Abstract
Deglycating enzymes, i.e. enzymes that reverse the initial stage of the Maillard reaction between glucose and primary amines, are known to occur in mammalian, fungal and other eukaryotic and prokaryotic cells. In this issue of Biochemical Journal, Wiame et al. now report the existence of bacterial enzymes and an operon that control the metabolism and deglycation of glucoselysine 6-phosphate, i.e. the phosphorylated condensation product of fructose and ϵ-aminolysine. The discovery has broad implications for bacterial metabolism and possibly for the repair of protein damage by fructose.
Keywords: advanced glycation end-product (AGE), diabetes, fructoselysine, glucoselysine, glycation, Maillard reaction
Glycation is the process by which reducing sugars react non-enzymatically with biological nucleophiles, such as lysine and arginine residues in proteins, and purine bases in DNA. Glycation is part of the broader concept of the Maillard reaction (see http://imars.case.edu, the homepage for the International Maillard Reaction Society), in which all sorts of reactive carbonyl compounds inflict cellular and tissue damage during aging and in age-related diseases. Diseases of ‘carbonyl stress’ include, among others, diabetes and end-stage renal disease, in which levels of AGEs (advanced glycation end-products) reach staggering concentrations. The end-products that result from glycation reactions are generally highly stable at physiological pH, but inflict molecular damage through cross-linking, conformational changes, blocking of proteolytic sites, or binding of redox active metals, to name but a few.
In view of the fact that glucose is the single major circulating carbonyl compound in most mammals, there has been considerable interest in understanding the role of glucose in the overall Maillard reaction in vivo. Of particular importance is the initial glycation product from the reaction of glucose and lysine, also called Amadori product, fructoselysine or fructosamine. Its structure is depicted in Figure 1. Fructoselysine is a major protein adduct in extracellular matrix proteins, haemoglobin, lens proteins and other cells in which glucose permeates freely. It is the precursor to glucosepane, a lysine–arginine protein cross-link that is increased severalfold in diabetes [1], as well as several glycoxidation products [2]. For this reason, considerable efforts are being deployed to find ways to block or reverse its formation.
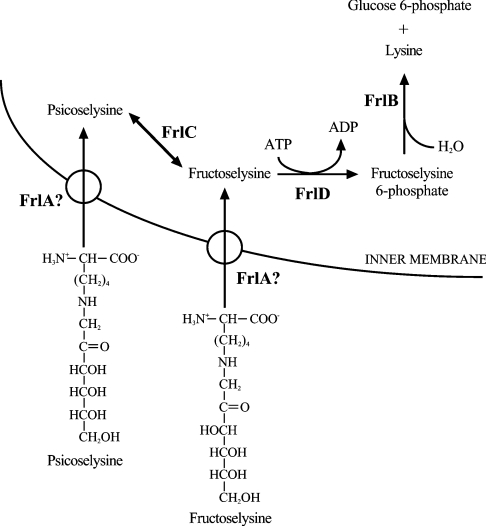
Figure 1. Metabolic machinery for fructoselysine and psicoselysine uncovered in E. coli by Van Schaftingen and colleagues [13].
The ability of the bacteria to take up the glycated substrates implies the existence of a putative cationic transporter (FrlA), an epimerase (FrlC) to convert psicoselysine into fructoselysine, a 6-phosphokinase (FrlD) and a deglycase (FrlB). Reproduced with permission from Wiame et al. [13]. © The Biochemical Society.
One question of major interest is how evolution has dealt with the formation of glycation products in prokaryotes and eukaryotes. To date, three groups of deglycating enzymes have been discovered. The first encompasses oxidases, specifically ‘fructosyl amine oxidase’ enzymes, which were first reported in Corynebacterium sp. and Aspergillus sp. by Horiuchi et al. [3,4]. These FAD enzymes were found to deglycate fructosyl amino acids into glucosone and the primary amine under formation of H2O2. These studies were then expanded to a large number of fungal enzymes with similar properties. Our own group cloned and fully characterized two ‘Amadoriase’ enzymes with high activity against glycated lysine in free form [5,6]. Unfortunately, these enzymes have little, or no, activity against glycated proteins, and no evidence for their presence in mammalian organisms has been found.
A second group of deglycating enzymes was discovered by Van Schaftingen and co-workers [7] based on earlier NMR studies by Szwergold et al. [8], who reported the existence of fructose 3-phosphate in the lens and an enzyme that can phosphorylate the Amadori product of glucose-modified protein into fructosamine 3-phosphate [9]. Van Schaftingen and co-workers [7] cloned and fully characterized the enzyme, and reported that it had deglycating activity, resulting in 3-deoxyglucosone formation while regenerating the free lysine residue. Thus the phosphorylating FN3K (fructosamine 3-kinase) enzyme is, in fact, a deglycating enzyme. Interestingly, the Km was found to be much lower for fructosamine than for fructose itself, thereby supporting its primary function as a deglycating enzyme. An FN3K-related protein, FN3K-RP, was recently discovered in vertebrates as far back as fish, which can phosphorylate ribuloselysine and erythruloselysine, and to a lesser degree psicoselysine, i.e. the condensation product of allose and lysine [10,11].
Further expanding on these landmark studies, a third group of deglycating enzymes was reported by Van Schaftingen and co-workers [12] based on the discovery that fructoselysine could sustain the growth of Escherichia coli, implying thereby the existence of a whole set of genes (i.e. a bacterial operon) that controls the uptake and metabolism of the glycated substrate. Database searches identified a kinase (YhfQ/FrlD) belonging to the PfkB/ribokinase family, a deglycase (YhfN/FrlB) that is homologous with the glucosamine-6-phosphate synthase, and a putative cationic amino acid transporter (YhfM/FrlA). This led to the identification of a fructoselysine-6-kinase enzyme, and the discovery of a deglycase enzyme, converting fructosamine 6-phosphate into glucose 6-phosphate and lysine [12]. Finally, since E. coli could also grow on psicoselysine, this implied the presence of a fructoselysine-3-epimerase (FrlC), which was indeed found and characterized [13]. These complex metabolic pathways are summarized in Figure 1.
In the current issue of Biochemical Journal, Van Schaftingen and colleagues [14] now describe the presence of a glucoselysine-6-phosphate deglycase in Enterococcus faecium. This may sound confusing, because glucoselysine is the condensation product of fructose with lysine leading to a Heyns product (Figure 2A), whereas, in contrast, the fructoselysine that was mentioned above is actually the condensation product of glucose and ϵ-amino-lysine (Figure 1). They hypothesized the existence of such an enzyme based on a bacterial genome search, which revealed operons encoding not only fructosamine-6-phosphate deglycase, but also a second homologue of the isomerase domain of glucosamine-6-phosphate synthase. For this operon, they coined the designation ‘gfr’, to indicate enzymes that degrade ‘glucation’ and ‘fructation’ products.
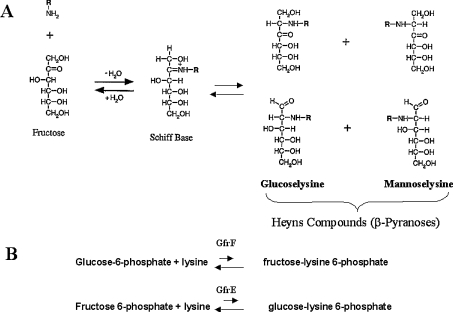
Figure 2. Examples of fructation products expected to form in vitro as a basis for structures resulting from the enzymatic action of GfrE in bacteria.
(A) The non-enzymatic reaction of fructose with ϵ-amino-lysine residues leads to the formation of various possible isomeric products (so-called Heyns rearrangement products), depending on whether the initial Schiff base isomerizes via the 1,2-, or 2,3-enolization pathway (B) Wiame et al. [14] have now identified not only enzymes that can catalyse and deglycate the formation of phosphorylated glycated lysine from glucose 6-phosphate, but also that of glucoselysine 6-phosphate from fructose 6-phosphate and ϵ-amino-lysine.
In order to clarify the biochemical function of this novel protein (GfrE), and that of the FrlB homologue GfrF (Figure 2B), Wiame et al. [14] PCR-amplified gfrF and gfrE from the genomic DNA of E. faecium, subcloned the products into pBluescript, expressed the whole open reading frame in E. coli, and purified GfrE and GfrF to homogeneity. These migrated as proteins of 39 kDa and 38 kDa in size, respectively, on an SDS gel. They also synthesized the radioactively labelled condensation product of fructose 6-phosphate and lysine (glucoselysine 6-phosphate), as well as fructoselysine 6-phosphate. Each enzyme was found to specifically degrade the substrates, as shown in Figure 2(B), without generating the other sugar. NMR spectroscopy was used to confirm the release of fructose 6-phosphate by GfrE. Interestingly, GfrE did not react with amino acids other than lysine. The equilibrium constant of 0.8 M suggests that the reaction is shifted towards deglycation. The Km for glucoselysine 6-phosphate was 0.4 mM, and the Vmax was 3 μmol/min per mg of protein, i.e. rather slow, and the enzyme could not catalyse the isomerization of glucoselysine 6-phosphate into fructoselysine 6-phosphate. A BLAST search revealed that enzyme GfrE is present in Enterococcus, Salmonella, Fusobacterium and Listeria, but not in E. coli species. In contrast, sequence similarities were found between GfrF or FrlB, i.e. the fructoselysine-6-phosphate deglycase in almost all bacterial species examined. The fructoselysine kinase was found in E. coli and selected other bacteria.
What is the significance of Van Schaftingen and colleagues work in the latest issue of Biochemical Journal? [14]. At first, it should be noted that it provides the first demonstration of an enzyme that participates in the metabolism of fructation products, and that it is a close homologue of glucosamine-6-phosphate synthase. The latter enzyme is known to catalyse the isomerization of the Schiff base resulting from the condensation of fructose 6-phosphate and ammonia. Both glucoselysine and fructoselysine are compounds that are abundantly present in free form in vegetable and fruits, where their concentration may reach approx. 7% of the fresh mass, i.e. 400 mM [15]. Thus it is not surprising that this enzyme and the operon that controls the metabolism of these modified amino acids are found in bacteria associated with the gastrointestinal tract or gastrointestinal tract infections, such as the Gram-positive Enterococcus, and the Gram-negative E. coli, Salmonella and Shigella. The precise significance of these enzymes for bacterial growth will have to be investigated using knockout approaches, but the presence of five deglycase-containing operons in E. faecium suggests that the glycation products could be an important energy source for this bacterium.
By analogy with FN3K and FN3K-RP, whose phosphorylating action destabilizes the Amadori product, another speculative role for these enzymes is protein repair. In that regard, GfrF and GfrE are true deglycating enzymes, but it is yet unknown whether they are active against modified proteins. If so, an obvious useful function would be to deglycate modified lysine-rich histones to make these available for acetylation and the control of gene expression. However, the cell appears to have potent mechanisms to protect its histones against the effects of high glucose or fructose from glycation [16], and thus more work will be needed to understand the biological role of histone glycation in cellular function. At any rate, the discovery of enzymes that deglycate phosphorylated Amadori and Heyns products of amino acids opens up a new and fascinating field of investigation, for which Van Schaftingen and colleagues should be congratulated.
Acknowledgments
I thank Dr Francois Collard for helpful discussions.
References
- 1.Sell D. R., Biemel K. M., Reihl O., Lederer M. O., Strauch C. M., Monnier V. M. Glucosepane is a major protein cross-link of the senescent human extracellular matrix. Relationship with diabetes. J. Biol. Chem. 2005;280:12310–12315. doi: 10.1074/jbc.M500733200. [DOI] [PubMed] [Google Scholar]
- 2.Baynes J. W., Thorpe S. R. Immunohistochemical and ELISA assays for biomarkers of oxidative stress in aging and disease. Diabetes. 1999;48:1–9. doi: 10.1111/j.1749-6632.1998.tb09909.x. [DOI] [PubMed] [Google Scholar]
- 3.Horiuchi T., Kurokawa T., Saito N. Purification and properties of fructosyl-amino acid oxidase from Corynebacterium sp. 2-4-1. Agric. Biol. Chem. 1989;53:103–110. [Google Scholar]
- 4.Horiuchi T., Kurokawa T. Purification and properties of fructosylamine oxidase from Aspergillus sp. 1005. Agric. Biol. Chem. 1991;55:333–338. [Google Scholar]
- 5.Takahashi M., Pischetsrieder M., Monnier V. M. Molecular cloning and expression of amadoriase isoenzyme (fructosyl amine:oxygen oxidoreductase, EC 1.5.3) from Aspergillus fumigatus. J. Biol. Chem. 1997;272:12505–12507. doi: 10.1074/jbc.272.19.12505. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Takahashi M., Chen S. G., Monnier V. M. Cloning of amadoriase I isoenzyme from Aspergillus sp.: evidence of FAD covalently linked to Cys342. Biochemistry. 2000;39:1515–1521. doi: 10.1021/bi992031g. [DOI] [PubMed] [Google Scholar]
- 7.Delpierre G., Rider M. H., Collard F., Stroobant V., Vanstapel F., Santos H., Van Schaftingen E. Identification, cloning, and heterologous expression of a mammalian fructosamine-3-kinase. Diabetes. 2000;49:1627–1634. doi: 10.2337/diabetes.49.10.1627. [DOI] [PubMed] [Google Scholar]
- 8.Szwergold B. S., Kappler F., Brown T. R. Identification of fructose 3-phosphate in the lens of diabetic rats. Science. 1990;247:451–454. doi: 10.1126/science.2300805. [DOI] [PubMed] [Google Scholar]
- 9.Szwergold B. S., Taylor K., Lal S., Su B., Kappler F., Brown T. R. Identification of a novel protein kinase activity specific for Amadori adducts on glycated proteins. Diabetes. 1997;46(Suppl. 1):108A. [Google Scholar]
- 10.Collard F., Wiame E., Bergans N., Fortpied J., Vertommen D., Vanstapel F., Delpierre G., Van Schaftingen E. Fructosamine 3-kinase-related protein and deglycation in human erythrocytes. Biochem. J. 2004;382:137–143. doi: 10.1042/BJ20040307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortpied J., Gemayel R., Stroobant V., Van Schaftingen E. Plant ribulosamine/erythrulosamine 3-kinase, a putative protein-repair enzyme. Biochem. J. 2005;388:795–802. doi: 10.1042/BJ20041976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiame E., Delpierre G., Collard F., Van Schaftingen E. Identification of a pathway for the utilization of the Amadori product fructoselysine in Escherichia coli. J. Biol. Chem. 2002;277:42523–42529. doi: 10.1074/jbc.M200863200. [DOI] [PubMed] [Google Scholar]
- 13.Wiame E., Van Schaftingen E. Fructoselysine 3-epimerase, an enzyme involved in the metabolism of the unusual Amadori compound psicoselysine in Escherichia coli. Biochem. J. 2004;378:1047–1052. doi: 10.1042/BJ20031527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiame E., Lamosa P., Santos H., Van Schaftingen E. Identification of glucoselysine-6-phosphate deglycase, an enzyme involved in the metabolism of the fructation product glucoselysine. Biochem. J. 2005;392:263–269. doi: 10.1042/BJ20051183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shallenberger R. S. Occurence of various sugars in foods. In: Sipple H. L., McNuttt K. W., editors. Sugars in Nutrition. New York: Academic Press; 1974. pp. 67–80. [Google Scholar]
- 16.Talasz H., Wasserer S., Puschendorf B. Nonenzymatic glycation of histones in vitro and in vivo. J. Cell. Biochem. 2002;85:24–34. [PubMed] [Google Scholar]