-L1 fibroblasts, the cells were first grown from low density to confluence, when cells should be in G0, and G0S2 mRNA expression was monitored. Interestingly, mRNA levels increased markedly when the cells reached full confluence, indicating that G0S2 expression is up-regulated in growth-arrested cells (Figure 8A). Subsequently, when cells were cell-cycle-synchronized by serum starvation, it was observed that expression of G0S2 was highest at the end of serum starvation, declined steeply in the next few hours after re-introducing serum, and reached a minimum after approx. 6–9 h, when cyclin E expression was maximal (Figure 8B). The peak of cyclin E expression is known to occur at the transition from the G1 to the S phase. G0S2 mRNA levels almost perfectly followed those of p27, which has been implicated previously in growth arrest in 3T3-L1 cells [23]. Expression of PPARγ did not change during serum starvation, suggesting that the fall in G0S2 mRNA is independent of PPARγ. These results provide strong evidence that, at least in 3T3-L1 cells, G0S2 expression is highest in growth-arrested cells and is minimal at the end of G1. Inasmuch as growth arrest is required for 3T3-L1 adipogenesis, G0S2 may thus be involved in adipogenesis by being implicated in growth arrest.
Figure 8. G0S2 mRNA is up-regulated during growth arrest.
(A) 3T3-L1 fibroblasts were plated out at low density and grown to full confluence. G0S2 expression was determined by RT-PCR. (B) 3T3-L1 fibroblasts at low confluence were serum-starved (0.2% foetal calf serum) for 33 h. After that, foetal calf serum was re-added to the cells at 10%, and cells were taken at regular intervals for RNA preparation. Expression of the genes indicated was determined by RT-PCR.
DISCUSSION
Using Affymetrix microarrays, we identified the G0S2 gene as being differentially expressed between livers of PPARα-null mice compared with wild-type mice. Follow-up analysis subsequently showed that G0S2 is a direct target gene of PPARγ, and probably PPARα and PPARβ/δ. Indeed, a functional PPRE could be identified in the human and mouse G0S2 promoter 1.4 kb upstream from the transcription start site.
However, some differences in the response to PPARα and PPARγ were observed. While PPARγ stimulated G0S2 promoter activity via the PPRE identified, the regulation by PPARα was a bit more complex. PPARα and Wy14643 failed to activate the full-length hG0S2 promoter, yet they decreased reporter activity after deleting the promoter to 1.0, 0.5 or 0.27 kb. The 0.27 kb promoter region thus appears to be able to mediate down-regulation of G0S2 promoter activity by PPARα. We hypothesize that this negative regulation is compensated for by positive regulation via the PPRE at −1.4 kb, causing the lack of responsiveness of the full G0S2 promoter to PPARα. A regulation very similar to that shown by PPARα was observed for PPARβ/δ (results not shown). Negative regulation by PPARα may be dominant in fed (male) mouse liver, where G0S2 is expressed at a somewhat higher level in PPARα-null mice compared with wild-type mice (Figure 1A). Currently, the mechanism behind this regulation is still unclear.
In the absence of PPARα, G0S2 expression declines during fasting. The mechanism behind this decrease is unclear, but may be due to decreased insulin signalling or increased glucagon or other hormonal changes during fasting, which are compensated for by PPARα.
Several lines of evidence suggest that G0S2 is also a target gene of PPARβ/δ in WAT. However, since the function of PPARβ/δ in WAT is debatable [7,24], the functional implications of this regulation remain unclear.
A limited number of genes are known to be dual targets of PPARα in liver and of PPARγ in adipose tissue. These include lipoprotein lipase, fatty acid transport protein, acyl-CoA synthase, FIAF (fasting-induced adipose factor)/ANGPTL4 (angiopoietin-like 4)/PGAR (PPARγ angiopoietin-related gene) and cytosolic GPDH [14,25–27]. As the roles of PPARα in liver and PPARγ in adipose tissue are almost completely opposite (PPARα: fatty acid oxidation=catabolism compared with PPARγ: adipo/lipo-genesis=anabolism), the pathways supported by the target genes in the respective organs are also likely to be different. This is true for cytosolic GPDH, fatty acid transport protein, acyl-CoA synthase and, to a lesser extent, lipoprotein lipase, which are part of different pathways in the two tissues. Accordingly, it is not unreasonable to suggest that G0S2, as a dual or even triple PPAR target, might participate in different pathways in liver and adipose tissue.
The dominant expression of G0S2 in BAT and WAT, combined with the dramatic (specific) up-regulation of G0S2 during mouse and human adipogenesis and the up-regulation of G0S2 during growth arrest in 3T3-L1 cells, which is required for 3T3-L1 adipogenesis, suggest that G0S2 may play a role in adipogenesis.
Adipogenesis describes the differentiation of pre-adipocytes into mature fat cells and has been extensively studied in vitro using 3T3-L1, 3T3-F442A and NIH-3T3 mouse fibroblasts. These studies have led to a generally accepted model of adipocyte differentiation in 3T3 cells, in which a sequential up- or down-regulation of several transcription factors, including E2Fs, GATAs and C/EBPs (CCAAT/enhancer-binding proteins) [28–30], brings about the emergence of an adipose phenotype via up-regulation of a large number of adipose-specific target genes. Perhaps the most important transcription factor is PPARγ, which was demonstrated to be both necessary and sufficient for induction of an adipose phenotype [31]. Up-regulation of target genes of PPARγ is connected with the acquisition of functions specific to adipocytes, such as fatty acid and triacylglycerol synthesis, insulin-dependent glucose transport and the synthesis of secreted factors such as resistin and adiponectin [32,33]. The differentiation of 3T3-L1 cells into adipocytes follows a well-studied sequence of events, each of which is essential for final differentiation and development of the adipocyte phenotype. One important event is cell-cycle withdrawal/growth arrest. According to our results, G0S2 may be associated with 3T3-L1 adipogenesis by its involvement in growth arrest.
Currently, the role of G0S2 in non-adipose tissues, such as liver, is not clear and, based on the previous argument, may diverge from its function in adipose tissue. Highest expression of G0S2 is found in adipose tissue, but mRNA levels are also reasonably high in liver, heart and other tissues. The very low expression of G0S2 in rapidly proliferating hepatoma cell lines (HepG2, FAO, Hepa1-6) in comparison with growth-arrested mouse liver suggests that the possible role of G0S2 in growth arrest/differentiation may extend beyond adipose tissue. At the same time, our studies in C2C12 cells clearly indicate that G0S2 is not a general marker of cell differentiation. Further studies are necessary to determine the role of G0S2 in non-adipose tissues.
G0S2 was initially discovered using differential hybridization in blood mononuclear cells as a gene that is very transiently induced after treatment with concanavalin A (a lectin), cycloheximide (a protein synthesis inhibitor) and the combination of PMA (a phorbol ester) and ionomycin (a calcium ionophore) [21]. This rapid and transient increase in expression was inhibited by cyclosporin A. These results led the authors to conclude that G0S2 expression is transiently induced upon re-entry of cells into the G1 phase of the cell cycle and would be required to commit cells to enter G1 [21]. In contrast, our results indicate that up-regulation of G0S2 is associated with cell-cycle withdrawal. The reason for this discrepancy is not clear, but it may point to a cell-type-specific function for G0S2. Alternatively, transient up-regulation of G0S2 in blood mononuclear cells by any of the compounds mentioned above may reflect a different event from re-entry into the cell cycle.
The limited information available about G0S2 before the present study included an in situ hybridization analysis of G0S2 expression in mice embryos. It was found that, at day 18.5, G0S2 expression is restricted to BAT and WAT [34]. The present study confirms that G0S2 is mainly expressed in WAT and BAT, but also indicates that G0S2 mRNA is reasonably well expressed in other tissues, such as lung, liver and heart. The reason for this discrepancy is not exactly clear, but it may be due to a difference in sensitivity between the techniques used to detect G0S2 mRNA (in situ hybridization compared with Q-PCR) or a difference in the age of the animal (embryonic day 18.5 compared with adult animal). The latter explanation would support a role for G0S2 in growth arrest, since, at the embryonic stage, tissues such as muscle and liver still display a high rate of cell proliferation, whereas, in the adult stage, liver and muscle cells are highly differentiated and arrested in G0, which would result in increased G0S2 expression.
In the present study, G0S2 protein was localized to the ER. Analysis of the primary sequence by the Internet-based PSORT II program predicted the N-terminal domain comprising amino acids 1–26 to be protruding into the cytoplasm, whereas the C-terminal domain comprising amino acids 43–103 is expected to be in the ER lumen. The molecular mechanism by which G0S2 may influence growth arrest and, accordingly, adipogenesis would probably involve some kind of protein–protein interaction via either of these domains. Future studies will have to address this in more detail.
Finally, our microarray experiment corroborated perfectly the concept that PPARα is an important regulator of fatty acid oxidation and ketogenesis, and that the function of PPARα becomes mainly evident during fasting. Possible new target genes of PPARα that emerged from our microarray screen include those for insulin-like growth factor-binding protein 2, folylpolyglutamate synthetase and LDL (low-density lipoprotein)-receptor-related protein 1. These results underscore the utility of microarray analysis in finding and characterizing novel potential target genes of nuclear hormone receptors.
In conclusion, we have identified the G0S2 as a novel direct target gene of PPARγ, and probably PPARα and PPARβ/δ, and present results suggesting that it is involved in adipocyte differentiation.
Acknowledgments
We are grateful to Dr Wilma Steegenga for the RT samples from C2C12 cells, and Dr M. Wabitsch for the gift of the SGBS cell line. We thank Erika Ferguson for excellent technical assistance, Yixin Wang for his support in microarray data analysis, Ken (Gang) Hu for his advice in GeneSpring program, Gary McMaster and Steven Hunt for their support to carry out the microarray study. This study was financed by the Netherlands Organization for Scientific Research (NWO), with additional support from the Royal Netherlands Academy of Art and Sciences (KNAW), the Wageningen Center for Food Sciences, the Swiss National Science Foundation and the Human Frontier Science Program (HFSP). We declare no conflict of interest.
References
- 1.Kersten S., Desvergne B., Wahli W. Roles of PPARs in health and disease. Nature (London) 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P., Hu E., Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 3.Barak Y., Nelson M. C., Ong E. S., Jones Y. Z., Ruiz-Lozano P., Chien K. R., Koder A., Evans R. M. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 4.Ren D., Collingwood T. N., Rebar E. J., Wolffe A. P., Camp H. S. PPARγ knockdown by engineered transcription factors: exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev. 2002;16:27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen E. D., Hsu C. H., Wang X., Sakai S., Freeman M. W., Gonzalez F. J., Spiegelman B. M. C/EBPα induces adipogenesis through PPARγ: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Way J. M., Harrington W. W., Brown K. K., Gottschalk W. K., Sundseth S. S., Mansfield T. A., Ramachandran R. K., Willson T. M., Kliewer S. A. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor gamma activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology. 2001;142:1269–1277. doi: 10.1210/endo.142.3.8037. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y. X., Zhang C. L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2004;2:E294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyama T. E., Lambert G., Nicol C. J., Matsusue K., Peters J. M., Brewer H. B., Jr, Gonzalez F. J. Peroxisome proliferator-activated receptor β/δ regulates very low density lipoprotein production and catabolism in mice on a Western diet. J. Biol. Chem. 2004;279:20874–20881. doi: 10.1074/jbc.M312802200. [DOI] [PubMed] [Google Scholar]
- 10.Di Poi N., Tan N. S., Michalik L., Wahli W., Desvergne B. Antiapoptotic role of PPARβ in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol. Cell. 2002;10:721–733. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- 11.Harman F. S., Nicol C. J., Marin H. E., Ward J. M., Gonzalez F. J., Peters J. M. Peroxisome proliferator-activated receptor-δ attenuates colon carcinogenesis. Nat. Med. 2004;10:481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 12.Gupta R. A., Wang D., Katkuri S., Wang H., Dey S. K., DuBois R. N. Activation of nuclear hormone receptor peroxisome proliferator-activated receptor-δ accelerates intestinal adenoma growth. Nat. Med. 2004;10:245–247. doi: 10.1038/nm993. [DOI] [PubMed] [Google Scholar]
- 13.Mandard S., Müller M., Kersten S. Peroxisome proliferator-activated receptor α target genes. Cell. Mol. Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patsouris D., Mandard S., Voshol P. J., Escher P., Tan N. S., Havekes L. M., Koenig W., Marz W., Tafuri S., Wahli W., et al. PPARα governs glycerol metabolism. J. Clin. Invest. 2004;114:94–103. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gervois P., Kleemann R., Pilon A., Percevault F., Koenig W., Staels B., Kooistra T. Global suppression of IL-6-induced acute phase response gene expression after chronic in vivo treatment with the peroxisome proliferator-activated receptor-α activator fenofibrate. J. Biol. Chem. 2004;279:16154–16160. doi: 10.1074/jbc.M400346200. [DOI] [PubMed] [Google Scholar]
- 16.Delerive P., Fruchart J. C., Staels B. Peroxisome proliferator-activated receptors in inflammation control. J. Endocrinol. 2001;169:453–459. doi: 10.1677/joe.0.1690453. [DOI] [PubMed] [Google Scholar]
- 17.Beigneux A. P., Moser A. H., Shigenaga J. K., Grunfeld C., Feingold K. R. The acute phase response is associated with retinoid X receptor repression in rodent liver. J. Biol. Chem. 2000;275:16390–16399. doi: 10.1074/jbc.M000953200. [DOI] [PubMed] [Google Scholar]
- 18.Moshage H., Casini A., Lieber C. S. Acetaldehyde selectively stimulates collagen production in cultured rat liver fat-storing cells but not in hepatocytes. Hepatology. 1990;12:511–518. doi: 10.1002/hep.1840120311. [DOI] [PubMed] [Google Scholar]
- 19.Wabitsch M., Brenner R. E., Melzner I., Braun M., Moller P., Heinze E., Debatin K. M., Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 20.Kersten S., Mandard S., Tan N. S., Escher P., Metzger D., Chambon P., Gonzalez F. J., Desvergne B., Wahli W. Characterization of the fasting-induced adipose factor FIAF, a novel peroxisome proliferator-activated receptor target gene. J. Biol. Chem. 2000;275:28488–28493. doi: 10.1074/jbc.M004029200. [DOI] [PubMed] [Google Scholar]
- 21.Russell L., Forsdyke D. R. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol. 1991;10:581–591. doi: 10.1089/dna.1991.10.581. [DOI] [PubMed] [Google Scholar]
- 22.Berger J., Leibowitz M. D., Doebber T. W., Elbrecht A., Zhang B., Zhou G., Biswas C., Cullinan C. A., Hayes N. S., Li Y., et al. Novel peroxisome proliferator-activated receptor (PPAR) γ and PPARδ ligands produce distinct biological effects. J. Biol. Chem. 1999;274:6718–6725. doi: 10.1074/jbc.274.10.6718. [DOI] [PubMed] [Google Scholar]
- 23.Morrison R. F., Farmer S. R. Role of PPARγ in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18INK4c and p21Waf1/Cip1, during adipogenesis. J. Biol. Chem. 1999;274:17088–17097. doi: 10.1074/jbc.274.24.17088. [DOI] [PubMed] [Google Scholar]
- 24.Matsusue K., Peters J. M., Gonzalez F. J. PPARβ/δ potentiates PPARγ-stimulated adipocyte differentiation. FASEB J. 2004;18:1477–1479. doi: 10.1096/fj.04-1944fje. [DOI] [PubMed] [Google Scholar]
- 25.Martin G., Schoonjans K., Lefebvre A. M., Staels B., Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J. Biol. Chem. 1997;272:28210–28217. doi: 10.1074/jbc.272.45.28210. [DOI] [PubMed] [Google Scholar]
- 26.Schoonjans K., Peinado-Onsurbe J., Lefebvre A. M., Heyman R. A., Briggs M., Deeb S., Staels B., Auwerx J. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 27.Mandard S., Zandbergen F., Tan N. S., Escher P., Patsouris D., Koenig W., Kleemann R., Bakker A., Veenman F., Wahli W., et al. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J. Biol. Chem. 2004;279:34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 28.Fajas L., Landsberg R. L., Huss-Garcia Y., Sardet C., Lees J. A., Auwerx J. E2Fs regulate adipocyte differentiation. Dev. Cell. 2002;3:39–49. doi: 10.1016/s1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 29.Tong Q., Dalgin G., Xu H., Ting C. N., Leiden J. M., Hotamisligil G. S. Function of GATA transcription factors in preadipocyte–adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 30.Christy R. J., Yang V. W., Ntambi J. M., Geiman D. E., Landschulz W. H., Friedman A. D., Nakabeppu Y., Kelly T. J., Lane M. D. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev. 1989;3:1323–1335. doi: 10.1101/gad.3.9.1323. [DOI] [PubMed] [Google Scholar]
- 31.Rosen E. D., Spiegelman B. M. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 32.Steppan C. M., Bailey S. T., Bhat S., Brown E. J., Banerjee R. R., Wright C. M., Patel H. R., Ahima R. S., Lazar M. A. The hormone resistin links obesity to diabetes. Nature (London) 2001;409:307–312. doi: 10.1038/35053000. [DOI] [PubMed] [Google Scholar]
- 33.Hu E., Liang P., Spiegelman B. M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 34.Bachner D., Ahrens M., Schroder D., Hoffmann A., Lauber J., Betat N., Steinert P., Flohe L., Gross G. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2) Dev. Dyn. 1998;213:398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]