Abstract
Activation of STAT3 (signal transducer and activator of transcription 3) plays a crucial role in cell survival and proliferation. The aim of the present study was to clarify the role of STAT3 signalling in the protection of polyamine-depleted intestinal epithelial cells against TNF-α (tumour necrosis factor-α)-induced apoptosis. Polyamine depletion by DFMO (α-difluoromethylornithine) caused phosphorylation of STAT3 at Tyr-705 and Ser-727. Phospho-Tyr-705 STAT3 was immunolocalized at the cell periphery and nucleus, whereas phospho-Ser-727 STAT3 was predominantly detected in the nucleus of polyamine-depleted cells. Sustained phosphorylation of STAT3 at tyrosine residues was observed in polyamine-depleted cells after exposure to TNF-α. Inhibition of STAT3 activation by AG490 or cell-membrane-permeant inhibitory peptide (PpYLKTK; where pY represents phospho-Tyr) increased the sensitivity of polyamine-depleted cells to apoptosis. Expression of DN-STAT3 (dominant negative-STAT3) completely eliminated the protective effect of DFMO against TNF-α-induced apoptosis. Polyamine depletion increased mRNA and protein levels for Bcl-2, Mcl-1 (myeloid cell leukaemia-1) and c-IAP2 (inhibitor of apoptosis protein-2). Significantly higher levels of Bcl-2 and c-IAP2 proteins were observed in polyamine-depleted cells before and after 9 h of TNF-α treatment. Inhibition of STAT3 by AG490 and DN-STAT3 decreased Bcl-2 promoter activity. DN-STAT3 decreased mRNA and protein levels for Bcl-2, Mcl-1 and c-IAP2 in polyamine-depleted cells. siRNA (small interfering RNA)-mediated inhibition of Bcl-2, Mcl-1 and c-IAP2 protein levels increased TNF-α-induced apoptosis. DN-STAT3 induced the activation of caspase-3 and PARP [poly(ADP-ribose) polymerase] cleavage in polyamine-depleted cells. These results suggest that activation of STAT3 in response to polyamine depletion increases the transcription and subsequent expression of anti-apoptotic Bcl-2 and IAP family proteins and thereby promotes survival of cells against TNF-α-induced apoptosis.
Keywords: caspase-3, α-difluoromethylornithine (DFMO), epithelium, putrescine, polyamine, STAT3
Abbreviations: BIR, baculovirus inhibitor of apoptosis protein repeat; CHX, cycloheximide; DFMO, α-difluoromethylornithine; DMEM, Dulbecco's modified Eagle's medium; dFBS, dialysed fetal bovine serum; IAP, inhibitor of apoptosis protein; IEC-6, intestinal epithelial cell-6; JAK, Janus kinase; Mcl-1, myeloid cell leukaemia-1; MMLV, Moloney murine leukaemia virus; NF-κB, nuclear factor-κB; PARP, poly(ADP-ribose) polymerase; RT-PCR, reverse transcription-PCR; siRNA, small interfering RNA; STAT3, signal transducer and activator of transcription 3; DN-STAT3, dominant negative-STAT3; TNF-α, tumour necrosis factor-α
INTRODUCTION
Apoptosis in the intestinal epithelium is a central regulator of mucosal homoeostasis. Apoptosis primarily occurs at the stem cell position in the crypt area and is responsible for the balance in cell number between newly divided and surviving cells [1,2]. The natural polyamines spermidine, spermine and their precursor putrescine are found in all eukaryotic cells. Polyamines are low-molecular-mass aliphatic amines with multiple-charged cations at physiological pH. Polyamines are synthesized from a common precursor, ornithine, which is decarboxylated by the rate-limiting enzyme ornithine decarboxylase. DFMO (α-difluoromethylornithine), an irreversible pharmacological inhibitor of ornithine decarboxylase, has been used extensively to study the function of polyamines in different cell systems [3,4]. Our laboratory and other investigators have studied the multiple regulatory roles of polyamines in the proliferation, differentiation, migration and apoptosis of intestinal epithelial cells [4–9]. In order to elucidate the function of polyamines in intestinal epithelium, we have used undifferentiated and untransformed intestinal crypt cells isolated from rat intestine [10]. Emerging evidence indicates a central role for cellular polyamines in influencing epithelial cell functions [5]. Disrupting polyamine homoeostasis using enzyme inhibitors or polyamine analogues supports the idea that polyamines are important determinants in the control of apoptotic cell death in multiple cell types [3]. Although polyamines have been implicated in the regulation of apoptosis in the intestinal epithelium, the mechanisms involved are unclear. Extensive studies have examined the role of polyamines in gastrointestinal mucosal homoeostasis both in cultured undifferentiated IEC (intestinal epithelial cell)-6 lines and in rats [4,11]. Previously, we have shown that polyamines are required for the normal progression of apoptosis in IEC-6 cells and we have identified the anti-apoptotic signal transduction pathways involved in the protection of these cells from TNF-α (tumour necrosis factor-α)- and camptothecin-induced apoptosis [4,12–14].
We have also shown that NF-κB (nuclear factor-κB) and STAT3 (signal transducer and activator of transcription 3) activation were the most immediate effects in response to short-term treatment of IEC-6 cells with DFMO [15,16]. In response to cytokine stimulation, JAKs (Janus kinases) activate STAT3 through phosphorylation of a single tyrosine residue (Tyr-705) resulting in dimerization and translocation of STAT3 to the nucleus where it binds to defined target sequences and activates transcription [17,18]. Additional phosphorylation of STAT3 at Ser-727 has been predicted to be responsible for increased transcriptional activity [19]. Constitutive activation of STAT3 has been described in a wide variety of primary tumours [20] and has also been reported to induce cell survival in association with the expression of survivin in gastric cancer cells [21]. Inhibition of STAT3 expression induces apoptosis in astrocytoma cells, large granular lymphocytes and human prostrate cancer cells [22–24]. STAT3 signalling is reported to mediate the survival of oncogenic Ras-transfected intestinal epithelial cells [25]. Furthermore, STAT3 activation results in the upregulation of various genes involved in cell survival and proliferation, such as those encoding Bcl-2, Bcl-XL, Mcl-1 (myeloid cell leukaemia-1), cyclin-D1 and c-Myc [25–27].
Bcl-2, an anti-apoptotic oncoprotein, plays a key role in regulating cell survival through protein–protein interactions with other Bcl-2-related family members, such as the cell death suppressors Bcl-XL, Mcl-1 and Bcl-w or the cell death agonists Bax, Bad (Bcl-2/Bcl-XL-antagonist) and Bid [28]. Although previous results from our laboratory demonstrated an increase in Bcl-2 protein levels in polyamine-depleted cells [29], the mechanism by which polyamines regulate the expression of Bcl-2 has not been elucidated. The recently identified human Mcl-1 early induction gene belongs to the pro-survival Bcl-2 family proteins [30,31]. Mcl-1 is regulated at the transcriptional and post-transcriptional levels resulting in alternative splicing [32]. Serine phosphorylation of STAT3 has been reported to mediate Mcl-1 expression and macrophage survival [33]. Mcl-1 expression has also been reported to be down-regulated during apoptosis in B-cells [34].
The IAP (inhibitor of apoptosis protein) family modulate apoptotic cell death and function by preventing activation of caspases, the principal effectors of cell death [35]. The IAPs are defined by a 70-amino-acid domain, the BIR (baculovirus IAP repeat) which binds directly to caspases and is required for IAPs to inhibit apoptosis. The IAP family includes c-IAP1, c-IAP2, X-linked IAP, survivin, neuronal apoptosis inhibitory protein and melanoma-IAP [36]. IAP2 has been shown to mediate the anti-apoptotic function of cAMP in normal and transformed intestinal epithelial cells [37,38].
Early activation of STAT3 in response to DFMO treatment suggests that STAT3 might be involved in the regulation of apoptosis in polyamine-depleted cells. Using microarray analysis we found that the genes encoding the anti-apoptotic proteins Bcl-2, Mcl-1, and c-IAP2 (also known as BIRC3), known to be upregulated by STAT3, have increased levels of expression in polyamine-depleted cells. In the present study we show that activation of these genes by STAT3 accounts for resistance to apoptosis following polyamine depletion.
EXPERIMENTAL
Reagents, constructs and antibodies
Cell culture ware was purchased from Corning Glass works (Corning, NY, U.S.A.). Media and other cell culture reagents were obtained from Gibco (Grand Island, MO, U.S.A.). dFBS (dialysed fetal bovine serum) and anti-actin antibody were purchased from Sigma (St. Louis, MO, U.S.A.). DN-STAT3 (dominant negative-STAT3) plasmid and empty vector were kindly given by Dr Lawrence Pfeffer, University of Tennessee Health Science Center, Memphis, TN, U.S.A. The Bcl-2 promoter–luciferase construct was kindly given by Dr Linda Boxer, Stanford University, Stanford, CA, U.S.A. The full-length Bcl-2 promoter was cloned into the BamHI and HindIII sites and the luciferase cDNA was cloned into the HindIII and KpnI sites of the pBS vector. The Bcl-2 promoter construct has been decribed previously [39]. FuGENE6™ transfection reagent was purchased from Roche Diagnostic Corporation, Indianapolis, IN, U.S.A. Geneticin (G418) was purchased from Gibco. TNF-α was obtained from PharMingen International, San Diego, CA, U.S.A. The luciferase assay kit was purchased from Promega, Madison, WI, U.S.A. The Enhanced Chemiluminescence (ECL) Western Blot detection system was purchased from PerkinElmer, Boston, MA, U.S.A. DFMO was a gift from ILEX Oncology™ Inc, San Antonio, TX, U.S.A. Mcl-1 and caspase-3 antibodies were from Santa Cruz Biotechnology Santa Cruz, CA, U.S.A. and Bcl-2, phospho-Ser-727 STAT3 and STAT3 antibodies were from BD Biosciences, San Diego, CA, U.S.A. Antibodies against phospho-Tyr-705-specific STAT3 and PARP [poly (ADP-ribose) polymerase], cleaved at Asp-214, were from Cell Signaling, Beverly, MA, U.S.A. cIAP-2 antibody was purchased from Novus Biologicals, Littleton, CO, U.S.A. and Santa Cruz Biotechnology. AG490 (JAK-2 inhibitor) and PpYLKTK (cell-membrane-permeant STAT3 inhibitor peptide) were purchased from Calbiochem, EMD Biosciences, La Jolla, CA, U.S.A. TRIzol® reagent was purchased from Invitrogen. Recombinant RNasin ribonuclease inhibitor, MMLV (Moloney-murine-leukaemia virus) reverse transcriptase, random primers, dNTP mix and PCR master mix were purchased from Promega Corporation, Madison, WI, U.S.A. Mcl-1 and Bcl-2 primer pairs were obtained from R&D Systems, Inc. (Minneapolis, MN, U.S.A.) and c-IAP2 primer was from Integrated DNA Technologies, Inc, Coralville, IA, U.S.A. Bcl-2, Mcl-1 and c-IAP2 siRNA (small interfering RNA) was obtained from Santa Cruz Biotechnology. The Cell Death Detection ELISA Plus kit was purchased from Roche Diagnostics. The nuclear fractionation kit was purchased from Active Motif, Carlsbad, CA, U.S.A. The IEC-6 line (A.T.C.C. CRL 1592) was obtained from American Type Culture Collection, Rockville, MD, U.S.A. at passage 13. The cell line was derived from normal rat intestine and was developed and characterized by Quaroni et al. [10]. IEC-6 cells originate from intestinal crypt cells as judged by morphological and immunological criteria. They are non-tumorigenic and retain the undifferentiated character of epithelial stem cells. Tests for mycoplasma were consistently negative. All chemicals were of the highest purity commercially available.
Cell culture
IEC-6 cell stock was maintained in T-150 flasks in a humidified incubator at 37 °C in an atmosphere of air/CO2 (90:10). The medium consisted of DMEM (Dulbecco's modified Eagle's medium), heat-inactivated 5% FBS, and 10 μg of insulin and 50 μg of gentamicin sulphate/ml. The stock flask was passaged weekly, cells were fed three times per week, and passages 15–22 were used. For experiments, cells were trypsinized with 0.05% trypsin and 0.53 mM EDTA and counted using a Beckman Coulter Counter (Model Z1). General protocols for experiments using IEC-6 cells have been described previously [4]. Briefly, IEC-6 cells were plated at 6.24×104 cells/cm2 in DMEM as described, fed every other day, and serum starved for 24 h before harvesting. Polyamines were depleted by incubation in 5 mM DFMO. We have previously reported that maximal polyamine depletion occurs after 4 days of treatment with 5 mM DFMO [4]. Within 6 h of DFMO treatment putrescine was undetectable, spermidine was absent after 24 h, and 40% of spermine remained after 4 days. One group of cells was given exogenous putrescine (10 μM) in addition to DFMO. This group acted as a control to indicate that all results were due to the depletion of polyamines and not to DFMO itself.
Transfection
The protocol for transfection of IEC-6 cells has been previously described [13]. Briefly, DN-STAT3 and pCEF (vector) DNA were prepared using the Qiagen (endotoxin-free) plasmid preparation kit (Valencia, CA, U.S.A). FuGENE-6 reagent was mixed with DNA (3 μl/2 μg) in serum-free medium, to a total volume of 100 μl and incubated for 30 min at room temperature. IEC-6 cells at a relatively early passage were grown to 70–80% confluence in 60 mm dishes. For transient transfection the cell monolayer was rinsed with serum-free medium and the DNA/FuGENE mixture was added dropwise on to the cell monolayers and incubated for a further 12 h at 37 °C.
The Bcl-2 promoter–luciferase reporter gene construct was transfected into IEC-6 cells using FuGENE-6 as described above. Reporter gene activity was determined on day 3 after transfection. Reporter gene activity was assayed using the Luciferase Assay System from Promega following the manufacturer's instructions. Activity was expressed as relative luciferase units per mg of protein (RLU/mg). All transient transfection results are presented as the average from at least six independent transfections.
RNA interference
IEC-6 cells at a relatively early passage were grown to 50–60% confluence in 12-well plates. For transfection, the cell monolayer was rinsed with serum-free medium and the siRNA/FuGENE-6 mixture (3 μl:3.4 μl) was added dropwise on to the cell monolayer and incubated for 24 h at 37 °C. Cells were serum starved for 24 h, treated with TNF-α/CHX (cycloheximide) for 3 h and apoptosis was determined by DNA fragmentation assay. One group of cells was transfected with control siRNA; the siRNA transfection method has previously been optimized for IEC-6 cells [14]. Efficiency of transfection was monitored using FITC-conjugated control siRNA.
Apoptosis
Cells were plated (day 0) at a density of 6.25×104 cells/cm2 in DMEM/dFBS with or without DFMO or DFMO plus putrescine, with triplicate samples for each group. Cells were fed on day 2; on day 3, the culture medium was removed and replaced with serum-free medium. On day 4, 20 ng/ml TNF-α or 25 μg/ml CHX was added to the serum-free medium for 3 h, with the vehicle DMSO added to controls, and cells were subjected to DNA fragmentation assay using the Cell Death Detection ELISA Plus kit as described previously [12,13].
Western blot analysis
Lysates were prepared as described earlier [14] and 25–50 μg of protein was trichloroacetic acid-precipitated and dissolved in 1× SDS-sample buffer for 5 min and separated on SDS/PAGE (10–15% gels), transferred overnight to Immobilon-P membranes (Millipore, Bedford, MA, U.S.A). and probed with the indicated antibodies. Membranes were subsequently incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature and the immunocomplexes were visualized using the ECL® detection system (Perkin Elmer). Blots were stripped and probed with the indicated antibodies to determine equal loading of the samples.
Immunocytochemistry
Cells were seeded on cover slips and grown for 2 days in DMEM/5% dFBS, washed with Dulbecco's PBS and fixed in PBS/4% paraformaldehyde for 15 min at room temperature. This was followed by permeabilization in 0.1% Triton X-100 for 10 min and rinsing with PBS. The cells were blocked in sterile PBS with 2% (w/v) BSA for 20 min. Subsequently, cells were rinsed with PBS/0.1% (w/v) BSA for 20 min and incubated with the primary antibody (1:200) for 1 h at room temperature. Cells were then rinsed five times with PBS/0.1% (w/v) BSA and incubated with the secondary antibody–FITC for 1 h at room temperature in the dark. They were rinsed with PBS three times and mounted on a slide and sealed. Images were captured using a microscope with a band pass filter to detect fluorescein.
RT-PCR (reverse transcription-PCR)
Total RNA was isolated using TRIzol® reagent according to the manufacturer's instructions. The quality of total RNA was tested by formaldehyde–agarose gel electrophoresis. Equal amounts of RNA (2 μg) were used to synthesize single-strand cDNA using 0.5 μg of random primers and MMLV reverse transcriptase following the manufacturer's instructions. The resulting cDNA was used as a template for PCR using gene-specific primers. Briefly, PCR primers were designed with a melting temperature of 59–61 °C. Forward and reverse primers spanning exon–exon junctions were selected to avoid amplification of genome sequences. Actin: forward primer, 5′-CGTGAAAAGATGACCCAGATCA-3′; reverse primer, 5′-CCAGCCTGGATGGCTACGT-3′; c-IAP2: forward primer, 5′-TCCATCAAATCCTGTAAACTCC-3′; reverse primer, 5′-AGCAAGCCACTCTGTCTCC-3′. Bcl-2 and Mcl-1 primer mixes were obtained commercially. The transcript of rat β-actin was used as an internal RNA control, and each sample was normalized on the basis of its β-actin content. The thermal cycling conditions involved an initial denaturing step at 94 °C for 45 s, followed by annealing at 55 °C for 45 s and 72 °C for 45 s and an annealing extension step at 72 °C for 10 min.
Statistical analysis
All data are expressed as means±S.E.M. All experiments were repeated three times, with triplicate samples for each. Analysis of variance and appropriate post-hoc testing determined the significance of the differences between means. Values of P<0.05 were regarded as significant.
RESULTS
Activation of STAT3 in polyamine-depleted cells
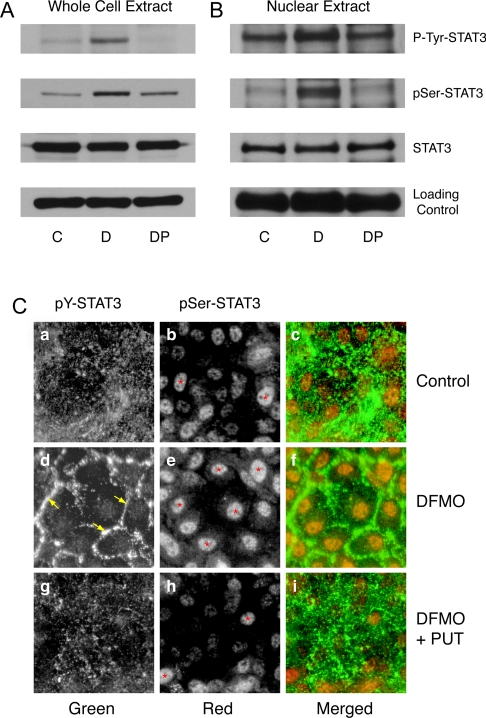
We determined the levels of phospho-Tyr-705 and phospho-Ser-727 STAT3 and total STAT3 from whole cell extracts and the nuclear fractions of cells grown in control, DFMO- and DFMO-plus-putrescine-containing medium for 4 days. Figure 1(A) shows significantly higher levels of phospho-Tyr-705 and phospho-Ser-727 STAT3 in cells grown in the presence of DFMO compared with those grown under control and DFMO plus putrescine conditions. The levels of total STAT3 protein remained the same under all three conditions. Although total STAT3 protein levels were the same in the nuclear fraction, significantly higher levels of phospho-Tyr-705 and phospho-Ser-727 STAT3 were present in DFMO-treated cells. Control cells and those treated with DFMO plus putrescine had relatively less nuclear phospho-Tyr-705 and phospho-Ser-727 STAT3 (Figure 1B). Immunolocalization of STAT3 was carried out using phospho-Tyr-705- and phospho-Ser-727-specific STAT3 antibodies. Figure 1(C) shows intense nuclear localization of phospho-Ser-727 STAT3 in DFMO-treated cells. Control cells and those treated with DFMO plus putrescine had relatively less nuclear phospho-Ser STAT3 (Figure 1C, panels b and h). Phospho-Tyr-705 STAT3 predominantly localized at the cell periphery, indicating membrane recruitment and activation in cells grown in the presence of DFMO (Figure 1C, panel d). Cells grown in control and DFMO-plus-putrescine-containing media had a punctate and scattered distribution of phospho-Tyr-705 STAT3 (Figure 1C, panels a and g). The merged images showed intense tyrosine and serine phosphorylation of STAT3 in polyamine-depleted cells (Figure 1C, panel f).
Figure 1. STAT3 phosphorylation in polyamine-depleted cells.
(A) Western blot analysis of whole cell extracts using antibodies against phospho-Tyr-705 STAT3 and phospho-Ser-727 STAT3. Blots were stripped and probed for total STAT3 protein, followed by β-actin as an internal loading control. (B) Western blot analysis of nuclear extracts probed with phospho-Tyr-705 and phospho-Ser-727 STAT3 antibodies. Membranes were stained with Ponceau-S as a loading control. IEC-6 cells were grown in DMEM/5% (w/v) dFBS (C) with or without 5 mM DFMO (D) or DFMO plus 10 μM putrescine (DP) for 3 days and serum-deprived for 24 h. Representative blots from three observations are shown. (C) Cells were grown on poly(L-lysine)-coated coverslips for 3 days and serum-deprived for 24 h. PUT, putrescine. Cells were stained using phospho-Tyr-705 and phospho-Ser-727 STAT3-specific antibodies followed by a secondary antibody conjugated to either Alexa-Fluor 488 (green) or Alexa-Fluor 568 (red). STAT3 phosphorylation at Tyr-705 (green) and Ser-727 (red) was visualized using a UV fluorescence microscope. ↓ and * indicate STAT3 localization at the cell periphery and nuclei respectively.
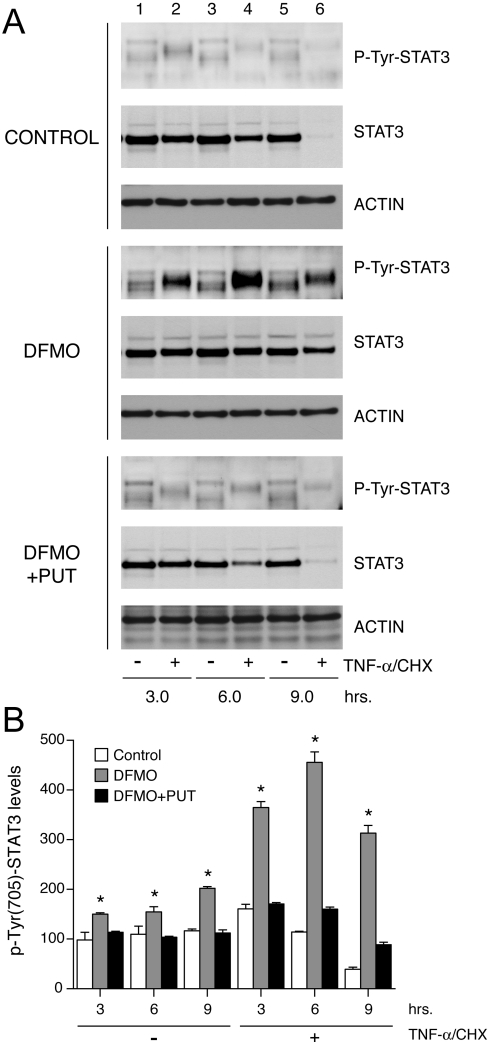
In order to investigate the role of STAT3 in apoptosis, we treated cells grown in control, DFMO- and DFMO-plus-putrescine-containing media with TNF-α for 3, 6 and 9 h, and determined STAT3 activation. Cell lysates were probed with phospho-Tyr-705 STAT3-specific antibody. Figure 2 shows that basal as well as TNF-α-induced phosphorylation of STAT3 at Tyr-705 increased in a time-dependent fashion in polyamine-depleted cells (Figure 2A, lanes 2, 4 and 6). Conversely, phosphorylation of STAT3 in cells grown in control, and DFMO plus putrescine containing media decreased in a time dependent manner following TNF-α treatment. The same lysates when probed with a STAT3 antibody showed a gradual decrease of STAT3 protein levels after 3 h of TNF-α treatment and its disappearance after 9 h both in control and DFMO plus putrescine groups (Figure 2A, lane 6). However, polyamine-depleted cells retained a significant amount of STAT3 protein even after 9 h of TNF-α treatment. Levels of β-actin, an internal loading control, showed no change in any of the three groups. Figure 2(B) depicts the densitometric analysis of the results shown in Figure 2(A).
Figure 2. STAT3 activation in response to TNF-α.
IEC-6 cells were grown as described in the Experimental section and treated with TNF-α/CHX for 3, 6 and 9 h. (A) Western blot analysis with antibodies against phospho-Tyr-705 STAT3 and total STAT3. Blots were stripped and probed with anti-actin antibody to confirm equal loading of samples. Representative blots from three independent observations are shown. (B) Densitometric analysis of STAT3 activation. *, P<0.005 compared with the respective untreated group. PUT, putrescine.
Effect of STAT3 inhibition on TNF-α-induced apoptosis in polyamine-depleted cells
Since polyamine depletion increased STAT3 activation (Figure 1), we determined the effect of STAT3 inhibition on apoptosis in these cells using a JAK inhibitor, tyrphostin AG490. We also used a highly selective cell-membrane-permeant peptide, PpYLKTK, containing a STAT3 SH2 (Src homology 2)-binding domain to block STAT3 activation. Inhibition of STAT3 dimerization and nuclear translocation by PpYLKTK did not increase spontaneous or TNF-α-induced apoptosis in control cells (Table 1). However, the inhibitory peptide significantly increased TNF-α-induced DNA fragmentation in the cells grown in presence of DFMO (Table 1). Addition of 50 μM AG490 increased basal, as well as TNF-α-induced, apoptosis in cells grown in control and in DFMO-containing media (Table 1). Although AG490 increased DNA fragmentation in control cells and significantly eliminated the protection conferred by DFMO in response to TNF-α, the degree of apoptosis was significantly lower compared with controls. The above results indicate that activation of STAT3 plays a significant role in the regulation of apoptosis. The constitutive activation of STAT3 (Figures 1 and 2) in polyamine-depleted cells contributes to the survival response against TNF-α in these cells.
Table 1. Inhibition of STAT3 and apoptosis.
IEC-6 cells were treated with TNF-α/CHX (plus/minus) either in the presence or absence of 50 μM PpYLKTK, a cell-permeant STAT3 inhibitory peptide, for 3 h or of 50 μM AG490, a JAK inhibitor. DNA fragmentation was assayed by cell death detection ELISA and expressed as absorbance 405 nm/mg of protein per min. *, P<0.005 compared with the respective untreated group (means±S.E.M., n=6).
| DNA fragmentation (A405/min per mg) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vehicle | PYLKTK | Vehicle | AG490 | ||||||
| TNF-α/CHX… | − | + | − | + | − | + | − | + | |
| Control | 6.6±0.2 | 351±8.3 | 11.7±2.8 | 401±4.2 | 14.7±1.3 | 192.2±8.5 | 117.2±5.9* | 287.4±23.5* | |
| DFMO | 1.8±0.5 | 44.5±1.3 | 2.1±0.6 | 209±11.9* | 16.6±3.2 | 76.3±6.7 | 41.6±5.4 | 141.8±14.3* | |
DN-STAT3 completely reverses the protective effect of polyamine depletion
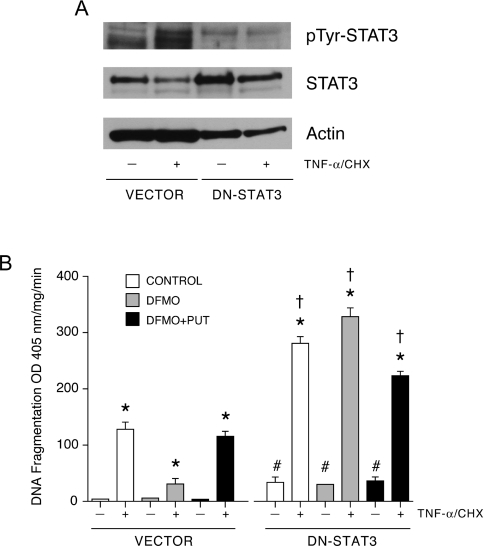
Since pharmacological agents may elicit multiple effects depending on dose and time of treatment, we validated our observations using IEC-6 cells transfected with empty vector and DN-STAT3 plasmids. Selected stable clones were analysed by Western blotting to determine the level of expression and phosphorylation of STAT3 at Tyr-705 (Figure 3A). The amount of STAT3 protein was significantly higher in cells transfected with DN-STAT3 compared with those transfected with empty vector. Basal phosphorylation of STAT3 at Tyr-705 was significantly lower in cells transfected with DN-STAT3, compared with cells transfected with empty vector. TNF-α increased STAT3 activation, as judged by the levels of Tyr-705-phosphorylated STAT3, in cells transfected with empty vector. In contrast, in cells expressing DN-STAT3, TNF-α failed to activate STAT3. TNF-α-induced STAT3 activation significantly decreased STAT3 protein in cells transfected with empty vector, while DN-STAT3 cells showed only a slight decrease in STAT3 protein in response to TNF-α. Exposure of DN-STAT3 cells to TNF-α significantly increased apoptosis compared with vector transfected cells and decreased the STAT3 protein. A simultaneous decrease in actin was also observed (Figure 3A). These results confirm that the selected clone expressed robust amounts of DN-STAT3 protein and that STAT3 activation was prevented in these cells. Inhibition of STAT3 activation by DN-STAT3 increased spontaneous apoptosis as evident by DNA fragmentation in all groups of cells (Figure 3B). Moreover, polyamine depletion in cells transfected with DN-STAT3 sensitized them to TNF-α compared with cells transfected with vector. Inhibition of STAT3 activation completely reversed apoptosis in polyamine-depleted cells.
Figure 3. DN-STAT3 prevents the protective effect of DFMO.
(A) Cells transfected with empty vector (vector) or DN-STAT3 were grown as described in the Experimental section and treated with TNF-α/CHX for 3 h. Western blot was performed with phospho-Tyr-705 STAT3-specific antibody. Membrane was stripped and first probed with total STAT3 specific antibody followed by β-actin antibody. (B) DNA fragmentation of vector- and DN-STAT3-expressing cells was determined in response to TNF-α/CHX. *, P<0.005 compared with untreated cells; †, P<0.005 compared with respective TNF-α/CHX-treated vector cells; #, P<0.005 compared with respective untreated vector cells (mean±S.E.M., n=6). PUT, putrescine.
Bcl-2 and c-IAP2 protein levels in polyamine-depleted cells
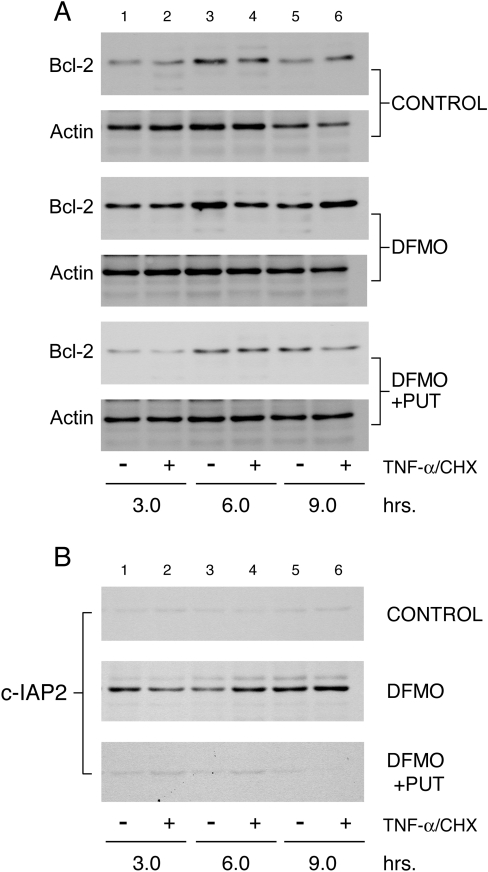
Previously we have shown that polyamine depletion increases Bcl-2 protein levels [29]. However, the upstream events regulating Bcl-2 expression in response to polyamine depletion are unknown. Microarray analysis of the genes involved in apoptosis, in cells grown in control and DFMO-containing medium in the presence and absence of TNF-α showed an upregulation of several anti-apoptotic genes in polyamine-depleted cells. Among these, Bcl-2 and Mcl-1 genes of the Bcl-2 protein family and the c-IAP2 gene were upregulated significantly in DFMO-treated cells (results not shown). Based on our findings that the inhibition of STAT3 sensitizes polyamine-depleted cells to TNF-α-induced apoptosis, and that STAT3 has been shown to activate the transcription of Bcl-2 and Bcl-XL [26], we determined the levels of Bcl-2 proteins during the induction of apoptosis. Figure 4(A) shows significantly higher basal levels of Bcl-2 protein in polyamine-depleted cells (Figure 4A, DFMO lane 1). The increase in Bcl-2 protein was maintained before and after 9 h of TNF-α treatment in DFMO-treated cells (Figure 4A, lanes 2, 4 and 6). Levels of β-actin, an internal loading control, showed no change in any group. Samples from the above experiment were probed with antibody specific for the c-IAP2 protein. Results shown in Figure 4(B) indicate that c-IAP2 protein was constitutively elevated in polyamine-depleted cells as compared with cells grown under control and DFMO plus putrescine conditions (Figure 4B, lane 1).
Figure 4. Expression of Bcl-2 and c-IAP2 proteins is elevated in polyamine-depleted cells.
IEC-6 cells were grown as described in the Experimental section and treated with TNF-α/CHX for 3, 6 and 9 h. Equal amounts of whole cell extract were separated by SDS/PAGE, and probed with antibodies specific for Bcl-2 (A) and c-IAP2 (B). Membranes were stripped and probed with β-actin antibody to determine equal loading of the samples. PUT, putrescine.
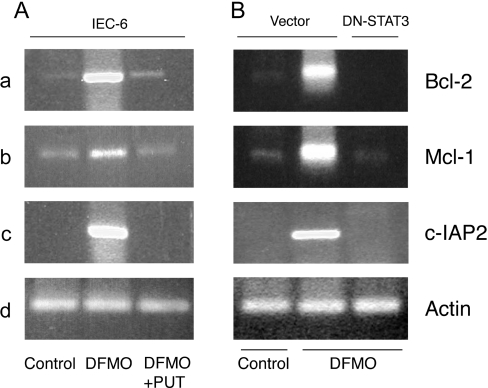
STAT3 regulates Bcl-2, Mcl-1 and c-IAP2 gene transcription in polyamine-depleted cells
Given the results in Figure 4, we predicted that polyamine-depleted cells should have higher basal levels of transcripts for the Bcl-2, Mcl-1 and c-IAP2 genes. Using gene specific primer pairs, semi-quantitative RT-PCR analysis was carried out to determine the levels of mRNAs for these genes. Polyamine depletion of IEC-6 cells significantly increased Bcl-2, Mcl-1 and c-IAP2 mRNAs (Figure 5A, lane 2) 2–3-fold compared with control cells and those grown in the presence of DFMO plus putrescine (Figure 5A, lanes 1 and 3). RT-PCR analysis of β-actin showed no change and served as an internal amplification as well as a loading control. Cells transfected with vector and grown in DFMO-containing media had significantly higher mRNA levels for Bcl-2, Mcl-1 and c-IAP2 (Figure 5B, lane 2) compared with control cells (Figure 5B, lane 1), which further confirmed the findings shown in Figure 5(A). Conversely, transfection with DN-STAT3 completely blocked the increase in mRNA levels of these genes that is normally seen in response to polyamine depletion (Figure 5B, lane 3). These observations strongly suggest that STAT3 regulates transcription of the Bcl-2, Mcl-1 and c-IAP2 genes in IEC-6 cells.
Figure 5. Bcl-2, Mcl-1 and c-IAP2 mRNA levels.
(A) IEC-6 cells were grown in DMEM/5% dFBS with or without 5 mM DFMO or DFMO plus 10 μM putrescine (PUT) for 3 days and serum-deprived for 24 h. RNA extraction, reverse transcription and PCR were carried out using gene-specific primers as described in the Experimental section. The PCR products were resolved by agarose gel electrophoresis and stained with ethidium bromide. Representative images from three observations are shown. (B) Vector- and DN-STAT3-transfected cells were grown, as described in the Experimental section for the IEC-6 cells. Total RNA was prepared, reverse-transcribed and PCR was carried out using primers specific for Bcl-2, Mcl-1, c-IAP2 and actin genes as described in the Experimental section. Representative images from three observations are shown.
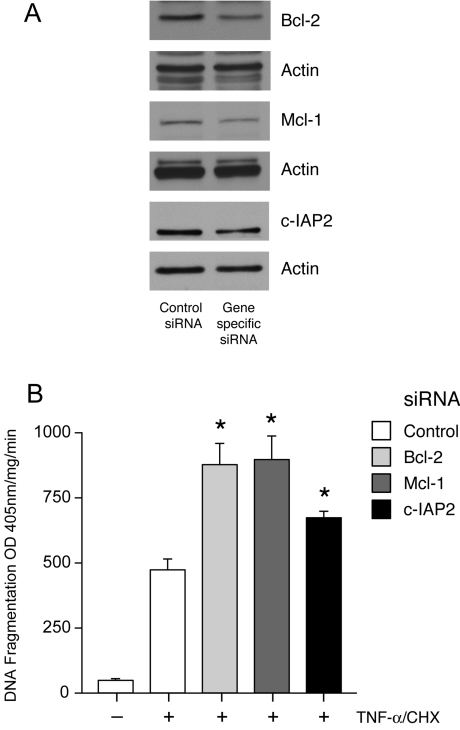
Inhibition of Bcl-2, Mcl-1 and c-IAP2 increases TNF-α-induced apoptosis
In order to determine the significance of the increased levels of Bcl-2 family and c-IAP2 proteins in relation to the decreased apoptotic response in polyamine-depleted cells, we inhibited the expression of these proteins using siRNA. Cells transfected with Bcl-2, Mcl-2 or c-IAP2 siRNA showed a significant decrease in the expression of these proteins compared with control siRNA transfection (Figure 6A). Transfection efficiency of siRNA in IEC-6 cells was monitored using a FITC-labelled control siRNA as described previously [14]. Transfection with Bcl-2, Mcl-1 and c-IAP2 siRNA significantly increased TNF-α-induced DNA fragmentation when compared with cells treated with transfection reagent alone (Figure 6B) indicating a direct role for these anti-apoptotic proteins in mediating cytoprotection against apoptosis. Induced expression of these proteins after polyamine depletion is therefore sufficient to confer protection against apoptosis.
Figure 6. siRNA-mediated inhibition of Bcl-2, Mcl-1 and c-IAP2 expression and apoptosis.
IEC-6 cells were transfected with either control or the respective siRNA as described in the Experimental section. (A) Expression of Bcl-2, Mcl-1 and c-IAP2. Western blot analysis of control and siRNA transfected cell lysates was performed using Bcl-2, Mcl-1 and c-IAP2 specific antibodies. Membranes were stripped and probed for actin as control for sample loading. Representative blots from three observations are shown. (B) DNA fragmentation. Cells were grown in control medium and transfected with either control siRNA or siRNA specific for Bcl-2, Mcl-1 and c-IAP2. After 48 h transfection the cells were serum-starved, and treated with TNF-α/CHX for 3 h and then subjected to DNA fragmentation analysis. Values are means±S.E.M. *P<0.05 compared with control siRNA transfected cells treated with TNF-α/CHX. OD, absorbance.
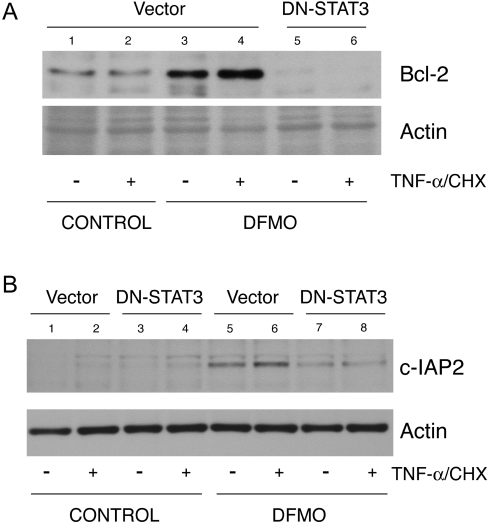
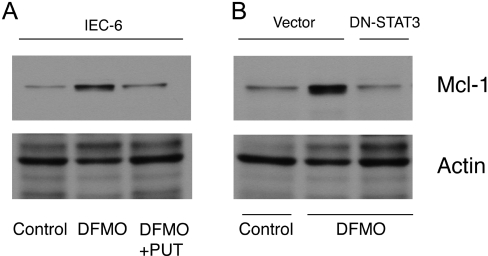
STAT3 regulates Bcl-2, Mcl-1 and c-IAP2 protein levels
We next determined whether inhibition of STAT3 activation decreases the levels of Bcl-2, Mcl-1 and c-IAP2 proteins, which are upregulated in cells depleted of polyamines. DFMO significantly elevated Bcl-2 (Figure 7A, lanes 3 and 4), c-IAP2 (Figure 7B, lanes 5 and 6) and Mcl-1 (Figure 8A, lane 2) protein levels in polyamine-depleted cells transfected with vector and either left untreated or treated with TNF-α, whereas in polyamine-depleted cells, DN-STAT3 completely prevented the increases in Bcl-2 (Figure 7A, lanes 5 and 6), c-IAP2 (Figure 7B, lanes 7 and 8) and Mcl-1 (Figure 8B, lane 3) protein levels. These findings suggest that STAT3 activation in response to polyamine depletion elevates Bcl-2, Mcl-1 and c-IAP2 proteins by transcriptional upregulation of these anti-apoptotic genes.
Figure 7. DN-STAT3 and Bcl-2 and c-IAP2 protein expression.
(A) Vector- and DN-STAT3-transfected cells were grown as described in the Experimental section and treated with TNF-α/CHX for 3 h. Lysates were prepared and separated by SDS/PAGE. Proteins were transferred to PVDF membrane and probed with Bcl-2 (A) and c-IAP2 (B) antibodies. Membrane was stripped and probed with β-actin antibody as an internal loading control. Representative blots from three observations are shown.
Figure 8. DN-STAT3 and Mcl-1 protein expression.
Parental IEC-6 cells (A) and cells transfected with vector and DN-STAT3 (B) were grown as described in the Experimental section. Lysates were prepared and separated by SDS/PAGE. Proteins were transferred to PVDF membrane and probed with Mcl-1 antibody. Membrane was stripped and probed with β-actin antibody as an internal loading control. Representative blots from three observations are shown.
STAT3 directly modulates transcription of Bcl-2
Since the inhibition of STAT3 decreased Bcl-2, Mcl-1 and c-IAP2 protein and mRNA levels, we determined whether STAT3 directly regulates the transcription of these genes. Transfection with the full-length Bcl-2 promoter–luciferase construct increased promoter activity when compared with empty vector-transfected cells. Inhibition of STAT3, either by the cell permeant JAK inhibitor AG490, or by DN-STAT3, significantly decreased Bcl-2 promoter activity (Table 2). Transfection efficiency was monitored by transfection with a plasmid expressing green fluorescent protein only (results not shown). These results indicate that induced levels of Bcl-2 mRNA and protein following polyamine depletion are due to the direct activation of the Bcl-2 gene by STAT3.
Table 2. STAT3 regulates Bcl-2 promoter activity.
IEC-6 cells were transfected with empty vector or a Bcl-2-promoter–luciferase construct (Bcl-2-Luc); DN-STAT3 cells were transfected with the Bcl-2-promoter–luciferase construct as described in the Experimental section. At 3 days after transfection, Bcl-2 promoter-transfected cells were treated with 50 μM AG490 for 30 min. Cells were lysed and luciferase activity was measured using the Promega luciferase assay kit. Values were expressed as RLU/min per mg (relative luciferase units/min per mg of protein). Values are means±S.E.M. *P<0.05 compared with Bcl-2-promoter-transfected cells.
| Reporter | Luciferase activity (RLU/min per mg) |
|---|---|
| Empty vector-Luc | 105.3±35.7 |
| Bcl-2-Luc | 702.0±50.2 |
| Bcl-2-Luc+AG490 | 193.7±83.1* |
| Bcl-2-Luc+DN-STAT3 | 387.0±91.3* |
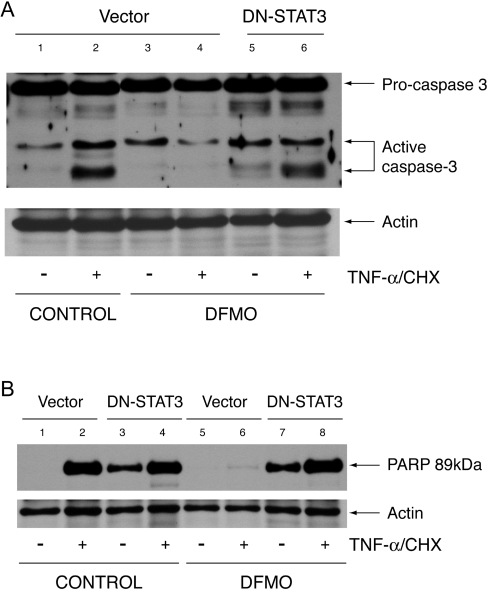
DN-STAT3 restores caspase-3 activation in polyamine-depleted cells
Since transfection with DN-STAT3 decreased the expression of anti-apoptotic genes, we predicted that inhibition of STAT3 activation would reverse the inhibition of caspase-3 activation observed in polyamine-depleted cells. We measured caspase-3 activation, and cleavage of its downstream target PARP, in cells transfected with vector or DN-STAT3 and grown in the presence or absence of DFMO. Vector cells treated with TNF-α had a significantly higher amount of activated caspase-3 as judged by the appearance of the small 16–18 kDa cleavage product (Figure 9A, lane 2) compared with untreated cells (Figure 9A, lane 1). Caspase-3 activation accompanied cleavage of PARP at Asp-214 in TNF-α-treated cells transfected with vector (Figure 9B). Consistent with our previous results, polyamine depletion of cells transfected with vector resulted in little caspase-3 activation and PARP cleavage in response to TNF-α (Figure 9A, lane 4, and Figure 9B, lane 6). Cells transfected with DN-STAT3 and grown under control conditions showed higher spontaneous PARP cleavage, which further increased in response to TNF-α (Figure 9B, lanes 3 and 4). Polyamine depletion of cells transfected with DN-STAT3 had increased basal caspase-3 processing (Figure 9A, lane 5) and PARP cleavage (Figure 9B, lane 7). Unlike polyamine-depleted cells transfected with vector (Figure 9B, lane 4), TNF-α treatment of polyamine-depleted DN-STAT3 cells showed significant caspase-3 activation and PARP cleavage (Figure 9A, lane 6, and Figure 9B, lane 8).
Figure 9. Caspase-3 activation and PARP cleavage.
Cells transfected with vector and DN-STAT3 were grown as described in the Experimental section and treated with TNF-α/CHX or left untreated. Protein (40 μg) from each sample was separated by SDS-gel electrophoresis, transferred to membrane and probed with caspase-3 antibody (A) and a cleaved PARP (Asp-214) rat-specific antibody (B). Membranes were stripped and probed with β-actin antibody as an internal control for equal loading. Representative blots from three experiments are shown.
DISCUSSION
Our previous results indicated that several anti-apoptotic pathways are activated in response to polyamine depletion in IEC-6 cells, including mitogen-activated protein kinase, protein kinase B and NF-κB [12–14]. We have also shown that after 3–6 h of incubation, in confluent IEC-6 cells with DFMO-containing medium, the tyrosine phosphorylation, nuclear translocation and DNA-binding activity of STAT3 increased significantly compared with untreated cells [16]. During this time intracellular putrescine completely disappeared. Exogenous addition of 5 μM putrescine to the DFMO-treated group completely prevented STAT3 activation [16]. However, the physiological significance of putrescine depletion and STAT3 activation remains unknown. Rapid depletion of putrescine subsequently leads to depletion of spermidine and spermine if cells are allowed to proliferate in the presence of DFMO [4]. Thus STAT3 might mediate some of the physiological effects of polyamine depletion. Polyamine depletion has been shown to regulate three fundamental processes, namely proliferation, migration and apoptosis, which are essential to homoeostasis of intestinal epithelium and to mucosal restitution in rats [7,8]. Polyamine depletion significantly inhibited all of these processes [4,7–9]. In the present study we have examined the effects of polyamine depletion on STAT3 activation and the mechanism by which STAT3 regulates the apoptotic pathway.
We report that polyamine depletion increased Tyr-705 and Ser-727 phosphorylation of STAT3 and subsequently altered localization of these proteins to the cell periphery and nucleus respectively, suggesting an increase in activation, nuclear translocation and transcriptional activity (Figure 1). Relatively smaller amounts of phospho-Ser-727 STAT3 in the nuclear fraction in control cells, and in those grown in DFMO plus putrescine, indicate weak transcriptional activation of target genes. Our previous report showed that nuclear localization and tyrosine phosphorylation of STAT3 was correlated with increased DNA binding in IEC-6 cells [16]. Therefore these observations indicate that polyamine depletion constitutively activates STAT3. In our present study, the correlation between sustained activation of STAT3 and decreased apoptosis in response to TNF-α in polyamine-depleted cells (Figure 2, DFMO, lanes 2, 4 and 6) indicates an anti-apoptotic role for STAT3. Inhibition of STAT3 activation, by PpYLKTK and AG490, or expression of DN-STAT3, increased the sensitivity of polyamine-depleted cells to TNF-α-induced apoptosis, supporting the idea that STAT3 activation protects cells from apoptosis (Table 1 and Figure 3). The increased basal levels of PARP cleavage in DN-STAT3 cells grown in control conditions, and with DFMO (Figure 9B), suggest that STAT3 activation is necessary for resistance to TNF-α-induced apoptosis. Polyamine depletion of vector-transfected IEC-6 cells showed inhibition of caspase-3 and PARP cleavage when treated with TNF-α, which is consistent with our previous observations (Figures 9A and 9B) [12]. However, TNF-α caused caspase-3 activation and PARP cleavage in DN-STAT3 cells depleted of polyamines, indicating a role for STAT3 as an anti-apoptotic cell survival factor.
We previously reported that polyamine depletion of IEC-6 cells inhibits Bax translocation to mitochondria and the subsequent release of cytochrome c [12,29]. In the present study, we demonstrate that polyamine depletion causes upregulation of transcription as well as translation of anti-apoptotic Bcl-2, Mcl-1 and c-IAP2 genes (Figures 4, 5, 7 and 8). Epling-Burnette et al. [23] showed that inhibition of STAT3 signalling leads to apoptosis in lymphocytes through decreased Mcl-1 expression. Downregulation of Mcl-1 expression is thought to be required for Bax translocation to mitochondria and cytochrome c release [40]. We show here that expression of DN-STAT3 increased basal levels of apoptosis and completely eliminated the protective effects of DFMO (Figure 3B). Prevention of STAT3 activation by expression of DN-STAT3 completely blocked the transcription (Figure 5B, panels a, b and c) and subsequent protein expression (Figure 7 and Figure 8B) of Bcl-2, Mcl-1 and c-IAP2 in response to polyamine depletion. Furthermore, RNA interference by siRNAs of Bcl-2, Mcl-1 and c-IAP2 significantly reduced the expression of these proteins, compared with control siRNA-treated cells, and increased TNF-α-induced apoptosis (Figures 6A and 6B). These results provide evidence that the induction of pro-survival genes by activated STAT3 is responsible for cell protection. Increases in mRNA and protein levels of these anti-apoptotic proteins are a direct consequence of STAT3-mediated transcriptional regulation. Transfection with the Bcl-2 promoter–luciferase construct increased reporter luciferase activity. The JAK inhibitor, AG490, and DN-STAT3 significantly decreased luciferase activity indicating that there is STAT3-dependent transcriptional activation of the Bcl-2 promoter (Table 2). A putative STAT3-binding site (TTACGGGAA) has been found in the murine Mcl-1 promoter sequence [23], which is identical with that of rat Mcl-1. Activated STAT3 has been shown to bind directly to a sis inducible element in the murine Mcl-1 promoter and regulate Mcl-1 expression [23]. STAT3 has also been shown to increase Bcl-2 and Bcl-XL promoter activity [41]. Sequence analysis of the rat c-IAP2 promoter also revealed a putative STAT3-binding region TTCCTTTAC. These findings emphasize that STAT3 regulates Bcl-2 and Mcl-1 gene expression and that constitutive activation of STAT3 in response to polyamine depletion protects cells by up-regulating transcription and subsequently the translation of these genes. Zou et al. [37] reported that NF-κB-mediated c-IAP2 and XIAP expression induces resistance to apoptosis in polyamine-depleted cells. A synergistic effect of several transcription factors could lead to increased expression of IAP proteins. This is the first report of the role of STAT3 in regulating cell survival in untransformed intestinal epithelial cells.
Our previous reports and the present findings suggest that polyamines regulate intestinal cell survival by inducing STAT3-mediated transcriptional upregulation of anti-apoptotic genes. Bcl-2, Mcl-1 and c-IAP2 are the downstream intracellular targets of activated STAT3. A decrease in intracellular polyamines activates STAT3 and stimulates the expression of the Bcl-2 and IAP families of anti-apoptotic proteins. Early activation of STAT3 in response to polyamine depletion results in the accumulation of anti-apoptotic proteins and inhibits the activation of caspases, protecting cells from TNF-α-induced apoptosis.
Acknowledgments
This study was supported by National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) grant DK-16505 and by the Thomas A. Gerwin Endowment. We sincerely acknowledge Mary Jane Viar for critically reading the manuscript, and Greg Short and Danny Morse for their help in preparation of the Figures. We also thank Dr Linda Boxer (Stanford University, CA, U.S.A.) for kindly providing us with the Bcl-2 promoter–luciferase construct and Dr Lawrence Pfeffer (University of Tennessee, IN, U.S.A.) for the DN mutant of STAT3.
References
- 1.Hall P. A., Coates P. J., Ansari B., Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract–the importance of apoptosis. J. Cell. Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 2.Potten C. S. Epithelial cell growth and differentiation 2: Intestinal apoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 1997;273:G253–G257. doi: 10.1152/ajpgi.1997.273.2.G253. [DOI] [PubMed] [Google Scholar]
- 3.Schipper R. G., Penning L. C., Verhofstad A. A. J. Involvement of polyamines in apoptosis. Facts and controversies: effectors or protectors? Semin. Cancer Biol. Rev. 2000;2:55–68. doi: 10.1006/scbi.2000.0308. [DOI] [PubMed] [Google Scholar]
- 4.Ray R. M., Viar M. J., Yuan Q., Johnson L. R. Polyamine depletion delays apoptosis of rat intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2000;278:C480–C489. doi: 10.1152/ajpcell.2000.278.3.C480. [DOI] [PubMed] [Google Scholar]
- 5.Tabor C. W., Tabor H. Polyamines. Annu. Rev. Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 6.Gerner E. W., Meyskens F. L. Polyamines and cancer: old molecules, new understanding. Nat. Rev. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 7.Wang J. Y., Johnson L. R. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology. 1991;100:333–343. doi: 10.1016/0016-5085(91)90200-5. [DOI] [PubMed] [Google Scholar]
- 8.Johnson L. R., McCormack S. A. Healing of gastrointestinal mucosa: involvement of polyamines. News Physiol. Sci. 1999;14:12–17. doi: 10.1152/physiologyonline.1999.14.1.12. [DOI] [PubMed] [Google Scholar]
- 9.Ray R. M., Zimmerman B. J., McCormack S. A., Patel T. B., Johnson L. R. Polyamine depletion arrests cell cycle and induces inhibitors of p21 (waf1/kip1) in IEC-6 cells. Am. J. Physiol. Cell Physiol. 1999;276:C684–691. doi: 10.1152/ajpcell.1999.276.3.C684. [DOI] [PubMed] [Google Scholar]
- 10.Quaroni A., Wands J., Trelstad R. L., Isselbacher K. J. Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobias K. E., Kahana C. Exposure to ornithine results in excessive accumulation of putrescine and apoptotic cell death in ornithine decarboxylase overproducing mouse myeloma cells. Cell Growth Differ. 1995;10:1279–1285. [PubMed] [Google Scholar]
- 12.Bhattacharya S., Ray R. M., Viar M. J., Johnson L. R. Polyamines are required for activation of c-Jun NH2-terminal kinase and apoptosis in response to TNF-α in IEC-6 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G980–G991. doi: 10.1152/ajpgi.00206.2003. [DOI] [PubMed] [Google Scholar]
- 13.Bhattacharya S., Ray R. M., Johnson L. R. Prevention of TNF-α-induced apoptosis in polyamine-depleted IEC-6 cells is mediated through the activation of ERK1/2. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G479–G490. doi: 10.1152/ajpgi.00342.2003. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharya S., Ray R. M., Johnson L. R. Decreased apoptosis in polyamine-depleted IEC-6 cells depends on Akt-mediated NF-κB activation but not GSK3β activity. Apoptosis. 2005;10:759–776. doi: 10.1007/s10495-005-2943-3. [DOI] [PubMed] [Google Scholar]
- 15.Pfeffer L. M., Yang C. H., Murti A., McCormack S. A., Viar M. J., Ray R. M., Johnson L. R. Polyamine depletion induces rapid NF-κB activation in IEC-6 cells. J. Biol. Chem. 2001;276:45909–45913. doi: 10.1074/jbc.M108097200. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer L. M., Yang C. H., Pfeffer S. R., Murti A., McCormack S. A., Johnson L. R. Inhibition of ornithine decarboxylase induces STAT3 tyrosine phosphorylation and DNA binding in IEC-6 cells. Am. J. Physiol. Cell Physiol. 2000;278:C331–C335. doi: 10.1152/ajpcell.2000.278.2.C331. [DOI] [PubMed] [Google Scholar]
- 17.Darnell J. E., Jr STATs and gene regulation. Science (Washington D.C.) 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 18.Zhong Z., Wen Z., Darnell J. E. Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science (Washington D.C.) 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 19.Mufson R. A. The role of serine phosphorylation in hematopoietic cytokine receptor signal transduction. FASEB J. 1997;11:37–44. doi: 10.1096/fasebj.11.1.9034164. [DOI] [PubMed] [Google Scholar]
- 20.Bowman T., Garcia R., Turkson J., Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 21.Kanda N., Seno H., Konda Y., Marusawa H., Kanai M., Nakajima T., Kawashima T., Nankin A., Sawabu T., Uenoyama Y., et al. STAT3 is constitutively activated and supports cell survival in association with survivin expression and in gastric cancer cells. Oncogene. 2004;23:4921–4929. doi: 10.1038/sj.onc.1207606. [DOI] [PubMed] [Google Scholar]
- 22.Konnikova L., Kotecki M., Kruger M. M., Cochran B. H. Knockdown of STAT3 expression by RNAi induces apoptosis in astrocytoma cells. BMC Cancer. 2003;3:23–31. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Epling-Burnette P. K., Liu J. H., Catlett-Falcone R., Turkson J., Oshiro M., Kothapalli R., Li Y., Wang J. M., Yang-Yen H. F., Karras J., Jove R., Loughran T. P. Inhibition of STAT3 signaling leads to apoptosis of leukemic granular lymphocytes and decreased Mcl-1 expression. J. Clin. Invest. 2001;107:351–362. doi: 10.1172/JCI9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S. O., Lou W., Qureshi K. M., Mehraein-Ghomi F., Trump D. L., Gao A. C. RNA interference targeting Stat3 inhibits growth and induces apoptosis of human prostate cancer cells. Prostate. 2004;60:303–309. doi: 10.1002/pros.20072. [DOI] [PubMed] [Google Scholar]
- 25.Zushi S., Shinomura Y., Kiyohara T., Matsuzawa Y. STAT3 mediates the survival signal in oncogenic ras-transfected intestinal epithelial cells. Int. J. Cancer. 1998;78:326–330. doi: 10.1002/(SICI)1097-0215(19981029)78:3<326::AID-IJC12>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Bromberg J. F., Wrzeszczynska M. H., Devgan G., Zhao Y., Pestell R. G., Albanese C., Darnell J. E., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 27.Rahaman S. O., Harbor P. C., Chernova O., Barnett G. H., Vogelbaum M. A., Haque S. J. Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene. 2002;21:8404–8413. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z. The chemical biology of apoptosis. Exploring protein–protein interactions and the life and death of cells with small molecules. Chem. Biol. 2002;9:1059–1072. doi: 10.1016/s1074-5521(02)00247-8. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Q., Ray R. M., Johnson L. R. Polyamine depletion prevents camptothecin-induced apoptosis by inhibiting the release of cytochrome c. Am. J. Physiol. Cell Physiol. 2002;282:C1290–C1297. doi: 10.1152/ajpcell.00351.2001. [DOI] [PubMed] [Google Scholar]
- 30.Kozopas K. M., Yang T., Buchan H. L., Zhou P., Craig R. W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams J. M., Cory S. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 2001;26:61–66. doi: 10.1016/s0968-0004(00)01740-0. [DOI] [PubMed] [Google Scholar]
- 32.Bingle C. D., Craig R. W., Swales B. M., Singleton V., Zhou P., Whyte M. K. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J. Biol. Chem. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- 33.Liu H., Ma Y., Cole S. M., Zander C., Chen K. H., Karras J., Pope R. M. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- 34.Lomo J., Smeland E. B., Krajewski S., Reed J. C., Blomhoff H. K. Expression of the Bcl-2 homologue Mcl-1 correlates with survival of peripheral blood B lymphocytes. Cancer Res. 1996;56:40–43. [PubMed] [Google Scholar]
- 35.Liston P., Young S. S., Mackenzie A. E., Korneluk R. G. Life and death decisions: the role of the IAPs in modulating programmed cell death. Apoptosis. 1997;2:423–441. doi: 10.1023/a:1026465926478. [DOI] [PubMed] [Google Scholar]
- 36.Deveraux Q. L., Reed J. C. IAP family proteins-suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 37.Zou T., Rao J. N., Guo X., Liu L., Zhang H. M., Strauch E. D., Bass B. L., Wang J. Y. NF-κB-mediated IAP expression induces resistance of intestinal epithelial cells to apoptosis after polyamine depletion. Am. J. Physiol. Cell Physiol. 2004;286:C1009–C1018. doi: 10.1152/ajpcell.00480.2003. [DOI] [PubMed] [Google Scholar]
- 38.Nishihara H., Kizaka-Kondoh S., Insel P. A., Eckmann L. Inhibition of apoptosis in normal and transformed intestinal epithelial cells by cAMP through induction of inhibitor of apoptosis protein (IAP)-2. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8921–8926. doi: 10.1073/pnas.1533221100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan H., Heckman C. A., Boxer L. M. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell. Biol. 2005;25:1608–1619. doi: 10.1128/MCB.25.5.1608-1619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nijhawan D., Fang M., Traer E., Zhong Q., Gao W., Du F., Wang X. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephanou A., Brar B. K., Knight R. A., Latchman D. S. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. 2000;7:329–330. doi: 10.1038/sj.cdd.4400656. [DOI] [PubMed] [Google Scholar]