Abstract
We previously found that EGF (epidermal growth factor) increases the EGFR (EGF receptor) kinase-binding affinity towards the major tyrosine phosphorylation sites in downstream adaptor proteins such as Gab1 (Grb2-associated binding protein 1) and Shc [Src homology 2 (SH2) domain and collagen containing protein], but not that towards EGFR autophosphorylation sites [Fan, Wong, Deb and Johnson (2004) J. Biol. Chem. 279, 38143–38150]. EGFR activation can also result in transphosphorylation of tyrosine resides in the C-terminal region of the related receptors ErbB2, ErbB3 and ErbB4 in heterodimers which are formed upon ligand stimulation. In the present study, we investigated the specificity of EGFR kinase by comparing the steady state kinetic parameters for peptides derived from all four ErbBs in the absence or presence of EGF. Our results demonstrated that (i) EGFR kinase can efficiently phosphorylate a broad range of diverse peptide sequences representing ErbB sites; (ii) certain ErbB2, ErbB3 and ErbB4 sites had higher specificity constants than any EGFR sequence and (iii) EGF stimulation consistently increases the kcat approx. 5-fold, but does not significantly alter the Km for any ErbB peptides. Furthermore, peptides containing lysine at position −2 or −3 N-terminal to the target tyrosine were found to be poor EGFR kinase substrates, and substitution of these lysines with glutamine decreased the Km and increased the kcat for these substrates. We conclude that EGFR kinase-mediated ErbB transphosphorylations are mostly controlled at the level of oligomerization, and not by a preference of the EGFR kinase for phosphorylation sites in any particular ErbB. The results also demonstrated that, unlike phosphorylation sites in select downstream targets, EGF does not regulate the recognition of phosphorylation sites in the C-terminal region of any of the ErbBs.
Keywords: autophosphorylation, epidermal growth factor receptor (EGFR), ErbB receptor, Michaelis–Menten, receptor tyrosine kinase (RTK) specificity, binding affinity
Abbreviations: EGF, epidermal growth factor; EGFR, EGF receptor; Gab1, Grb2-associated binding protein 1; MALDI–TOF, matrix-assisted laser desorption ionization–time-of-flight; PLC, phospholipase C; PI3K, phosphoinositide 3-kinase; RP-HPLC, reverse-phase HPLC; RTK, receptor tyrosine kinase; Shc, Src homology 2 (SH2) domain and collagen containing protein
INTRODUCTION
EGFR (epidermal-growth-factor receptor, ErbB1, HER1) is the prototypical member of the ErbB family of RTKs (receptor tyrosine kinases), which also includes ErbB2/HER2/neu, ErbB3/HER3 and ErbB4/HER4. ErbBs consist of an extracellular ligand-binding domain, a transmembrane spanning segment, an intracellular tyrosine kinase domain and a C-terminal region that contains multiple tyrosines. Among the ErbB receptor family, ErbB2 has no identified ligand and ErbB3 does not possess kinase activity. The ErbBs mediate signalling by a large number of growth factors that are structurally related to EGF such as transforming growth factor-α and amphiregulin. This family of receptor kinases function in the proliferation, migration, survival and differentiation of mammalian cells [1,2], and dysregulation of signalling by ErbBs has been implicated in cell transformation and cancer [3,4]. Engagement of the ligand induces the formation of receptor–receptor homo- and hetero-dimerization and activation of the intrinsic kinase. The heterodimer formation diversifies ErbB-mediated signalling, and almost every possible dimeric combination of ErbBs has been observed [5,6]. The kinase activation results in phosphorylation of specific tyrosine residues in their C-terminal tails (autophosphorylation), in other ErbBs and in cytosolic downstream targets such as Gab1 (Grb2-associated binding protein 1) [7], Shc [Src homology 2 (SH2) domain and collagen containing protein] [8], and PLCγ-1 (phospholipase Cγ-1) [9,10].
Receptor kinase specificity towards various targets is believed to be critical for selective EGFR cellular signalling. The use of synthetic peptides in detailed studies investigating protein kinase specificity and catalysis is widely recognized and supported by crystallographic observations that peptide bound to the active site appears to exist in an extended conformation [11,12]. By using a degenerate-peptide library it has been shown previously that each tyrosine kinase has its own optimal peptide substrate, demonstrating that specificity is predominantly based on the residues immediately surrounding the phosphoacceptor tyrosine. The identified optimal peptide for EGFR kinase was EEEEYFELV [13]. The validity of using synthetic peptides in detailed investigations of kinase specificity and catalysis has also been supported by our previous study, which examined the phosphorylation of specific sites in full-length proteins relative to phosphorylation of these sites in peptides derived from the proteins [14]. This work evaluated the effects of EGF on the steady state kinetic parameters of EGFR-catalysed phosphorylation of peptides derived from its C-terminal region and on cellular targets. We found that EGF-stimulation consistently increases kcat values for all tested peptides, but selectively increases the binding affinities for the major phosphorylation sites in Gab1 (Y627) and Shc (Y317) in peptides, and the full-length proteins. The results indicated that the ligand alters EGFR kinase specificity towards physiological targets [14].
Upon the formation of a heterodimer, EGFR kinase phosphorylates tyrosine sites in other ErbB partner in trans. Each ErbB receptor possesses a distinct pattern of tyrosine phosphorylation sites in its C-terminus. It has been demonstrated that phosphorylation of the ErbB receptors upon growth factor stimulation is dependent on the dimerization partner, and the differential receptor phosphorylation is suggested to be the basis for the differences in the cellular signalling elicited by homodimerization and heterodimerization [15]. To investigate EGFR kinase specificity for tyrosine sites from various ErbBs, and whether the relative specificity is altered upon EGF-stimulation, in the present study we have compared the phosphorylation of peptides derived from ErbBs by EGFR kinase. The results indicated that EGFR kinase efficiently phosphorylates ErbB peptides with a broad diversity of sequences. For all ErbB sites tested, EGF-stimulation increased the kcat approx. 5-fold without significant effects on the Km.
MATERIALS AND METHODS
Expression and purification of EGFR
Full-length human EGFR with a 25-amino-acid sequence containing Myc and hexahistidine epitope sequences at the C-terminus was expressed in a sTable 32D cell line and the tagged EGFR was purified by Co2+-based immobilized metal affinity chromatography as described previously [14]. Briefly, cells were grown in 2 l stirring flasks to a density of approx. 1.5×106 cells/ml, harvested by centrifugation at 2000 g and pellets were extracted for 20 min at 4 °C with lysis buffer [20 mM Hepes (pH 7.4), 1% Triton X-100, 10% glycerol, 200 mM NaCl, 5 mM β-glycerophosphate, 0.1 mM PMSF, 0.2 μg/ml pepstain A, 0.2 μg/ml aprotinin, 0.2 μg/ml leupeptin, 1 mM sodium orthovanadate and 5 mM imidazole]. After centrifugation at 15000 g for 40 min, the supernatant was incubated with Talon metal affinity resin (Clontech) at 4 °C for 1 h. EGFR was eluted with lysis buffer containing 50 mM imidazole. EGFR concentration was measured by silver staining and Western blotting of SDS/PAGE gels using BSA and Myc–GST (glutathione S-transferase)–Grb2 as standards respectively.
Preparation of peptides
Synthetic peptides were prepared by solid-phase synthesis using standard Fmoc (N-9-fluorenyl methoxycarbonyl) chemistry on an Applied Biosystems 432A peptide synthesizer and purified using preparative RP (reverse-phase)-HPLC. The identity and purity of peptides were confirmed by MALDI–TOF (matrix-assisted laser desorption ionization–time-of-flight)-MS and analytical RP-HPLC.
Tyrosine kinase assay
All kinase reactions were performed in 25 mM Hepes (pH 7.4), 10 mM MgCl2, 2.5 mM MnCl2, 50 μM sodium orthvandate, 0.5 mM dithiothreitol, 0.2% Triton X-100, 40 μg/ml BSA, 25 μM ATP with 62.5 μCi/ml [γ-32P]ATP (Amersham) and 9 nM EGFR in a total volume of 40 μl at 25 °C. EGFR was pre-incubated with or without 1 μM EGF for 10 min at 25 °C. Reactions were initiated by addition of peptide and were stopped using 8.5% H3PO4. Terminated reaction mixtures were transferred to P30 Filtermats (Wallac), washed 3× with 0.85% H3PO4, 3× with water and bound radioactivity was quantified with a Trilux Macrobeta scintillation counter (Perkin Elmer). Initial rates were determined from the linear portion of the reaction. Replicates (4) were measured in all experiments and kinetic parameters were calculated using the Enzyme Kinetic Module of Sigma Plot (Jandel Scientific).
RESULTS
Steady-state enzyme kinetic studies provide measurements of relative substrate-binding affinity (Km) and the catalytic-centre activity (kcat, ‘turnover number’). The combined parameter, kcat/Km, which is referred to as the specificity constant, is the best measurement for comparison of catalytic efficiencies towards different substrates. EGFR possesses three major and two minor tyrosine autophosphorylation sites located at Y1068, Y1148, Y1173, and at Y992 and Y1086 respectively. In addition, EGFR Y1114 is preceded by glutamic acid (Figure 1), which should be preferred by the EGFR kinase as indicated in previous work [16]. Our previous study demonstrated that human EGFR peptides representing these sites exhibit specificity constants >50 min−1·mM−1 in the presence of EGF, with the exception of the Y1086 peptide (12 min−1·mM−1). EGF activation does not alter the affinity binding of EGFR peptides to the kinase, but consistently increases the kcat and specificity constants approx. 5-fold for the EGFR peptides [14].
Figure 1. C-terminal sequence alignment of human ErbB family receptors.
ErbB amino acid residue numbers are presented on left. Identical residues are denoted by dots and gaps which were introduced are represented by hyphens. The major and minor phosphorylation of tyrosine residues in EGFR are marked by asterisks and triangles respectively, with position number above. The tyrosine residues are highlighted in bold and the surrounding sequences included in synthetic peptides containing conserved tyrosines or non-conserved phosphorylation sites are underlined with single or double lines respectively.
The C-terminal domain is the most variable region among the ErbB family receptors. As aligned in Figure 1, most of the identified phosphorylation tyrosines in this region are conserved in human EGFR, ErbB2 and ErbB4. However, the short sequences surrounding these tyrosines are not identical. The C-terminal domain of ErbB3 has much less consensus relative to other ErbB members and has only one tyrosine (Y1309) which is conserved relative to the other three ErbBs. In addition, Y1035 in ErbB3 is conserved in ErbB4, and Y1140 of ErbB3 is conserved in ErbB2 and ErbB4 (Figure 1).
To explore EGFR substrate selectivity towards identified or potential phosphorylation sites contained within the C-terminal regions of other human ErbBs, and whether ligand affects this selectivity, we synthesized and carried out phosphorylation kinetic analyses of the peptides representing these sites. The synthetic 20 mer peptides used in the present study contain sequences identical to 17 amino acids surrounding the phosphoacceptor tyrosine and were named by origin+tyrosine position (i.e. ErbB2-Y1023). The peptides also contained two or one lysine residues at the N- and C-termini respectively, allowing them to bind strongly to phosphocellulose filters upon acidification [17].
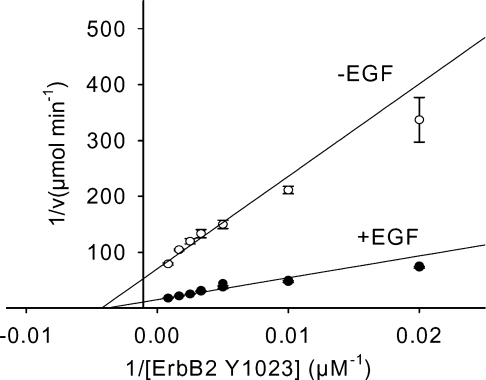
Although no ligand has been identified for ErbB2, it appears to be the preferred heterodimerization partner for all other ErbBs [18]. Five conserved tyrosines in ErbB2 compared with EGFR have been identified as autophosphorylation sites [19,20] and peptides were prepared representing these ErbB2 tyrosines. Additionally, the peptide corresponding to ErbB2-Y1127 was prepared and studied because this site is conserved in ErbB3 and ErbB4 (Figure 1). Figure 2 contains representative Lineweaver– Burk plots of ErbB2-Y1023 phosphorylation at a fixed ATP concentration of 25 μM in the presence or absence of EGF. The double reciprocal plots in the presence and absence of EGF intersected with the abscissa at the same point, indicating that EGF did not influence the Km. However, EGF increased the kcat value approx. 4.7-fold as evidenced by distinct intersections of the plots with ordinate. As summarized in Table 1, the ErbB2 peptides exhibited specificity constants ≥40 min−1·mM−1 in the presence of EGF with the exception of the ErbB2-Y1127 peptide (10.2 min−1·mM−1). Clearly peptides representing the five identified phosphorylation tyrosines were phosphorylated much more efficiently by the EGFR kinase than was the Y1127 peptide. Surprisingly, the peptide ErbB2-Y1221/Y1222 that contains tandem tyrosines, but does not have an acidic residue at position −1 N-terminal to the target tyrosine, possessed the highest specificity constant among the ErbB2 peptides (159 min−1·mM−1 in the presence of EGF). This specificity constant was at least 2-fold greater than that previously determined for any of the EGFR autophosphorylation site peptides [14]. EGF had no significant (<40%) effects on the Km values for any of the ErbB2 peptides, but enhanced the kcat values 3.8–4.8-fold.
Figure 2. Lineweaver–Burk plots of ErbB2-Y1023 peptide phosphorylation by EGFR.
Purified EGFR (7 nM) was pre-incubated with or without 1 μM EGF for 10 min at 25 °C followed by the addition of kinase reaction buffer containing 25 μM radiolabelled ATP. The reactions were initiated by the addition of various concentrations of peptide and terminated using 8.5% phosphoric acid. The results represent the means±S.E.M. for four replicates.
Table 1. Steady-state kinetic constants of EGFR tyrosine kinase with peptide substrates derived from ErbB2.
Results represent means±S.E.M.
| Km (μM) | kcat (min−1) | kcat/Km (min−1·mM−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| ErbB2 tyrosine position | Sequence | EGF… | − | + | − | + | − | + |
| 1023 | GDLVDAEEYLVPQQGFF | 215±28 | 228±22 | 2.5±0.1 | 11.7±0.2 | 11.7±1.7 | 52.2±5.0 | |
| 1127 | PLPSETDGYVAPLTCSP | 516±51 | 450±44 | 1.1±0.1 | 4.6±0.1 | 2.1±0.5 | 10.2±1.8 | |
| 1139 | LTCSPQPEYVNQPDVRP | 220±20 | 149±12 | 4.3±0.1 | 18.8±0.2 | 19.5±3.8 | 126±13 | |
| 1196 | GGAVENPEYLTPQGGAA | 280±31 | 168±13 | 2.1±0.1 | 8.4±0.1 | 7.6±1.0 | 49.8±5.1 | |
| 1221/1222 | SPAFDNLYYWDQDPPER | 140±14 | 104±11 | 4.4±0.1 | 16.5±0.1 | 31.8±3.7 | 159±18 | |
| 1248 | TPTVAENPEYLGLDVPV | 197±15 | 122±12 | 1.6±0.1 | 6.5±0.1 | 8.1±1.2 | 53.3±6.7 | |
Using an EGFR/ErbB3 chimaera, tyrosine binding sites in ErbB3 for the p85 subunit of PI3 kinase and Shc binding have been identified by phosphopeptide competition [21]. Whereas Y1039 of ErbB3 is responsible for Shc binding, 6 YXXM motifs are found to interact with the p85 subunit of PI3K. Of the ErbB3 peptides, three corresponding to Y1178/Y1180, Y1203/Y1205 and Y1241/Y1243 contain two tyrosine residues (Table 2). Peptides representing these sites were synthesized and their phosphorylation by EGFR kinase was analysed. ErbB3-Y1140 was also included because this tyrosine is conserved in ErbB2 and ErbB4. The results are summarized in Table 2. As was observed for ErbB2-Y1221/Y1222, the three ErbB3 peptides containing dual tyrosines (Y1178/Y1180, Y1203/Y1205 and Y1241/Y1243) were found to be highly preferred by the EGFR kinase with specificity constants >100 min−1·mM−1 in the presence of EGF. Peptides ErbB3-Y1140, -1257 and -1309 were also effectively phosphorylated by EGFR kinase with specificity constants ≥20 min−1·mM−1. ErbB3-Y1035 and -Y1270 exhibited relatively low specificity constants (<10 min−1·mM−1) probably due to the lack of acidic residue in position −1. By contrast, the ErbB3-Y1140 peptide does not contain an acidic residue at position −1, but had a high specificity constant of approx. 92 min−1·mM−1. As was found for EGFR peptides [14], EGF had little demonstrable effect on the Km values for ErbB3 peptides but consistently increased the kcat values approx. 5-fold.
Table 2. Steady-state kinetic constants of EGFR tyrosine kinase with peptide substrates derived from ErbB3.
Results represent means±S.E.M.
| Km (μM) | kcat (min−1) | kcat/Km (min−1·mM−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| ErbB3 tyrosine position | Sequence | EGF… | − | + | − | + | − | + |
| 1035 | SLLSPSSGYMPMNQGNL | 528±91 | 841±87 | 0.1±0.1 | 0.5±0.1 | 0.2±0.1 | 0.6±0.1 | |
| 1140 | LEEEDVNGYVMPDTHLK | 152±15 | 176±13 | 2.5±0.1 | 16.2±0.4 | 16.1±2.7 | 92.1±10 | |
| 1178/1180 | EEEDEDEEYEYMNRRRR | 129±10 | 114±8 | 3.0±0.1 | 14.5±0.3 | 23.1±2.6 | 128±12 | |
| 1203/1205 | PSSLEELGYEYMDVGSD | 111±11 | 153±10 | 3.1±0.1 | 15.8±0.3 | 28.3±3.5 | 103±9 | |
| 1241/1243 | AGTTPDEDYEYMNRQRD | 118±13 | 109±6 | 3.2±0.1 | 19.4±0.3 | 27.5±3.9 | 179±13 | |
| 1257 | DGGGPGGDYAAMGACPA | 343±29 | 460±20 | 1.4±0.1 | 9.2±0.4 | 4.1±0.7 | 20.0±1.8 | |
| 1270 | ACPASEQGYEEMRAFQG | 931±99 | 836±64 | 1.2±0.1 | 7.6±0.3 | 1.3±0.23 | 9.1±1.1 | |
| 1309 | DSAFDNPDYWHSRLFPK | 245±16 | 232±10 | 2.8±0.1 | 15.7±0.2 | 11.3±1.1 | 67.8±4.0 | |
ErbB4 displays the highest sequence homology with EGFR and has 6 conserved tyrosines (Figure 1). In addition, Y1056 of ErbB4 was identified as a binding site of PI3K [22]. Peptides representing these tyrosines were prepared and enzymatically analysed as EGFR kinase substrates. Peptides corresponding to Y1202 and Y1258 were also studied because of the presence of a −1 acidic residue. The results are summarized in Table 3. In general, results obtained for peptides derived from ErbB4 were similar to those of other ErbB peptides. Most of the peptides were efficiently phosphorylated by EGF-activated EGFR, whereas some, such as ErbB4-Y1056, -Y1162 and -Y1221, exhibited relatively low specificity constants (<20 min−1·mM−1). EGF did not significantly influence the Km values for ErbB4 peptides but consistently increased the kcat values approx. 5-fold.
Table 3. Steady-state kinetic constants of EGFR tyrosine kinase with peptide substrates derived from ErbB4.
Results represent means±S.E.M.
| Km (μM) | kcat (min−1) | kcat/Km (min−1·mM−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| ErbB3 tyrosine position | Sequence | EGF… | − | + | − | + | − | + |
| 1022 | EDMMDAEEYLVPQAFNI | 279±13 | 260±20 | 3.1±0.1 | 17.0±0.4 | 11.1±0.9 | 65.3±7.4 | |
| 1056 | IGHSPPPAYTPMSGNQF | 1072±120 | 1752±234 | 0.7±0.1 | 3.0±0.2 | 0.7±0.1 | 1.7±0.4 | |
| 1150 | RGELDEEGYMTPMRDKP | 865±44 | 579±32 | 4.1±0.1 | 15.5±0.4 | 4.7±0.4 | 26.8±2.3 | |
| 1162 | MRDKPKQEYLNPVEENP | 1100±154 | 1093±81 | 2.0±0.1 | 12.5±0.5 | 1.9±0.4 | 11.4±1.4 | |
| 1188 | LQALDNPEYHNASNGPP | 291±23 | 286±16 | 2.4±0.2 | 15.3±0.3 | 8.2±0.9 | 53.5±3.0 | |
| 1202 | ASNGPPKAEDEYVNEPL | 284±19 | 194±10 | 2.9±0.1 | 14.8±0.2 | 10.1±1.1 | 76.3±4.9 | |
| 1221 | ANTLGKAEYLKNNILSM | 1805±365 | 1567±198 | 1.4±0.2 | 8.1±0.3 | 0.8±0.4 | 5.2±0.7 | |
| 1242 | KKAFDNPDYWNHSLPPR | 276±18 | 172±16 | 4.4±0.2 | 21.1±0.6 | 15.9±2.0 | 122.7±3.4 | |
| 1258 | HSLPPRSTLQHPDYLQE | 377±19 | 271±14 | 3.5±0.2 | 19.5±0.3 | 9.3±0.1 | 72.0±2.8 | |
| 1284 | PIVAENPEYLSEFSLKP | 335±36 | 287±25 | 2.7±0.1 | 15.7±0.3 | 8.1±0.8 | 54.7±4.6 | |
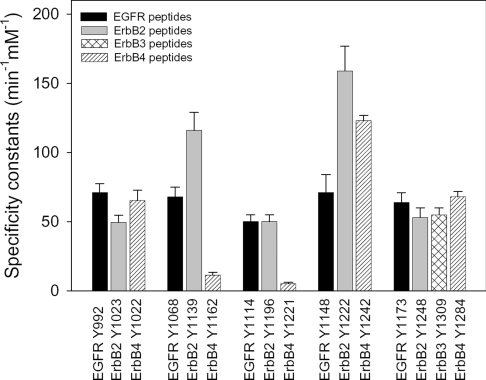
The specificity constants in the presence of EGF for peptides representing the conserved tyrosines in each of the ErbBs are compared in Figure 3. Peptides containing tyrosine conserved relative to EGFR-Y992 or -Y1173 have similar specificity constants, whereas ErbB2-Y1139, ErbB2-Y1221/Y1222 and ErbB4-Y1242 have higher specificity constants than their EGFR homologues, EGFR-Y1068 and -Y1148 respectively. ErbB4-Y1162 and -Y1221 peptides, which are conserved relative to EGFR-Y1068 and -Y1114 respectively, and contain glutamic acid residues at position −1, had relatively low specificity constants. This was mainly due to weak binding affinities for EGFR kinase (Km values of approx. 1 and 1.6 mM for ErbB4-Y1162 and -Y1221 respectively). Taken together, these results demonstrated that the EGFR kinase was incapable of efficiently phosphorylating certain tyrosine-containing peptides, but exhibited a broad catalytic specificity towards substrate primary sequences. In addition, certain ErbB2, ErbB3 and ErbB4 peptides had higher specificity constants than their EGFR counterparts.
Figure 3. Comparison of EGFR kinase specificity constants for peptide substrates derived from conserved regions of the ErbBs.
The specificity constants were determined in the presence of EGF. The values for EGFR peptides were taken from [14].
As described above, the relatively low specificity constants for the peptides corresponding to ErbB4-Y1162 and -Y1221 were unexpected considering the presence of a glutamic acid at position −1. This is similar to the scenario found for a peptide derived from the EGFR activation loop (Y845) in our previous work [14]. Even this target tyrosine is preceded by glutamate residues at positions −1, −3 and −4, which have been determined to be optimal for the EGFR substrate [13]. This peptide has a relatively low specificity constant (∼6 min−1·mM−1 in the presence of EGF) due to a high Km (∼1 mM) and a modest kcat (∼6 min−1) (Table 4). We proposed that the lysine residue in position −2 or −3 in these peptides may exert negative effects and be unfavourable. To test this hypothesis we replaced the lysine residue of EGFR-Y845 at position −2 and that of ErbB4-Y1221 at position −3 with a glutamine. Consistent with our proposal, the substitution in EGFR-Y845 peptide resulted in an approx. 4-fold reduction in the Km and an approx. 2-fold increase in the kcat, and the substitution in ErbB4-Y1221 decreased the Km approx. 3-fold and increased the kcat approx. 1.4-fold. Furthermore, the kcat values for both peptides containing lysine to glutamine substitutions were increased approx. 5-fold upon EGF stimulation without obvious change in the Km values, indicating that the lysine substitution did not alter the ligand regulation of the kinase activity towards these peptides.
Table 4. Effects of amino acid substitution on enzymatic parameters for EGFR-Y845 and ErbB4-Y1221 peptides.
The substituted residues are shown in italic bold. Results represent means±S.E.M.
| Km (μM) | kcat (min−1) | kcat/Km (min−1·mM−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| Peptide | Sequence | EGF… | − | + | − | + | − | + |
| EGFR-Y845 | LLGAEEKEYHAEGGKVP | 931±56 | 943±42 | 1.4±0.1 | 5.6±0.1 | 1.5±0.1 | 5.9±0.7 | |
| EGFR-Y845/K843Q | LLGAEEQEYHAEGGKVP | 266±18 | 213±10 | 2.1±0.1 | 11.2±0.2 | 7.1±0.9 | 52.6±3.6 | |
| ErbB4-Y1221 | ANTLGKAEYLKNNILSM | 1805±365 | 1567±198 | 1.4±0.2 | 8.1±0.3 | 0.8±0.4 | 5.2±0.7 | |
| ErbB4-Y1221/K1218Q | ANTLGQAEYLKNNILSM | 567±63 | 521±40 | 1.8±0.2 | 11.3±0.3 | 3.2±0.2 | 21.7±1.0 | |
DISCUSSION
We have previously compared the EGFR kinase-catalysed phosphorylation of peptides representing its autophosphorylation sites with that of peptides derived from other cellular substrates [14]. We found that EGF consistently enhances the kcat for all peptides approx. 5-fold, but selectively increases the binding affinity for peptides representing the major EGFR-mediated phosphorylation sites of adaptor proteins, such as Gab1 Y627 and Shc Y317, and the EGFR optimal peptide substrate which contains the EEEEYFELV sequence. In the present study, we investigated EGFR kinase-catalysed phosphorylation of peptides derived from ErbB2, ErbB3 and ErbB4 receptors. Our result indicated that the EGFR kinase was able to efficiently phosphorylate a broad range of diverse ErbB peptide sequences. EGF-stimulation resulted in approx. 5-fold increases in the kcat values with no obvious change in Km values. This is similar to the results found for EGFR peptides, but different from those for peptides derived from Gab1 and Shc [14]. It seems striking that of the many physiologically relevant substrate sequences tested to date, including all of the ErbBs, we have only observed ligand-induced increased binding affinity (i.e. recognition) in sequences derived from downstream cellular targets.
Whereas the EGFR kinase is able to efficiently phosphorylate peptide substrates with a broad diversity of sequences, the broad range of specificity constants indicates the preference of the kinase for certain peptides. Interestingly, the ErbB2-Y1139 and -Y1221/Y1222 peptides had higher specificity constants than their homologous counterparts in other ErbBs (Figure 3). This is consistent with the previous observation that EGFR kinase phosphorylates an EGFR/ErbB2 C-terminal domain chimaera to a higher level than EGFR autophosphorylation both in vivo and in vitro [23]. It has been recognized for a long time that ErbB2 is the preferred dimerization partner for the ErbB receptors (for reviews see [5,6]). Our findings in the present study suggest that ErbB2 possesses preferred phosphorylation target sites for EGFR kinase which may partially account for the enhanced transforming activity in cells coexpressing ErbB2 and EGFR. It would be of interest to determine whether these ErbB2 sites represent preferred substrates for the ErbB2 and ErbB4 kinases.
In previous investigations, Y1035 in ErbB3 and Y1056 in ErbB4 have been identified as PI3K binding sites via SH2 interactions and are important in signalling mediated by these receptors [21,22,24]. However, we found that peptides representing these sites had the lowest specificity constants among all the peptides tested with EGFR kinase. This suggests that these sites are not efficiently phosphorylated by EGFR in oligomeric complexes and that ErbB2 or ErbB4 kinases may mediate direct phosphorylation of the ErbB3 Y1035 or ErbB4 Y1056 sites. Another possibility is that these sites are phosphorylated by other cytosolic protein tyrosine kinases such as Src and Abl, which can be activated upon EGF stimulation. We believe that this latter possibility may be more plausible because the EGFR, ErbB2 and ErbB4 kinases have similar substrate preferences [25].
Relative selectivity for each specific position in peptide substrates has been assigned for many tyrosine kinases using a degenerate-peptide library [13]. We calculated an additive score for each peptide tested in our previous [14] and present studies using the published relative EGFR position selectivity. We found no correlation between these predicted peptide scores and either the kcat, Km or kcat/Km values (results not shown), suggesting a complicated synergy of amino acids flanking the target tyrosine with respect to peptide-binding and catalysis. The optimal EGFR peptide had a specificity constant of approx. 2500 min−1·mM−1 in the presence of EGF, which is much higher than those of all the physiologically relevant substrates tested, mainly due to its very low Km (Km of 5.6 μM in the presence of EGF) [14]. However, many peptides tested had significantly higher kcat values than the optimal EGFR peptide.
Early studies indicated that EGFR kinase prefers to phosphorylate substrates with an acidic residue at position −1 relative to the target tyrosine [16,26]. However, we found that some peptides such as EGFR-Y845, ErbB4-Y1162 and ErbB4-Y1221 contain a glutamic acid at position −1 but were relatively poor substrates for the EGFR kinase, mainly due to the low binding affinities (Tables 3 and 4). Our amino acid substitution experiments indicated that a lysine at position −2 or −3 exerted a dominant negative effect. On the other hand, a −1 acidic residue in the peptide was not a requisite for efficient phosphorylation by EGFR. Among the 6 peptides which lack a glutamic or aspartic acid at position −1, ErbB2-Y1221/Y1222 and ErbB3-Y1140 were efficient receptor kinase substrates (Tables 1 and 2). However, these peptides contain one or more acidic residues at positions −2, −3 or −4, which may play a role in EGFR recognition and binding of peptides in the absence of a −1 acidic side chain [27].
The broad specificity of EGFR kinase may be essential to the phosphorylation of diverse in vivo substrates and activation of multiple signalling pathways. This diversity has been exemplified by our finding that the EGFR kinase was able to efficiently phosphorylate a broad diversity of peptides representing ErbB phosphorylation sites. However, high fidelity signal transduction also requires phosphorylation of specific signalling proteins, implying that EGFR specificity should be controlled by other mechanisms in addition to the primary target sequence. Numerous studies have demonstrated that EGFR-mediated phosphorylation of ErbB receptors is dictated by dimerization partners, which in turn is determined by ligands [6]. Furthermore, for individual ErbB dimers, different ligands induce differential receptor phosphorylation patterns and therefore elicit differential cell signalling [28]. Among the ErbBs, only EGFR and ErbB4 have both ligands and kinase activity. Previous investigations have demonstrated that EGFR can be a dimerization partner of and be phosphorylated by ErbB4 upon neuregulin1-β stimulation. However, there is no evidence for EGF-stimulated EGFR/ErbB4 dimers or ErbB4 tyrosine phosphorylation [22,29]. Thus ErbB4 is unlikely to be phosphorylated by EGFR in cells even though several ErbB4 peptides are phosphorylated by EGFR efficiently in vitro. However, it is possible that other ErbB ligands, such as betacellulin, may induce EGFR kinase-mediated phosphorylation of ErbB4 [30,31]. Ligand binding and subsequent dimerization may induce conformational changes in the kinase domain. Slight conformational adjustments within the active site can result in dramatic changes in enzymatic activity and specificity of an enzyme [32,33]. In addition, it has been suggested that secondary and tertiary structures are important for target recognition [34]. We previously found that EGFR-Y1114 peptide was phosphorylated efficiently by EGFR with a specificity constant of ∼50 min−1·mM−1 [14], but this site in the intact EGFR was not found to be phosphorylated either in autophosphorylation reactions in vitro or in cells after EGF stimulation [35]. Access of these tyrosine residues to the kinase catalytic site may be prohibited by the secondary or tertiary structure. Taken together, we conclude that whereas the primary sequences near the target tyrosine residues play an important role in receptor kinase specificity, but EGFR kinase-mediated ErbB transphosphorylation is mostly controlled at the level of oligomerization.
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Jorissen R. N., Walker F., Pouliot N., Garrett T. P., Ward C. W., Burgess A. W. Epidermal growth factor receptor: mechanisms of activation and signaling. Exp. Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 3.Hynes N. E., Lane H. A. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer. 2001;37:S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 5.Riese D. J., II, Stern D. F. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays. 1998;20:41–48. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 6.Olayioye M. A., Neve R. M., Lane H. A., Hynes N. E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holgado-Madruga M., Emlet D. R., Moscatello D. K., Godwin A. K., Wong A. J. A Grb2-associated docking protein in EGF- and insulin-receptor signalling. Nature (London) 1996;379:560–564. doi: 10.1038/379560a0. [DOI] [PubMed] [Google Scholar]
- 8.Pelicci G., Lanfrancone L., Grignani F., McGlade J., Cavallo F., Forni G., Nicoletti I., Pawson T., Pelicci P. G. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter G. Receptor tyrosine kinase substrates: src homology domains and signal transduction. FASEB J. 1992;6:3283–3289. doi: 10.1096/fasebj.6.14.1385243. [DOI] [PubMed] [Google Scholar]
- 10.Kim J., Sim S., Kim U., Nishibe S., Wahl M., Carpenter G., Rhee S. Tyrosine residues in bovine phospholipase C-gamma phosphorylated by the epidermal growth factor receptor in vitro. J. Biol. Chem. 1990;265:3940–3943. [PubMed] [Google Scholar]
- 11.Hubbard S. R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science (Washington, D.C.) 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 13.Songyang Z., Carraway K. L., 3rd, Eck M. J., Harrison S. C., Feldman R. A., Mohammadi M., Schlessinger J., Hubbard S. R., Smith D. P., Eng C., et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature (London) 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 14.Fan Y. X., Wong L., Deb T. B., Johnson G. R. Ligand regulates epidermal growth factor receptor kinase specificity: activation increases preference for GAB1 and SHC versus autophosphorylation sites. J. Biol. Chem. 2004;279:38143–38150. doi: 10.1074/jbc.M405760200. [DOI] [PubMed] [Google Scholar]
- 15.Olayioye M. A., Graus-Porta D., Beerli R. R., Rohrer J., Gay B., Hynes N. E. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol. Cell. Biol. 1998;18:5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter T., Cooper J. A. Protein-tyrosine kinases. Annu. Rev. Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 17.Casnellie J. E., Harrison M. L., Pike L. J., Hellstrom K. E., Krebs E. G. Phosphorylation of synthetic peptides by a tyrosine protein kinase from the particulate fraction of a lymphoma cell line. Proc. Natl. Acad. Sci. U.S.A. 1982;79:282–286. doi: 10.1073/pnas.79.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graus-Porta D., Beerli R. R., Daly J. M., Hynes N. E. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazan R., Margolis B., Dombalagian M., Ullrich A., Zilberstein A., Schlessinger J. Identification of autophosphorylation sites of HER2/neu. Cell Growth Differ. 1990;1:3–7. [PubMed] [Google Scholar]
- 20.Segatto O., Lonardo F., Pierce J. H., Bottaro D. P., Di Fiore P. P. The role of autophosphorylation in modulation of erbB-2 transforming function. New Biol. 1990;2:187–195. [PubMed] [Google Scholar]
- 21.Prigent S., Gullick W. Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 1994;13:2831–2841. doi: 10.1002/j.1460-2075.1994.tb06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen B. D., Green J. M., Foy L., Fell H. P. HER4-mediated biological and biochemical properties in NIH 3T3 cells. Evidence for HER1-HER4 heterodimers. J. Biol. Chem. 1996;271:4813–4818. doi: 10.1074/jbc.271.9.4813. [DOI] [PubMed] [Google Scholar]
- 23.Di Fiore P. P., Segatto O., Lonardo F., Fazioli F., Pierce J. H., Aaronson S. A. The carboxy-terminal domains of erbB-2 and epidermal growth factor receptor exert different regulatory effects on intrinsic receptor tyrosine kinase function and transforming activity. Mol. Cell. Biol. 1990;10:2749–2756. doi: 10.1128/mcb.10.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elenius K., Choi C. J., Paul S., Santiestevan E., Nishi E., Klagsbrun M. Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidylinositol 3-kinase. Oncogene. 1999;18:2607–2615. doi: 10.1038/sj.onc.1202612. [DOI] [PubMed] [Google Scholar]
- 25.Brignola P. S., Lackey K., Kadwell S. H., Hoffman C., Horne E., Carter H. L., Stuart J. D., Blackburn K., Moyer M. B., Alligood K. J., Knight W. B., Wood E. R. Comparison of the biochemical and kinetic properties of the type 1 receptor tyrosine kinase intracellular domains. Demonstration of differential sensitivity to kinase inhibitors. J. Biol. Chem. 2002;277:1576–1585. doi: 10.1074/jbc.M105907200. [DOI] [PubMed] [Google Scholar]
- 26.Patschinsky T., Hunter T., Esch F. S., Cooper J. A., Sefton B. M. Analysis of the sequence of amino acids surrounding sites of tyrosine phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 1982;79:973–977. doi: 10.1073/pnas.79.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knighton D. R., Cadena D. L., Zheng J., Ten Eyck L. F., Taylor S. S., Sowadski J. M., Gill G. N. Structural features that specify tyrosine kinase activity deduced from homology modeling of the epidermal growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5001–5005. doi: 10.1073/pnas.90.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney C., Carraway K. L., III Ligand discrimination by ErbB receptors: differential signaling through differential phosphorylation site usage. Oncogene. 2000;19:5568–5573. doi: 10.1038/sj.onc.1203913. [DOI] [PubMed] [Google Scholar]
- 29.Tzahar E., Waterman H., Chen X., Levkowitz G., Karunagaran D., Lavi S., Ratzkin B. J., Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beerli R. R., Hynes N. E. Epidermal growth factor-related peptides activate distinct subsets of ErbB receptors and differ in their biological activities. J. Biol. Chem. 1996;271:6071–6076. doi: 10.1074/jbc.271.11.6071. [DOI] [PubMed] [Google Scholar]
- 31.Riese D. J., II, Bermingham Y., van Raaij T. M., Buckley S., Plowman G. D., Stern D. F. Betacellulin activates the epidermal growth factor receptor and erbB-4, and induces cellular response patterns distinct from those stimulated by epidermal growth factor or neuregulin-β. Oncogene. 1996;12:345–353. [PubMed] [Google Scholar]
- 32.Fan Y. X., Ju M., Zhou J. M., Tsou C. L. Activation of chicken liver dihydrofolate reductase by urea and guanidine hydrochloride is accompanied by conformational change at the active site. Biochem. J. 1996;315:97–99. doi: 10.1042/bj3150097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y. X., McPhie P., Miles E. W. Guanidine hydrochloride exerts dual effects on the tryptophan synthase α2β2 complex as a cation activator and as a modulator of the active site conformation. Biochemistry. 1999;38:7881–7890. doi: 10.1021/bi990307e. [DOI] [PubMed] [Google Scholar]
- 34.Geahlen R. L., Harrison M. L. Protein tyrosine kinases. In: Kemp B. E., editor. Peptides and Proteins Phosphorylation. Boca Raton, FL, U.S.A.: CRC; 1990. pp. 239–253. [Google Scholar]
- 35.Margolis B. L., Lax I., Kris R., Dombalagian M., Honegger A. M., Howk R., Givol D., Ullrich A., Schlessinger J. All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. Identification of a novel site in EGF receptor. J. Biol. Chem. 1989;264:10667–10671. [PubMed] [Google Scholar]