Abstract
Reduced insulin-mediated glucose transport in skeletal muscle is a hallmark of the pathophysiology of T2DM (Type II diabetes mellitus). Impaired intracellular insulin signalling is implicated as a key underlying mechanism. Attention has focused on early signalling events such as defective tyrosine phosphorylation of IRS1 (insulin receptor substrate-1), a major target for the insulin receptor tyrosine kinase. This is required for normal induction of signalling pathways key to many of the metabolic actions of insulin. Conversely, increased serine/threonine phosphorylation of IRS1 following prolonged insulin exposure (or in obesity) reduces signalling capacity, partly by stimulating IRS1 degradation. We now show that IRS1 levels in human muscle are actually increased 3-fold following 1 h of hyperinsulinaemic euglycaemia. Similarly, transient induction of IRS1 (3-fold) in the liver or muscle of rodents occurs following feeding or insulin injection respectively. The induction by insulin is also observed in cell culture systems, although to a lesser degree, and is not due to reduced proteasomal targeting, increased protein synthesis or gene transcription. Elucidation of the mechanism by which insulin promotes IRS1 stability will permit characterization of the importance of this novel signalling event in insulin regulation of liver and muscle function. Impairment of this process would reduce IRS1 signalling capacity, thereby contributing to the development of hyperinsulinaemia/insulin resistance prior to the appearance of T2DM.
Keywords: diabetes, human muscle, insulin, insulin receptor substrate-1 (IRS1), mammalian target of rapamycin (mTOR)
Abbreviations: BMI, body-mass index; GSK3, glycogen synthase kinase 3; IRS, insulin receptor substrate; LDM, low-density membrane; mTOR, mammalian target of rapamycin; PDK1, 3-phosphoinositide-dependent kinase; PI3K, phosphoinositide 3-kinase; PK(B/C), protein kinases B and C respectively; S6K, S6 ribosomal protein kinase; TOR, target of rapamycin; T2DM, Type II diabetes mellitus
INTRODUCTION
T2DM (Type II diabetes mellitus) is characterized by cellular resistance to insulin, as well as abnormal insulin secretion [1,2]. This reduced responsiveness of cells to insulin is due to defective intracellular signalling processes [1–6]. Insulin induces multiple signalling pathways in all tissues that express the transmembrane insulin receptor (e.g. muscle, liver and adipose tissue). The association of insulin with its receptor activates the intrinsic tyrosine kinase activity of the receptor, leading to its autophosphorylation as well as phosphorylation of a series of tyrosine residues on IRS (insulin receptor substrate) proteins such as IRS1, IRS2 and IRS4 (see [5,7] for reviews). Subsequently the phosphotyrosine motifs on these proteins recruit signalling proteins to the plasma membrane, initiating a number of signalling pathways that ultimately alter glucose metabolism. For example, recruitment and activation of PI3K (phosphoinositide 3-kinase) promotes the generation of 3-phosphoinositides that induce the activity of protein kinases such as PDK1 (3-phosphoinositide-dependent kinase) and PKB (protein kinase B) [8,9]. PKB then phosphorylates GSK3 (glycogen synthase kinase-3) at an N-terminal serine residue (Ser-21 on GSK-3α and Ser-9 on GSK-3β), rendering it inactive [10,11]. This PKB-mediated inhibition of GSK3 contributes to insulin activation of glycogen and protein synthesis [11,12]. PDK1 and PKB also contribute to the insulin stimulation of the rapamycin-sensitive mTOR (mammalian target of rapamycin)–S6K (S6 ribosomal protein kinase) pathway [13]. This pathway is also regulated by nutrients through an interaction with the TSC (tuberous sclerosis complex)–Rheb complex [14], and regulates, among other things, the rate of protein synthesis [15].
These signalling pathways are turned off by the removal of stimuli and subsequent dephosphorylation of the various signalling molecules. Dephosphorylation of the insulin receptor and IRS is performed by the phosphotyrosine phosphatases PTP1B and TCPTP (T-cell protein tyrosine phosphatase) [16], whereas the lipid phosphatase PTEN (phosphatase and tensin homologue deleted on chromosome 10) reduces 3-phosphoinositide levels [9]. In addition to inactivation by dephosphorylation, IRS1 and IRS2 have been identified as major sites of feedback inhibition. IRS1 phosphorylation on specific serine/threonine residues reduces signalling in a number of ways, including reduced tyrosine phosphorylation and increased proteasomal degradation of IRS1 [5]. However, serine/threonine phosphorylation may also have positive effects on insulin action, presumably dependent on the residues that are specifically phosphorylated [17]. A number of IRS1 kinases have been proposed to regulate these events. These include several isoforms of PKC (protein kinase C), JNK (Jun N-terminal kinase), IKK (IκB kinase) and p70S6K (see [18] for a review).
It is assumed that this reduction in IRS1 protein leads to lower signalling capacity in the system. Therefore overactive negative feedback of IRS molecules could promote intracellular insulin resistance, and evidence is accumulating that this may occur in the liver, muscle and adipose tissue prior to the onset of T2DM [1,5,19]. In particular, increased accumulation of fatty acids in tissues (as seen in obesity) promotes serine/threonine phosphorylation of IRS proteins [4].
These repressive effects on IRS1 expression have only been observed after prolonged insulin exposure (4–48 h) [20], but very little is known about acute regulation of IRS1 expression. We now present evidence that a significant increase in IRS1 protein is observed following acute insulin treatment of muscle or liver, and propose that the previously reported reduction in IRS1 levels occurs subsequent to an initial up-regulation of this key signalling molecule.
EXPERIMENTAL
Inhibitors and antibodies
Rabbit polyclonal antibody to IRS1 (raised against 14 C-terminal amino acids) was purchased from Upstate Biotechnology (Lake Placid, NY, U.S.A.), anti-GLUT4 antibody was generously given by Professor Gwyn Gould (Faculty of Biomedical and Life Sciences, University of Glasgow, Scotland, U.K.), anti-phospho-S6 was purchased from Cell Signalling Technology (Beverly, MA, U.S.A.) whereas anti-Mdm2 and MG132 were gifts from Dr Mark Saville (Department of Surgery, Molecular Oncology, University of Dundee, Scotland, U.K.), and the antibodies against β-actin (C-terminal fragment) and cycloheximide were purchased from Sigma-Aldrich, Inc. (St Louis, MO, U.S.A.). Puromycin was purchased from Invitrogen (San Diego, CA, U.S.A.). Rapamycin and LY294002 were purchased from Calbiochem (Nottingham, U.K.). Actrapid (human insulin) was purchased from Novo Nordisk A/S (Bagsværd, Denmark).
Hyperinsulinaemic euglycaemic clamp
Following favourable review by the Tayside Committee on Medical Ethics, 10 non-diabetic male subjects [aged<40 years; BMI (body-mass index) range 19–31 kg/m2] were recruited by advertisement. These volunteers had no family history of diabetes. After giving fully informed consent, they were studied following an overnight fast according to the protocol of the European Group for the Study of Insulin Resistance [21]. Two 18-gauge intravenous cannulae were inserted: the first retrogradely into the right dorsal hand vein for blood sampling, and the second antegradely into the left antecubital fossa for infusions. Three-way taps enabled easy sampling and simultaneous infusion of insulin and dextrose. Soluble human insulin (Actrapid; NovoNordisk A/S) was prepared in 45 ml (10%, v/v) of each patient's own blood in 0.9% saline in order to minimize adsorption of insulin to the plastic surfaces of syringes and infusion lines (40 m-units·min−1·m−2), and a continuous infusion was administered for 2 h. A variable 20% glucose infusion (mg·kg−1·min−1) was started 2 min after the beginning of the experiment and was corrected, if necessary, to maintain euglycaemia at 5.2 mM. Blood samples for determination of plasma glucose were obtained at 5 min intervals throughout the study to adjust the glucose infusion rate (Yellow Springs Instruments, Yellow Springs, OH, USA).
Calculation of insulin sensitivity from the euglycaemic hyperinsulinaemic clamp
During hyperinsulinaemia with steady-state plasma glucose concentrations (the last 30 min of a 120 min procedure), the glucose infused is equal to that being removed from the glucose space. The M-value is a measure of total body glucose metabolism, and reflects the ability of insulin to enhance tissue glucose disposal, assuming suppression of hepatic glucose production. M-values were calculated according to the procedures of the European Group for the Study of Insulin Resistance [21].
Human skeletal muscle extraction
Forceps biopsies were taken according to established procedures from the right vastus lateralis muscle 5 min before the start of the clamp (control samples) [22]. A second biopsy was taken after 1 h of the clamp (insulin-stimulated sample). Muscle biopsies were directly frozen in liquid nitrogen and kept at −80 °C until the moment they were analysed.
Cell culture
The rat hepatoma cell line H4IIE was maintained in DMEM (Dulbecco's modified Eagle's medium) containing 1000 mg/l glucose, 5% (v/v) foetal bovine serum and 1% (v/v) penicillin/streptomycin at 37 °C with 5% CO2. Rat L6 skeletal muscle cells were cultured to myotubes in α-MEM (minimal essential medium) containing 2% (v/v) FBS and 1% (v/v) antimycotic/antibiotic solution (100 units/ml penicillin, 100 μg/ml streptomycin and 250 ng/ml amphotericin B) at 37 °C with 5% CO2, as described previously [23]. In brief, cells were seeded in 10 cm dishes at the density of 5.5×103 cell/cm2. Fused multinuclear myotubes can be observed 5–6 days post-seeding. Cells were cultured in 60 mm dishes and treated with hormones for the times and concentrations shown in the Figure legends prior to lysis. Cells were pre-incubated with inhibitors for 30 min (LY294002, rapamycin, puromycin and cycloheximide) or for 4 h (MG132) before hormone and inhibitor treatments, as described in the Figure legends.
Preparation of protein extracts for Western blotting or immunoprecipitation
Muscle biopsies were thawed on ice and homogenized (using a Dounce homogenizer; 10–15 strokes) in 0.5 ml of ice-cold lysis buffer [25 mM Tris/HCl (pH 7.4)/50 mM NaF/100 mM NaCl/1 mM sodium vanadate/5 mM EGTA/1 mM EDTA/1% (v/v) Triton X-100/10 mM sodium pyrophosphate/0.27 M sucrose/Complete™ Protease inhibitor cocktail tablets (1 tablet/10 ml)/0.1% (v/v) 2-mercaptoethanol]. Lysates were obtained from the supernatant fraction after a 10 min centrifugation at 13000 g, and pre-cleared for 1 h at 4 °C with Protein G–Sepharose in PBS (50%, v/v). This was to remove contaminating antibodies present due to the variable amount of blood in the samples.
H4IIE rat hepatoma cells and L6 skeletal muscle cells were incubated in serum-free medium with hormones and inhibitors for the times and at the concentrations indicated in the Figure legends after overnight serum-free starvation. Cells were then scraped into ice-cold lysis buffer. Tissue and cell debris were removed by centrifugation at 4 °C for 10 min at 13000 g, and the protein concentration was determined by the method of Bradford [24] using BSA as an internal standard. To analyse the Triton-X-100-insoluble pellet, the cell debris was sonicated for 10 s in 1% (w/v) SDS containing Mops–NuPAGE buffer (Novex, Invitrogen, UK), before addition of 2% LDS loading buffer (Novex, Invitrogen, Carlsbad, CA, U.S.A.), incubation at 70 °C for 15 min and separation by SDS/PAGE.
Animals
Mice were fasted overnight and injected intraperitoneally with insulin (150 m-units/g) or saline solution, as described previously [25]. Skeletal muscle was extracted at the indicated time points and frozen in liquid nitrogen. Liver samples were obtained from mice fasted overnight, and maintained in the fasted state (‘fasted’) or re-fed (‘fed’) for 1 h. Livers were excised and frozen immediately in liquid nitrogen. They were then pulverized to a powder, a 10-fold mass excess of ice-cold lysis buffer was added, the solution was centrifuged and the supernatants were snap-frozen and stored at −80 °C.
Western blot analysis
Lysates from tissues (30–40 μg from muscle; 15 μg from liver) and cell lines (10–20 μg from H4IIE; 50 μg from L6) were separated on Novex SDS/4–12% polyacrylamide gels. Following transfer to nitrocellulose, blots were blocked with 5% (w/v) non-fat milk in TBST [Tris-buffered saline containing 0.1% (v/v) Tween 20] for 1 h, and incubated with primary antibodies at 4 °C overnight before incubation for 1 h at room temperature with the secondary antibody and development using an ECL (enhanced chemiluminescence) kit (Amersham Biosciences, Inc.).
Quantification
Protein bands were scanned and quantified by densitometry using AIDA Image Analyzer software. Band densities were expressed relative to those obtained from β-actin for each sample.
Immunoprecipitation and phosphatase treatment
Pre-cleared human muscle lysates and L6 cell extracts (50–200 μg) were immunoprecipitated with 2 μg of anti-IRS-1 antibody coupled with Protein G–Sepharose. Lysates were incubated at 4 °C on a shaking platform for 1 h. Immune complexes were washed once with 1 ml of 0.5 M NaCl in lysis buffer and then twice with wash buffer [25 mM Tris/HCl (pH 7.4)/0.1 mM EGTA/5% (v/v) glycerol/0.1% (v/v) 2-mercaptoethanol]. Immunoprecipitates were split, and one-half was analysed directly by Western blotting (control); the other half was incubated with protein phosphatase 1 (10 m-units) for 30 min at 30 °C (dephosphorylated samples) prior to Western blotting.
Statistical analysis
Data were compared by Student's t test analysis. Differences were considered to be statically significant at P<0.05.
RESULTS
Insulin induces IRS1 protein levels in humans and rodents
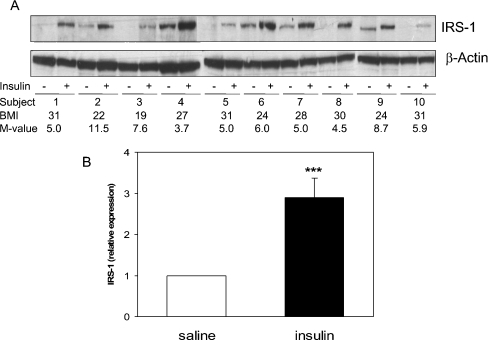
Basal IRS1 levels vary in muscle from different individuals (Figure 1A); however, IRS1 protein increased approx. 3-fold in all 10 subjects studied following 1 h of euglycaemic hyperinsulinaemia (Figure 1B). In this small group of male volunteers, no obvious association was observed between insulin regulation of IRS1 and either the BMI or M-value, an estimate of whole-body insulin sensitivity (Figure 1A). Although standardized reference ranges for measures of insulin sensitivity have not yet been clearly defined (the M-value is known to vary quite widely even amongst healthy subjects of similar age and BMI), the variation we observed was in keeping with that expected in non-diabetic male subjects [26]. It has been known for some time that insulin treatment (>3 h) can lead to down-regulation of IRS1 expression; however, this is the first demonstration that insulin acutely induces IRS1 expression. This induction is not due to changes in phosphorylation recognized by the IRS1 antibody, since immunoprecipitated IRS1 from human muscle is increased in the insulin-exposed samples even following incubation with protein phosphatases (results not shown). Therefore increases in signal strength are related to increased protein, rather than a post-translational modification.
Figure 1. Acute induction of IRS1 levels by insulin in human muscle.
Muscle biopsies were obtained from 10 male healthy adult subjects with a BMI between 19 and 31 kg/m2 before and after 1 h of hyperinsulinaemic, euglycaemic clamp. (A) Muscle samples were separated by SDS/PAGE and IRS1 expression was analysed by Western blotting. Expression was quantified (B) by densitometry and is presented (means±S.E.M., n=10), after correction for β-actin expression, relative to basal expression (saline). ***P<0.001, saline compared with insulin. The M-value (mg/kg per min) is a measure of total body glucose metabolism, and is calculated as described in the Experimental section.
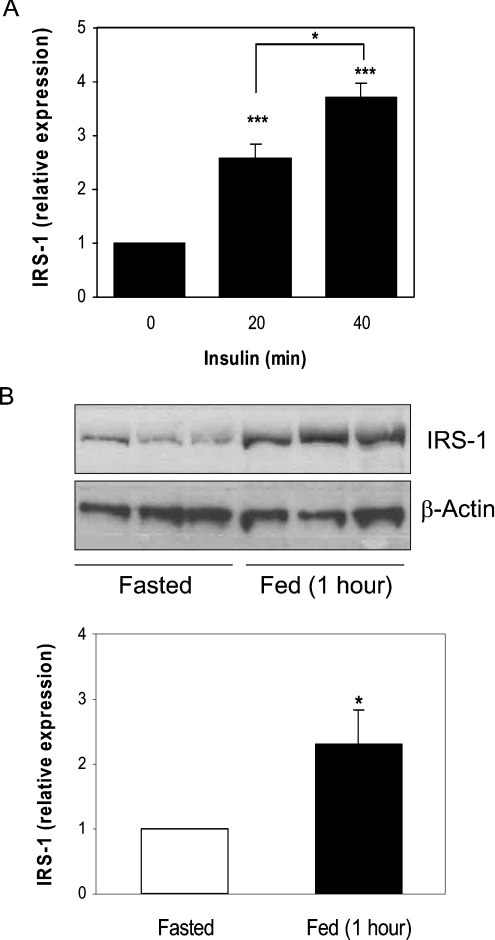
Significant induction of IRS1 expression in rodent muscle also occurs within 20 min of a single insulin injection (Figure 2A), and increases further (to 3.5-fold over basal) after 40 min. Therefore the increase is not unique to euglycaemic hyperinsulinaemia. Similarly, hepatic IRS1 expression is induced 2.2-fold in re-fed mice (Figure 2B). This demonstrates that IRS1 increases in three different in vivo models of insulin exposure, whereas the effect is not muscle- or species-specific, and occurs at insulin concentrations found in response to normal feeding.
Figure 2. Acute induction of IRS1 levels by insulin and feeding in mice.
(A) Mice were fasted overnight and injected intraperitoneally with insulin (150 m-units/g) or saline solution. Skeletal muscle extracts (30–40 μg) were analysed for IRS1 expression by Western blotting, and blots were quantified by densitometry. (B) Mice were fasted for 16 h, and maintained in the fasted state (Fasted) or re-fed (Fed) for 1 h. Liver extracts were analysed for IRS1 expression by Western blotting (upper panel) and quantified (lower panel). Values (means±S.E.M., n=3) are expressed, after correction for β-actin expression, relative to basal expression (saline for muscle and fasted state for liver). *P<0.05 (20 min compared with 40 min, and fasted compared with fed); ***P<0.001 (experimental compared with no insulin).
Insulin induces IRS1 protein in tissue culture
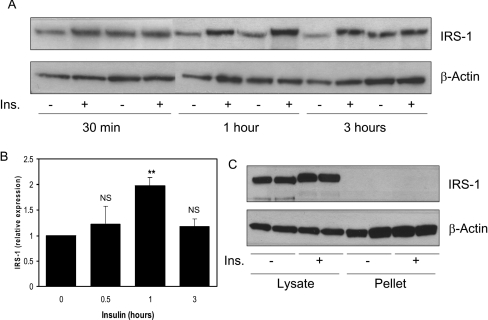
L6 myotubes are often employed as an insulin-responsive culture model of intact muscle. Insulin treatment of L6 myotubes for 1 h increases IRS1 protein levels (Figure 3A). This increase is not as substantial as that seen in intact human muscle, possibly because these cells have a high basal expression of IRS1. The induction is transient, with IRS1 levels returning to baseline after 3 h (Figure 3B). Previous work has demonstrated that IRS1 protein can exist in more than one intracellular pool. Indeed, three groups have found that insulin induces IRS1 translocation from LDMs (low-density membranes) to the cytosol in adipocytes [27–29]. However, in our experiments the lysis method does not distinguish between cytosol and membrane fractions. For example, GLUT4 translocates from LDMs to plasma membranes following insulin treatment. However, we observe no difference in GLUT4 levels following insulin treatment using our lysis method (Figure 3C). Therefore the LDM fraction must be present in our lysates, and the increased IRS1 observed in our experiments is not due to translocation of IRS1 from LDM to cytosol. In addition, no IRS1 is detectable in the Triton X-100-insoluble fraction following lysis of human muscle samples before or after insulin treatment (results not shown).
Figure 3. Induction of IRS1 protein by insulin in L6 myotubes.
(A) Rat L6 myotubes were serum-starved overnight and treated with 100 nM insulin for 1, 2 or 3 h. Western blot analysis was performed on 50 μg of cell lysates to assess IRS-1 and β-actin protein levels. (B) Expression was quantified by densitometric scan of the blots. Values are the ratio of IRS1 to β-actin and are presented relative to serum-starved cells (average±S.E.M. of two experiments performed in duplicate). **P<0.01 (0 compared with 1 h); NS, not significant. (C) GLUT4 extraction following L6 cell lysis was assessed by Western blot analysis.
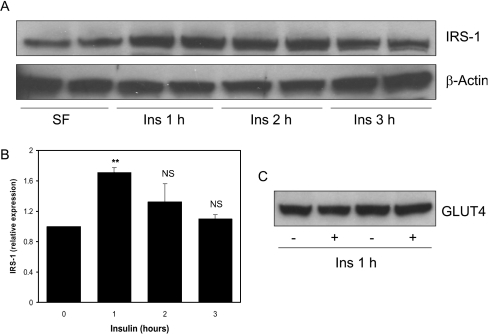
H4IIE is a rat hepatoma cell line that is exquisitely sensitive to insulin [30,31]. Insulin treatment of these cells also induces a transient induction of IRS1 protein levels (Figure 4A), peaking at around 1 h (2-fold) and returning to basal levels following 3 h treatment (Figure 4B). Once more, there is no IRS1 in the Triton X-100-insoluble fraction of the cells before or after insulin treatment (Figure 4C), demonstrating an increase in protein, rather than translocation, from an insoluble pool. We decided to use the H4IIE rather than L6 cells to investigate the mechanism by which insulin induces IRS1 expression, since induction following insulin is higher in the H4IIE cells and they do not require differentiation.
Figure 4. Induction of IRS1 protein by insulin in hepatoma cells.
H4IIE cells were serum-starved overnight and then treated with or without 10 nM insulin for the indicated times. (A) Western blot analysis was performed on 15 μg of cell lysates to assess the IRS1 and β-actin protein expression. (B) Blots were quantified by densitometry. Values are the ratio of IRS1 to β-actin expressed relative to serum-starved cells and are the average±S.E.M. of three experiments performed in duplicate. **P<0.01 (0 compared with 1 h); NS, not significant; Ins., insulin. (C) IRS1 and β-actin expression was visualized in the Triton-X-100-insoluble fraction and directly compared with the levels in the soluble fraction (an equivalent amount of each fraction was loaded, and the blot overexposed).
Preliminary investigation of the mechanism of induction of IRS1 expression
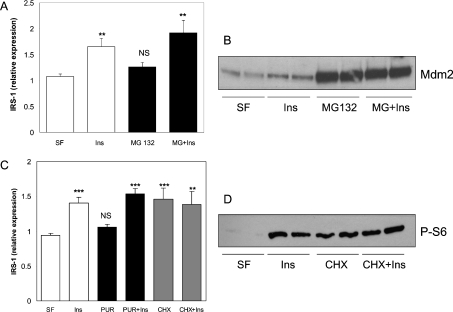
IRS1 mRNA levels are relatively low in the H4IIE cells, and do not change following insulin treatment for 30 min or 1 h (results not shown); therefore the rapid induction of IRS1 protein is not due to increased gene transcription. Proteasomal degradation of IRS1 is a well-characterized process [6,32–34] and its regulation represents a potential mechanism by which insulin could increase as well as decrease IRS1 expression. Therefore IRS1 expression was assessed in the H4IIE cells following 1 h exposure to insulin in the presence or absence of the proteasome inhibitor MG132 (Figure 5A). Interestingly, inhibition of the proteasome for this time has very little effect on IRS1 levels, suggesting that proteasomal degradation is not a major acute regulator of IRS1 protein levels in resting H4IIE cells. Meanwhile, insulin induces IRS1 expression to a similar extent in the presence or absence of MG132 (Figure 5A), demonstrating that insulin does not inhibit the proteasome to induce IRS1 levels. MG132 treatment of H4IIE cells dramatically increases Mdm2 levels (Figure 5B). This protein is normally rapidly degraded by the proteasome, and therefore the dramatic increase in Mdm2 levels indicates that inhibition of degradation was achieved in our experiments [35].
Figure 5. Induction of IRS1 protein by insulin is not mediated by proteasomal inhibition or translational activation.
(A) Starved H4IIE cells (SF) were pre-incubated with or without 10 μM MG132 (proteasome inhibitor) for 4 h prior to a 1 h incubation in the absence or presence of 10 nM insulin (Ins) and Western blot analysis of IRS1 and β-actin expression. (B) Mdm2 expression was assessed in the samples from (A). (C) After overnight serum deprivation, H4IIE cells were pre-incubated with or without 10 μg/ml cycloheximide (CHX) or puromycin (PUR) for 30 min, and then treated with 10 nM insulin±each drug for 1 h prior to Western blot analysis and quantification. (D) Phosphorylation of ribosomal S6 protein (P-S6) was determined by Western blot analysis of protein lysates from starved, cycloheximide (CHX; 10 μg/ml)- or insulin (10 nM)-treated cells (1 h incubations). For (A) and (C), values are the ratio of IRS1 to β-actin, and are the average±S.E.M. of at least two experiments performed in duplicate. ***P<0.001; **P<0.01 (experimental compared with serum-free); NS, not significant. For (B) and (D), representative blots from two experiments are shown.
Finally, inhibition of protein synthesis (using puromycin) has no effect on basal or insulin-induced IRS1 expression (Figure 5C). This suggests that insulin is not inducing IRS1 expression through increased protein translation, and confirms that the mechanism does not involve increased transcription. A distinct protein synthesis inhibitor, cycloheximide, induces IRS1 expression in the absence of insulin (Figure 5C), and its effects are not additive to insulin. It is likely that this is related to its activation of the S6K pathway rather than through general effects on protein synthesis (Figure 5D, and below). Interestingly, hepatic S6K was originally purified from cycloheximide-treated rats, and this molecule is the most potent activator of hepatic S6K reported to date in vivo [36,37].
The mTOR pathway is required for insulin regulation of IRS1 expression
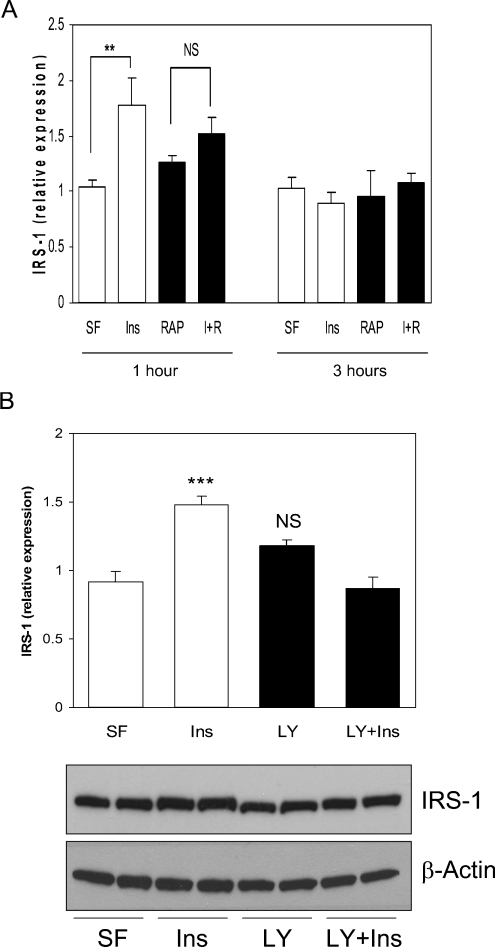
The mTOR–S6K pathway is known to negatively regulate IRS1 levels by increasing IRS1 serine/threonine phosphorylation, thereby promoting degradation. This negative-feedback pathway can be blocked by treatment of cells with the mTOR inhibitor rapamycin; therefore rapamycin is predicted to stabilize IRS1 protein levels [38]. Interestingly, in our acute incubations, rapamycin alone has little effect, whereas insulin does not significantly induce IRS1 in the presence of rapamycin (Figure 6A). A similar effect is seen in the presence of the PI3K inhibitor LY294002 (Figure 6B). This implicates both PI3K and mTOR activity in the acute induction of IRS1 levels following insulin treatment of cells. A common downstream target for both molecules is S6K [39,40]. Meanwhile, cycloheximide induces IRS1 expression (Figure 5C), and activates the phosphorylation of S6 [36,37], a downstream target of the mTOR pathway, to a similar extent as that observed with insulin (Figure 5D). This is consistent with acute activation of the S6K pathway being sufficient to induce IRS1 levels.
Figure 6. Insulin activation of PI3K and mTOR regulates the induction of IRS1 protein.
(A) H4IIE cells were serum-starved overnight (SF), pre-incubated with 10 nM rapamycin (RAP) or vehicle for 30 min prior to a 1 or 3 h treatment with 10 nM insulin (Ins), RAP or both (I+R), followed by lysis and Western blot analysis. Data are presented as the ratio of IRS1 to β-actin, and are the average±S.E.M. of at least two experiments performed in duplicate. **P<0.01 (insulin compared with serum-free). (B) H4IIE cells were serum-starved overnight (SF), pre-incubated with 100 μM LY294002 (LY) or vehicle for 30 min prior to a 1 h incubation with 10 nM insulin (Ins), LY or both (LY+Ins), followed by lysis and Western blot analysis. The lower panels contain a representative blot, whereas the upper panel contains the ratio of IRS1 to β-actin as the average±S.E.M of three experiments performed at least in duplicate. ***P<0.001 (insulin compared with serum-free); NS, not significant.
Another signalling molecule downstream of PI3K that is reported to negatively regulate IRS1 levels and activity through serine/threonine phosphorylation is GSK3 [41,42]. Therefore pharmacological inhibition of GSK3 should stabilize IRS1 protein if GSK3 phosphorylation of IRS1 represents a major pathway to its degradation. However, IRS1 levels do not change following treatment of H4IIE cells with the GSK3 inhibitor CT99021 for up to 3 h (results not shown). In addition, hepatic IRS1 protein is induced following feeding of S21A/S9A GSK3 knock-in mice to a similar extent as that observed in wild-type mice (results not shown). These animals express a form of GSK3 that is insensitive to insulin [25]. Therefore inhibition of GSK3 does not acutely induce IRS1 protein, and inhibition of GSK3 is not required for insulin induction of IRS1 protein (at least 1 h post-prandium).
DISCUSSION
Degradation of IRS1 is controlled by negative-feedback signalling pathways that may be overactive in T2DM. However, we now demonstrate that regulation of IRS1 expression is a biphasic process, with an initial increase in protein levels prior to the previously reported reduction in IRS1 protein. As this is a completely novel observation, we have focused on establishing the validity of the event in three distinct in vivo models (euglycaemic hyperinsulinaemia, insulin bolus and physiological feeding) and two cell-culture systems. Although this process is seen in all of these various scenarios covering two organs and two species, it is most evident in human muscle. The reasons why this has not been identified in previous studies on IRS1 expression are likely to be two-fold. First, most studies have been conducted in cell lines (e.g. L6 myotubes and 3T3-L1 adipocytes) where the induction is weakest owing to high basal IRS1 expression. Secondly, almost all previous studies have examined much longer incubation periods, where the reduction in IRS1 levels is clear and substantial. Therefore we propose that insulin induction of IRS1 levels occurs transiently (within 20 min), peaking by 1 h and returning to basal levels by 3 h. Longer incubation times lead to a reduction in IRS1 levels below basal.
Extending the argument that reduction in IRS1 levels following prolonged insulin exposure results in reduced signalling and may contribute to insulin resistance, we would propose that the acute induction in levels actually increases signalling capacity. The inability to produce this acute induction, for whatever reason, would reduce capacity and thus constitutes a potential ‘insulin-resistance-causing’ event. Obviously, confirmation of the importance of the induction in health and disease requires detailed knowledge of the mechanism by which insulin signals to IRS1 acutely.
Pharmacological and biochemical analysis has successfully ruled out regulation of IRS1 transcription and translation, or the need for ongoing protein synthesis, for the acute induction by insulin. We have preliminary evidence that a non-proteasome-mediated stabilization of IRS1 is occurring, suggesting insulin inhibition of an IRS1 protease. There are many proteases that could be investigated as mediators of this action of insulin. For example, insulin-like growth factor-1 activation of PKB promotes inhibition of neuronal caspase 3 [43]. However, addition of caspase inhibitor I (purchased from Calbiochem) to H4IIE cells has no effect on basal or insulin-induced IRS1 expression (results not shown). Oxidative stress promotes IRS1 degradation by a proteasome-independent mechanism, although the protease involved is not known [44]. Identification of the protease responsible, as well as the molecular mechanism by which IRS1 stability is regulated, will be required to ascertain the importance of this regulation in health and disease.
Finally, the mTOR–S6K pathway promotes serine phosphorylation of IRS1, leading to reduced insulin signalling [34,45]. Hence the mTOR inhibitor rapamycin should enhance insulin sensitivity through reduced IRS1 serine phosphorylation. However, we have now found that full induction of IRS1 expression by acute insulin exposure also requires mTOR activity (as well as PI3K activity); therefore these short-term increases in signalling capacity/sensitivity can actually be blocked by rapamycin. Takano et al. [28] have previously found that rapamycin can block the translocation of IRS1 between a cytosolic and a Triton X-100-insoluble fraction in adipocytes. We did not observe any IRS1 in the Triton X-100-insoluble fraction in H4IIE cells or human muscle; therefore this mechanism could not account for the increase in IRS1 in our studies. However, it appears that there are at least three different mechanisms by which mTOR regulates IRS1 activity. These are translocation, increased proteasomal degradation (after long-term insulin stimulation) and now proteasome-independent stabilization immediately following stimulation. Rapamycin protects IRS1 from oxidative-stress-induced degradation [44], and therefore the mTOR pathway also regulates this non-proteasomal destruction of IRS1. Identification of this protease may provide a common mTOR target linking oxidative destruction and hormonal stabilization of IRS1.
Proteasomal degradation of IRS1 may be controlled by mTOR through activation of S6K [45] or PKC [46,47], but currently these protein kinases are only known to induce proteasomal degradation of IRS1. Whether they regulate IRS1 stability by an additional, more acute mechanism remains to be elucidated. The increased stability occurs simultaneously with increased tyrosine phosphorylation of IRS1. Therefore mTOR may control the interaction of a stabilizing protein with phosphotyrosine motifs on IRS1, synergistically increasing the capacity of IRS1 signalling. Activation of the mTOR–S6K pathway is dependent on two distinct signalling inputs, namely insulin and nutrients (such as amino acids). Therefore IRS1 induction, translocation and degradation are all linked to both nutrient and insulin availability through the mTOR pathway, although we still observed acute induction of IRS1 in rodent muscle following an insulin bolus (which promotes hypoglycaemia), suggesting the effect is independent of glucose.
In summary, the increased degradation of IRS1 associated with hyperphosphorylation on serine/threonine residues is a potential mechanism that could lead to insulin resistance. Indeed, evidence is accumulating that both serine phosphorylation and basal IRS1 levels are altered in insulin-resistant states, including diabetes [29,48,49]. However, recent work in tissue culture has suggested that the increase in degradation of IRS1 actually occurs subsequent to defects in insulin action [20]. Therefore loss of the ability to induce IRS1 protein acutely (as shown herein) is an alternative mechanism by which signalling capacity could be reduced, contributing to insulin resistance. We propose that this mechanism may be of equal importance for determining insulin-signalling capacity/sensitivity, and it will be of interest to examine the effect of short-term insulin treatment on IRS1 up-regulation in tissue from insulin-resistant humans.
Acknowledgments
C. L. and E. J. M. are recipients of a BBSRC (Biotechnology and Biological Sciences Research Council) CASE (Co-operative Awards in Science and Engineering) studentship and an MRC Predoctoral Fellowship respectively, while C. S. is a recipient of the Diabetes UK Senior Fellowship (02/0002473). This work was supported by Diabetes UK Project Grant number (03/0002583) and CSO project grant number (CZB/4/125). We thank Dr Yvonne Woods and Dr Mark Saville for technical advice.
References
- 1.Withers D. J., White M. Perspective: the insulin signaling system-a common link in the pathogenesis of Type 2 diabetes. Endocrinology. 2000;141:1917–1921. doi: 10.1210/endo.141.6.7584. [DOI] [PubMed] [Google Scholar]
- 2.Lazar M. A. How obesity causes diabetes: not a tall tale. Science. 2005;307:373–375. doi: 10.1126/science.1104342. [DOI] [PubMed] [Google Scholar]
- 3.Kahn C. R., White M. F. The insulin receptor and the molecular mechanism of insulin action. J. Clin. Invest. 1988;82:1151–1156. doi: 10.1172/JCI113711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shulman G. I. Cellular mechanisms of insulin resistance. J. Clin. Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White M. F. IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 2002;283:E413–E422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y. F., Herschkovitz A., Boura-Halfon S., Ronen D., Paz K., Leroith D., Zick Y. Serine phosphorylation proximal to its phosphotyrosine binding domain inhibits insulin receptor substrate 1 function and promotes insulin resistance. Mol. Cell. Biol. 2004;24:9668–9681. doi: 10.1128/MCB.24.21.9668-9681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White M. F. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol. Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 8.Avruch J. Insulin signal transduction through protein kinase cascades. Mol. Cell. Biochem. 1998;182:31–48. [PubMed] [Google Scholar]
- 9.Leslie N. R., Biondi R. M., Alessi D. R. Pi-regulated kinases and PI phosphatases. Chem. Rev. 2001;101:2365–2380. doi: 10.1021/cr000091i. [DOI] [PubMed] [Google Scholar]
- 10.Sutherland C., Leighton I. A., Cohen P. Inactivation of glycogen synthase kinase-3β by phosphorylation; new kinase connections in insulin and growth factor signalling. Biochem. J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross D. A. E., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. Inhibition of GSK3 by insulin mediated by protein kinase B. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 12.Welsh G. I., Foulstone E. J., Young S. W., Tavare J. M., Proud C. G. Wortmannin inhibits the effects of insulin and serum on the activities of GSK3 and MAPK. Biochem. J. 1994;303:15–20. doi: 10.1042/bj3030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lizcano J. M., Alessi D. R. The insulin signalling pathway. Curr. Biol. 2002;12:R236–R238. doi: 10.1016/s0960-9822(02)00777-7. [DOI] [PubMed] [Google Scholar]
- 14.Manning B. D., Cantley L. C. Rheb fills a GAP between TSC and TOR. TIBS. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Shigemitsu K., Tsujishita Y., Hara K., Nanahoshi M., Avruch J., Yonezawa K. Regulation of translational effectors by amino acid and mammalian target of rapamycin signaling pathways. J. Biol. Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 16.Galic S., Hauser C., Kahn B. B., Haj F. G., Neel B. G., Tonks N. K., Tiganis T. Coordinated regulation of insulin signaling by the protein tyrosine phosphatases PTP1B and TCPTP. Mol. Cell. Biol. 2005;25:819–829. doi: 10.1128/MCB.25.2.819-829.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene M. W., Garofalo R. S. Positive and negative regulatory role of insulin receptor substrate 1 and 2 (IRS-1 and IRS-2) serine/threonine phosphorylation. Biochemistry. 2002;41:7082–7091. doi: 10.1021/bi015992f. [DOI] [PubMed] [Google Scholar]
- 18.Tanti J. F., Gual P., Gremeaux T., Gonzalez T., Barres R., Le Marchand-Brustel Y. Alteration in insulin action: role of IRS-1 serine phosphorylation in the retroregulation of insulin signalling. Ann. Endocrinol. (Paris) 2004;65:43–48. doi: 10.1016/s0003-4266(04)95629-6. [DOI] [PubMed] [Google Scholar]
- 19.Kaburagi Y., Yamauchi T., Yamamoto-Honda R., Ueki K., Tobe K., Akanuma Y., Yazaki Y., Kadowaki T. The mechanism of insulin-induced signal transduction mediated by the insulin receptor substrate family. Endocrinol. J. 1999;46:S25–S34. doi: 10.1507/endocrj.46.suppl_s25. [DOI] [PubMed] [Google Scholar]
- 20.Renstrom F., Buren J., Eriksson J. W. IRS1 and 2 are both depleted but by different mechanisms in rat adipocytes following downregulation of glucose transport. Endocrinology. 2005;146:3044–3051. doi: 10.1210/en.2004-1675. [DOI] [PubMed] [Google Scholar]
- 21.Hills S. A., Balkau B., Coppack S. W., Dekker J. M., Mari A., Natali A., Walker M., Ferrannini E. The EGIR-RISC Study (the European Group for the Study of Insulin Resistance: relationship between insulin sensitivity and cardiovascular disease risk): I. methodology and objectives. Diabetologia. 2004;47:566–570. doi: 10.1007/s00125-004-1335-5. [DOI] [PubMed] [Google Scholar]
- 22.Cuthbertson D., Smith K., Babraj J., Leese G., Waddell T., Atherton P., Wackerhage H., Taylor P. M., Rennie M. J. Anabolic signaling deficits underlie amino acid resistance of wasting, aging muscle. FASEB J. 2005;19:422–424. doi: 10.1096/fj.04-2640fje. [DOI] [PubMed] [Google Scholar]
- 23.Hajduch E., Alessi D. R., Hemmings B. A., Hundal H. S. Constitutive activation of protein kinase B alpha by membrane targeting promotes glucose and system A amino acid transport, protein synthesis, and inactivation of glycogen synthase kinase 3 in L6 muscle cells. Diabetes. 1998;47:1006–1013. doi: 10.2337/diabetes.47.7.1006. [DOI] [PubMed] [Google Scholar]
- 24.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., Alessi D. R. Role that phosphorylation of GSK3 plays in insulin and Wnt-signalling defined by knockin analysis. EMBO J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrannini E., Natali A., Capaldo B., Lehtovirta M., Jacob S., Yki-Jarvinen H. Insulin resistance, hyperinsulinemia, and blood pressure: role of age and obesity. European Group for the Study of Insulin Resistance (EGIR) Hypertension. 1997;30:1144–1149. doi: 10.1161/01.hyp.30.5.1144. [DOI] [PubMed] [Google Scholar]
- 27.Clark S. F., Martin S., Carozzi A. J., Hill M. M., James D. E. Intracellular localization of phosphatidylinositide 3-kinase and insulin receptor substrate-1 in adipocytes: potential involvement of a membrane skeleton. J. Cell Biol. 1998;140:1211–1225. doi: 10.1083/jcb.140.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takano A., Usui I., Haruta T., Kawahara J., Uno T., Iwata M., Kobayashi M. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptor substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol. Cell. Biol. 2001;21:5050–5062. doi: 10.1128/MCB.21.15.5050-5062.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tremblay F., Gagnon A., Veilleux A., Sorisky A., Marette A. Activation of the mammalian target of rapamycin pathway acutely inhibits insulin signaling to Akt and glucose transport in 3T3-L1 and human adipocytes. Endocrinology. 2005;146:1328–1337. doi: 10.1210/en.2004-0777. [DOI] [PubMed] [Google Scholar]
- 30.Sutherland C., O'Brien R. M., Granner D. K. Phosphatidylinositol 3-kinase, but not p70/p85 ribosomal S6 protein kinase, is required for the regulation of phosphoenolpyruvate carboxykinase gene expression by insulin. J. Biol. Chem. 1995;270:15501–15506. doi: 10.1074/jbc.270.26.15501. [DOI] [PubMed] [Google Scholar]
- 31.Finlay D., Patel S., Dickson L. M., Shpiro N., Marquez R., Rhodes C. J., Sutherland C. Glycogen synthase kinase-3 regulates IGFBP-1 gene transcription through the thymine-rich insulin response element: inhibition is required for full regulation of this promoter element by insulin. BMC Mol. Biol. 2004;5:15. doi: 10.1186/1471-2199-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greene M. W., Sakaue H., Wang L., Alessi D. R., Roth R. A. Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by serine 312 phosphorylation. J. Biol. Chem. 2003;278:8199–8211. doi: 10.1074/jbc.M209153200. [DOI] [PubMed] [Google Scholar]
- 33.del Rincon S. V., Guo Q., Morelli C., Shiu H. Y., Surmacz E., Miller W. H. Retinoic acid mediates degradation of IRS-1 by the ubiquitin-proteasome pathway, via a PKC-dependant mechanism. Oncogene. 2004;23:9269–9279. doi: 10.1038/sj.onc.1208104. [DOI] [PubMed] [Google Scholar]
- 34.Shah O. J., Wang Z., Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 35.Saville M. K., Sparks A., Xirodimas D. P., Wardrop J., Stevenson L. F., Bourdon J. C., Woods Y. L., Lane D. P. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J. Biol. Chem. 2004;279:42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 36.Price D. J., Nemenoff R. A., Avruch J. Purification of a hepatic S6 kinase from cycloheximide-treated rats. J. Biol. Chem. 1989;264:13825–13833. [PubMed] [Google Scholar]
- 37.Kozma S., Lane H. A., Ferrari S., Luther H., Siegmann M., Thomas G. A stimulated S6 kinase from rat liver: identity with the mitogen activated S6 kinase of 3T3 cells. EMBO J. 1989;8:4125–4132. doi: 10.1002/j.1460-2075.1989.tb08597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher T. L., White M. F. Signaling pathways: the benefits of good communication. Curr. Biol. 2004;14:R1005–R1007. doi: 10.1016/j.cub.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 39.Gao X., Zhang Y., Arrazola P., Hino O., Kobayashi T., Yeung R. S., Ru B., Pan D. Tsc proteins antagonize amino acid-TOR signalling. Nat. Cell. Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 40.Potter C. J., Pedraza L. G., Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell. Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 41.Eldar-Finkelman H., Krebs E. G. Phosphorylation of IRS-1 by glycogen synthase kinase-3 impairs insulin action. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9660–9664. doi: 10.1073/pnas.94.18.9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberman Z., Eldar-Finkelman H. Serine 332 phosphorylation of insulin receptor substrate-1 by glycogen synthase kinase-3 attenuates insulin signaling. J. Biol. Chem. 2005;280:4422–4428. doi: 10.1074/jbc.M410610200. [DOI] [PubMed] [Google Scholar]
- 43.Zhong J., Deng J., Phan J., Dlouhy S., Wu H., Yao W., Ye P., D'Ercole A. J., Lee W. H. Insulin-like growth factor-I protects granule neurons from apoptosis and improves ataxia in weaver mice. J. Neurosci. Res. 2005;80:481–490. doi: 10.1002/jnr.20490. [DOI] [PubMed] [Google Scholar]
- 44.Potashnik R., Bloch-Damti A., Bashan N., Rudich A. IRS1 degradation and increased serine phosphorylation cannot predict the degree of metabolic insulin resistance induced by oxidative stress. Diabetologia. 2003;46:639–648. doi: 10.1007/s00125-003-1097-5. [DOI] [PubMed] [Google Scholar]
- 45.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parekh D., Ziegler W., Yonezawa K., Hara K., Parker P. J. Mammalian TOR controls one of two kinase pathways acting upon nPKCd and nPKCe. J. Biol. Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 47.Sarbassov D. D., Ali S. M., Kim D.-H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 48.Catalano P. M., Nizielski S. E., Shao J., Preston L., Qiao L., Friedman J. E. Downregulated IRS-1 and PPAR in obese women with gestational diabetes: relationship to FFA during pregnancy. Am. J. Physiol. Endocrinol. Metab. 2002;282:E522–E533. doi: 10.1152/ajpendo.00124.2001. [DOI] [PubMed] [Google Scholar]
- 49.Beeson M., Sajan M. P., Dizon M., Grebenev D., Gomez-Daspet J., Miura A., Kanoh Y., Powe J., Bandyopadhyay G., Standaert M. L., Farese R. V. Activation of protein kinase C-zeta by insulin and phosphatidylinositol-3,4,5-(PO4)3 is defective in muscle in type 2 diabetes and impaired glucose tolerance: amelioration by rosiglitazone and exercise. Diabetes. 2003;52:1926–1934. doi: 10.2337/diabetes.52.8.1926. [DOI] [PubMed] [Google Scholar]