Abstract
IP3K (inositol 1,4,5-trisphosphate 3-kinase) catalyses the Ca2+-regulated phosphorylation of the second messenger Ins(1,4,5)P3, thereby inactivating the signal to release Ca2+ and generating Ins(1,3,4,5)P4. Here we have investigated the localization and activity of IP3KB and its modulation by proteolysis. We found that the N- and C-termini (either side of residue 262) of IP3KB localized predominantly to the actin cytoskeleton and ER (endoplasmic reticulum) respectively, both in COS-7 cells and in primary astrocytes. The functional relevance of this was demonstrated by showing that full-length (actin-localized) IP3KB abolished the histamine-induced Ca2+ response in HeLa cells more effectively than truncated constructs localized to the ER or cytosol. The superior efficacy of full-length IP3KB was also attenuated by disruption of the actin cytoskeleton. By transfecting COS-7 cells with double-tagged IP3KB, we show that the translocation from actin to ER may be a physiologically regulated process caused by Ca2+-modulated constitutive proteolysis in intact cells.
Keywords: calcium (Ca2+); cytoskeleton; endoplasmic reticulum (ER); inositol 1,4,5-trisphosphate 3-kinases A and B (IP3KA and IP3KB); proteolysis; subcellular localization
Abbreviations: AFF, 400 mM ammonium formate+0.1 M formic acid; CaM, calmodulin; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; (e)GFP, (enhanced) green fluorescent protein; ER, endoplasmic reticulum; Fura Red™ AM, Fura Red™ acetoxymethyl ester; HIFBS, heat-inactivated fetal-bovine serum; IP3K, inositol 1,4,5-trisphosphate 3-kinase; mRFP1, monomeric red fluorescent protein; PLC, phospholipase C
INTRODUCTION
Receptor-mediated activation of PtdIns-specific PLC (phospholipase C) results in the hydrolysis of PtdIns(4,5)P2 to liberate membrane-bound diacylglycerol and soluble Ins(1,4,5)P3 [1]. Ins(1,4,5)P3 mediates many cellular functions by releasing Ca2+ from internal Ca2+ pools through binding to its protein receptor [2]. It is inactivated either by 5-phosphatases to produce Ins(1,4)P2 [3] or by phosphorylation by IP3K (inositol 1,4,5-trisphosphate 3-kinase) to generate the putative signalling molecule Ins(1,3,4,5)P4 [4]. Three isoforms of IP3KB occur in mammals, which are distinguished by different molecular masses, sensitivity to Ca2+/CaM (calmodulin), intracellular distribution and tissue expression [4–7]. IP3KA is abundant in neuronal cells and associated with the filamentous actin (F-actin) cytoskeleton in dendritic spines [8]. Its involvement in brain function is reinforced by the observation that long-term potentiation is altered in IP3KA-deficient mice [9]. IP3KB is more widely expressed, and recent studies with knock-out mice suggest that IP3KB and Ins(1,3,4,5)P4 play an essential role in T-cell precursor responsiveness and development [10,11]. IP3KB is also highly expressed in the brain, but whereas the IP3KA is neuronal-specific, the B isoform is specific to glial cells [12].
Recently, the localization of IP3KB has been studied in more detail [13], including the identification of a binding domain for F-actin [14] and the determination of putative proteolytic cleavage sites [15]. With regard to the subcellular location, early expression studies using an N-terminally truncated enzyme showed IP3KB to be associated with the ER (endoplasmic reticulum) membrane [16], whereas more recent studies have suggested an association with F-actin on the plasma membrane [14] or a mixed localization [15,17]. Calpain, a Ca2+-dependent protease, has been proposed to be responsible for the proteolytic processing of IP3KB [15]. However, whether this processing occurs in intact cells, and the consequences for localization or function, have not been determined.
Here we have investigated the cellular control of the IP3KB localization and its functional consequences. We provide evidence for the limited proteolysis of IP3KB in cells, which produces domains with unique subcellular localizations, and we demonstrate an influence of the changed localization on the Ca2+ response induced by the generation of Ins(1,4,5)P3.
EXPERIMENTAL
cDNA constructs
The cDNA for full-length or truncated forms of rat IP3KB was amplified from 0.1 μg of the cDNA clone, which was a gift from Professor George Banting (Department of Biochemistry, School of Medical Sciences, University of Bristol, Bristol, U.K.). This cDNA contained a single base-pair mutation (T220C) in the open reading frame, which was corrected using the Quikchange Kit (Stratagene) with the following primers: sense primer, 5′-GGAGTGCTGGGGGTTGGCGCAGCGGC-3′, and antisense primer, 5′-GCCGCTGCGCCAACCCCCAGCACTCC-3′. The cloning was performed using PCR. The reaction was initiated by the addition of the Pfu polymerase and allowed to proceed for 35 cycles with cycling segments of 30 s/95 °C, 1 min/60 °C and 3.5 min/72 °C. The presence of DMSO (10%, v/v) in the PCR mixture was required to generate amplified DNA of the correct size. The cDNA for mRFP1 (monomeric red fluorescent protein) was a gift from Dr Roger Y. Tsien (Departments of Pharmacology and Chemistry and Biochemistry, University of California at San Diego, San Diego, CA, U.S.A.) [18]. The cDNA clone was amplified and digested with XhoI and ApaI restriction enzymes before subcloning into pCMV2A vector (Stratagene). A linker sequence coding for two glycine residues was added to the upstream primer in order to increase the flexibility of the mRFP1 protein. All PCR products were digested with appropriate restriction endonucleases, purified, and cloned into pEGFPN1 or C1 expression vectors (Clontech). All PCR experiments used the Pfu polymerase (Stratagene), and all constructs were verified by restriction-endonuclease mapping followed by DNA sequencing.
Cell culture and transfection
Both COS-7 and HeLa cells were cultured routinely in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (w/v) HIFBS (heat-inactivated fetal-bovine serum), 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were maintained at 37 °C under a humidified atmosphere containing 5% CO2. COS-7 cells were transfected with various plasmids using FuGENE 6 (Roche) according to the protocol supplied by the manufacturer. HeLa cells were transfected using the standard calcium phosphate precipitation method [8]. Control cells were transfected with the empty expression vector only.
Primary cultures of astrocytes were derived from neonatal (0–24 h old) rat cerebral cortex. Briefly, cortices were removed, washed and manually dissociated after trypsinization. The cells were then grown in plastic flasks and maintained at 37 °C under a humidified atmosphere supplied with 5% CO2 until reaching confluence. The flasks were shaken to remove microglia, and the culture medium was discarded and replaced with fresh medium. After 2 h in the incubator at 37 °C, the flask was again shaken vigorously overnight. The type 1 astrocytes, which remain firmly attached to the culture flasks, were removed by treatment with trypsin/EDTA, centrifuged, and resuspended in culture medium. The astrocytes were then grown in DMEM supplemented with 10% HIFBS, 100 units/ml penicillin and 100 μg/ml streptomycin. Culture medium was replaced every 3 or 4 days. Astrocytes were seeded on poly-D-lysine-coated glass coverslips 24 h before the transfection using calcium phosphate precipitation. Cell confluency, amount of DNA and incubation time were optimized such that transfection efficiencies of 50 and 10% were obtained from cell lines and primary culture respectively.
Immunostaining and confocal microscopy
Cells grown on glass coverslips were rinsed three times with PBS and then fixed with 4% formaldehyde/0.1 M sodium phosphate, pH 7.4, for 20 min at room temperature. Cells were permeabilized in 0.1% Triton X-100 for 5 min, washed briefly, and blocked for 1 h in 2% (v/v) normal donkey serum/2% (w/v) fish skin gelatin (Sigma). Coverslips were then incubated with primary antibodies for 1 h, washed three times in PBS and then incubated with appropriate fluorophore-Alexa-conjugated secondary antibodies (Molecular Probes) for 30 min. After three PBS washes, the coverslips were mounted using ProLong antifade reagent (Molecular Probes). Cells incubated with secondary antibodies only served as controls for image analysis (results not shown). Confocal microscopy was performed using a Zeiss LSM 510 microscope using a ×63 Plan-Apochromatic oil-immersion objective. The images were processed with Adobe Photoshop software. Polyclonal antibody recognizing the ER marker calreticulin was obtained from Calbiochem. Simultaneous quantitative co-localization analysis was performed using the analysis provided by Zeiss LSM software (Version 3.2). This function presents a comparison of the two colours, pixel by pixel, such that two identical images would produce a clean diagonal line running from bottom left to top right with R2 (correlation coefficient)=1. Any differences in localization between the colours causes spots deviating from this line, so that if the two colours had no co-localization, a value of R2=0 would result. A statistical analysis in the program gives an estimate of the confidence of the R2 value. See Figure 6(B) below for illustrations of the data obtained.
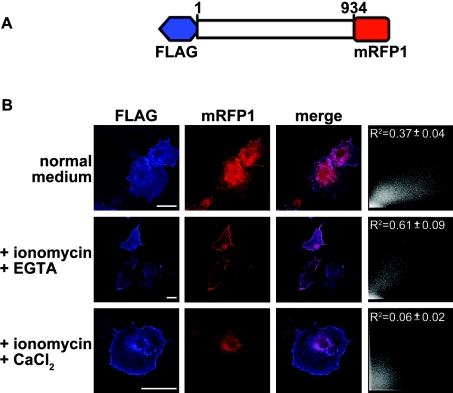
Figure 6. Differential association of IP3KB with different compartments is affected by the intracellular Ca2+ level.
(A) Schematic representation of IP3KB construct in modified pCMV2A vector. The red box represents mRFP1 and blue box represents FLAG tag. (B) COS-7 cells expressing mRFP1–IP3KB1–934–FLAG were loaded for 24 h with 1 μM ionomycin and EGTA (500 μM) or CaCl2 (10 mM). Cells were then fixed, permeabilized and stained with anti-FLAG M2 antibody, followed by fluorophore-Cy5-conjugated secondary antibody. Cells were observed by confocal immunofluorescence microscopy and only representative cells are shown. Co-localization analysis of FLAG and mRFP1 expression was performed as described in the Experimental section, and representative scatter diagrams of the images shown are shown on the right of each set. Results from three separate experiments (n>35 cells) are expressed as means±S.E.M. The scale bar represents 10 μm.
IP3K assays
IP3K assays were performed as described in [19], using 20000 d.p.m. of [3H]Ins(1,4,5)P3. The eGFP (enhanced green fluorescent protein)-tagged protein was immunoprecipitated from transfected COS-7 cells and re-suspended with 2×assay buffer [100 mM Hepes/NaOH adjusted to pH 7.5, with 200 mM KCl, 10 mM MgCl2, 2 mM EGTA and 12 mM DTT (dithiothreitol)]. Incubations were carried out at 37 °C for 20 min in the presence of 1 mM NaATP, and stopped by the addition of AFF (400 mM ammonium formate+0.1 M formic acid). Samples were loaded on to AG 1-X8 Dowex (Bio-Rad) anion-exchange columns that were then washed twice with 10 ml of 400 mM AFF, once with 10 ml of 800 mM AFF, and residual radiolabelled total Ins(1,3,4,5)P4 was eluted with 10 ml of 1.2 M AFF. Ins(1,3,4,5)P4 accumulation was then quantified by liquid-scintillation counting.
Ca2+ imaging
HeLa cells grown on 25-mm-diameter glass coverslips were incubated with the indicator Fura Red™ AM (Fura Red™ acetoxymethyl ester; 2.5 μM; Molecular Probes) for at least 20 min at 37 °C in DMEM as described previously [20,21]. Cells were then placed on a 37 °C heated microscope stage for imaging of the intracellular Ca2+ response. The extracellular solution for these experiments was imaging buffer (121 mM NaCl/5.4 mM KCl/0.8 mM MgCl2/1.8 mM CaCl2/6 mM NaHCO3/5.5 mM glucose/25 mM Hepes, pH 7.3, at 37 °C). Florescence images were obtained using a Zeiss 510 confocal microscope using a ×20 Plan-Apochromatic objective. Data were collected at a rate of 2 Hz before during and after the bath application of 100 μM histamine. Parallel experiments, comparing directly different IP3KB constructs and their ability to alter Ca2+ signals, were performed to ensure that different levels of expression was not a contributory factor to any differences seen. In experiments designed to stabilize the actin cytoskeleton structure, the cells were pretreated with 1 μM jasplakinolide (Molecular Probes), a fungal toxin that stabilizes F-actin [25], for 30 min. Alternatively, to collapse actin filaments, the cells were treated with 1 μM cytochalasin D (Sigma) for 10 min.
Immunoprecipitation and Western blotting
COS-7 cells were lysed on ice in RIPA buffer (50 mM Tris/HCl/0.1% SDS/2 mM DTT/0.5% Nonidet P40) containing 1 mM PMSF, 40 μg/ml aprotinin and 20 μg/ml pepstatin A. Cells were homogenized by passing them through 0.2-mm-diameter needles. The homogenate was centrifuged at 15000 g for 20 min and the resulting supernatants were incubated with monoclonal anti-GFP antibody (Clontech) for 2 h at 4 °C. The antibody–protein complexes were adsorbed on to Protein G–agarose beads (Amersham Biosciences) and the immunoprecipitated protein or total cell lysates were resolved on SDS/10%-(w/v)-PAGE gels and then transferred to nitrocellulose membrane. Blocking and antibody dilution were in 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20. All Western blots were carried out with a polyclonal anti-GFP antibody (ab290; Abcam) or monoclonal anti-actin antibody (clone AC-74; Sigma). Immunoreactive bands were detected using an enhanced-chemiluminescence kit (Pierce) and visualized on film. For examination of proteins bound to the cytoskeleton, we modified a published protocol [8] to extract soluble proteins and preserve the cytoskeleton. At 48 h after transfection, COS-7 cells plated on to poly-D-lysine-coated coverslips were washed once in PBS and incubated for 5 min at room temperature in 80 mM Pipes/KOH/1 mM MgCl2/1 mM EGTA containing 1% Triton X-100. Cells were washed once in the same buffer without Triton X-100, followed by whole-cell lysis and processing for Western blotting. Densitometric analysis was performed using the integrated density analysis provided by the Adobe Photoshop software.
Data analysis
To calculate the normalized fluorescence intensity (F/F0) of the cytosolic region, the average fluorescence intensity (F) for all pixels in the selected region and F0, the initial value, were used. Data are presented as means±S.E.M. For simple statistical comparisons, such as treatment compared with control, ANOVA was carried out to determine the significance, followed by Dunnett's t test. A P value of less than 0.01 was considered significant.
RESULTS AND DISCUSSION
Intracellular localization of IP3KB
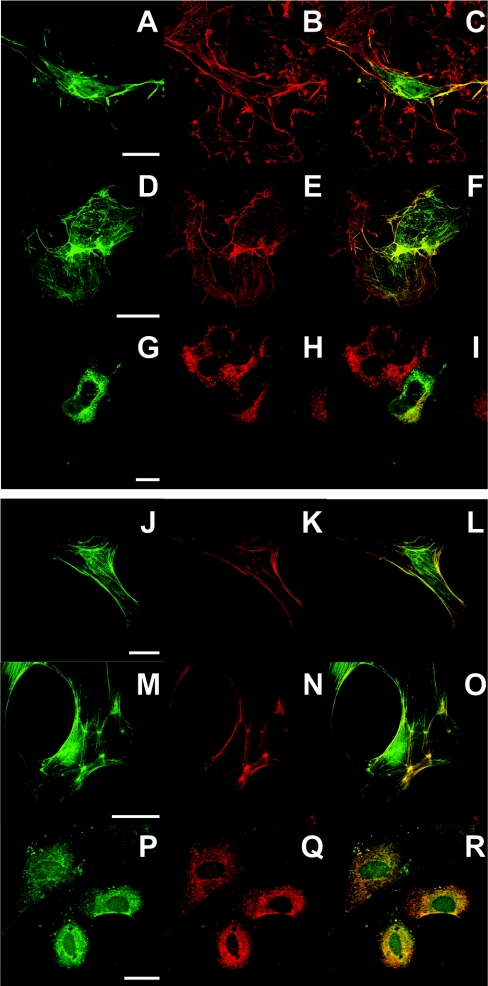
In these experiments, different IP3KB expression plasmids containing eGFP tags (Figure 1) were transfected into cells prior to fixation and immunostaining. We used two different cell types [COS-7 (Figures 2A–2I) and primary rat astrocytes (Figures 2J–2R)] to control for possible cell-type-specific localizations of IP3KB. Both cell types showed similar distributions, but the results from astrocytes in general showed clearer distinctions between the localizations. The localization of full-length IP3KB (IP3KB1–934) to different cellular compartments is shown in Figures 2(A)–2(C) and Figures 2(J)–2(L). Three localizations could be seen, namely cytoskeleton, plasma membrane and cytoplasm (which is mostly ER; see below). It should be noted here that the tendency of some IP3KB constructs to localize apparently near the plasma membrane is probably due to a binding to cortical F-actin [6], and for all subsequent discussion we focus on the binding or non-binding of the enzyme to F-actin.
Figure 1. Schematic representation of the IP3KB domain structure and the constructs used in the present study.
Numbers represent the amino acid positions in rat IP3KB. CAMKII, Ca2+/calmodulin-dependent protein kinase II.
Figure 2. Intracellular localization of full-length and truncated forms of IP3KB.
COS-7 cells (A–I) or primary cultured astrocytes (J–R) overexpressing full-length (IP3KB1–934; A–C, J–L), C-terminally truncated (IP3KB1–261; D–F, M–O) or N-terminally truncated (IP3KB262–934; G–I, P–R) were fixed and labelled as described in the Experimental section. Cells transfected with the various eGFP constructs were then labelled either for F-actin using Alexa 568/phalloidin (B, E, K and N) or for ER using anti-calreticulin followed by Alexa 568-labelled secondary antibody (H and Q). Cells were observed by confocal immunofluorescence microscopy. Co-localization of the two labels is shown in merged images on the right of each set. The scale bar represents 10 μm.
To investigate the roles of different structural domains of IP3KB in determining subcellular localization, we transfected cells with various truncated forms of IP3KB. In contrast with the full-length IP3KB, which localizes to several compartments within the cells (Figures 2C and 2L), the N-terminal domain, corresponding to amino acids 1–261 (IP3KB1–261), was concentrated only with F-actin (Figures 2F and 2O). These findings are consistent with data reported by other groups [14,17], and Dewaste et al. [17] have localized the probable F-actin-binding region to amino acids 107–180. On the other hand, the remaining C-terminal portion of the enzyme, corresponding to amino acids 262–934 (IP3KB262–934), accumulated in the perinuclear region of the cell, indicative of ER localization (Figures 2G and 2P). This was confirmed by its co-localization with the ER marker calreticulin (Figures 2I and 2R) and is in agreement with a previous study showing the association of a truncated version of IP3KB with the cytosolic face of the extended ER network [16].
More truncated C-terminal constructs (IP3KB399–934 or IP3KB633–934) did not show ER localizations, but rather were largely cytosolic (results not shown), suggesting that the domain that determines ER localization of the protein requires the central or entire C-terminal region of the protein (see also [15]).
Enzymatic activity of IP3KB constructs in vitro
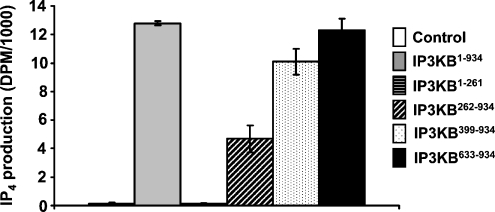
After determining the intracellular localizations of IP3KB constructs, we were next interested in exploring the consequences of the truncations for enzymatic activity. For this, we generated eGFP-fusion proteins containing different fragments of IP3KB attached to eGFP (Figure 1) and immunoprecipitated them from transfected COS-7 cells. After measuring the GFP signal on Western blots to calibrate uniform amounts of enzyme, we directly compared the ability of the full-length IP3KB and its fragments to phosphorylate Ins(1,4,5)P3. We found that all of the constructs that contain the catalytic region (Figure 1) were active (Figure 3). Surprisingly, we found that the fragment (IP3KB262–934), which co-localized to the ER region (Figure 2), showed less kinase activity than both longer or shorter fragments (Figure 3).
Figure 3. Effect of truncation on IP3K activity.
Expression plasmids for full-length IP3K or its peptide fragments were transfected into COS-7 cells lysed, immunoprecipitated with anti-GFP antibody and assayed for IP3K activity as described in the Experimental section. Representative results are shown (n≥4).
Effects of over-expressed IP3KB constructs on Ca2+ mobilization in intact cells
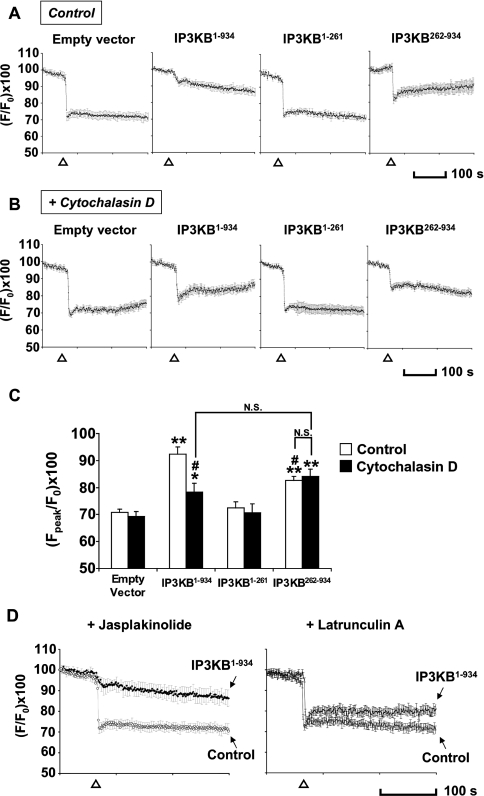
The agonist histamine, when applied to HeLa cells, results in the production of intracellular Ins(1,4,5)P3, which in turn stimulates the release of Ca2+ from the ER [22,23]. We evaluated the efficacy of IP3KB enzyme activity in reducing the Ca2+ response in intact cells using the Ca2+ indicator Fura Red™, following a protocol similar in principle to that described by Dewaste et al. [17]. Histamine (100 μM) induced an approx. 30% decrease in normalized Fura Red fluorescence (indicating an increase in cytosolic calcium) in HeLa cells transfected with empty vector only (n=21 cells; Figure 4A, empty vector) or non-transfected cells (Figure 4D, control). This response was almost completely blocked in cells overexpressing the full-length IP3KB (IP3KB1–934). These results indicate that IP3KB attenuates the Ca2+ response induced by histamine, consistent with previous reports [24]. We then evaluated the effect of the different fragments of IP3KB on the histamine-induced Ca2+ response. As expected, the N-terminal fragment of IP3KB (IP3KB1–261), which lacks of the catalytic region of the protein, had no effect (Figure 4A, third panel). The C-terminal fragment (IP3KB262–934), which localizes primarily to the ER (Figure 2), had a much decreased effect compared with the full-length construct [Figure 4A (fourth panel) and Figure 4C]. This implies that the localization of the enzyme to closer to the site of Ins(1,4,5)P3 generation significantly increases its efficacy in intact cells.
Figure 4. Cytosolic Ca2+ levels in HeLa cells monitored in Fura-Red-loaded cells.
(A) Cells expressing empty vector (n=21), full-length (IP3KB1–934; n=19), N- or C-terminal domains (IP3KB1–261, n=28; IP3KB262–934, n=24 respectively) of IP3KB were incubated with 2.5 μM Fura Red™ before Ca2+ imaging. A decrease in Fura Red™ fluorescence indicates an increase in intracellular Ca2+ concentration. The application of histamine (100 μM; Δ) resulted in a sustained Ca2+ signal in control cells. Full-length and C-terminal domain blocked histamine-induced Ca2+ response completely and partially respectively. (B) Cytochalasin D (1 μM), when added to the medium before imaging, compromised the ability of transfected full-length IP3KB to block the Ca2+ response (n=24), but not other constructs (empty vector, n=21; IP3KB1–261, n=23; IP3KB262–934, n=19). (C) Data are expressed as means±S.E.M. for the ‘peak’ of Fura Red™ decrease. *P<0.01, **P<0.001 versus ‘empty vector’; #P<0.001 versus ‘IP3KB1–934 Control’; N.S., not significant. (D) Jasplakinolide (1 μM), when added to the medium before imaging, showed no effect on the Ca2+ signals in transfected (n=17) or non-transfected cells (n=25), whereas latrunculin A (5 μM) showed a similar effect to cytochalasin D (n=17).
This latter conclusion is subject to the caveat that the IP3KB262–934 construct has a lower catalytic activity in vitro (Figure 3), which might account for some or all of the decreased efficacy. To obtain independent evidence for the importance of the F-actin binding of IP3KB, we manipulated the actin cytoskeleton itself and examined the consequences for the ability of the full-length enzyme to attenuate Ca2+ signals. We first promoted filament disassembly by treating cells with cytochalasin D. Treatment of cytochalasin D (1 μM) for 10 min induced a disruption of F-actin [26], as judged by an alteration in the localization of the full-length IP3KB (result not shown). Ca2+-imaging experiments revealed that, in cytochalasin D-treated cells, full-length IP3KB now had a much smaller effect on the Ca2+ signal, whereas (as a control for non-specific effects of cytochalasin D) the effect of IP3KB262–934 was unaltered by cytochalasin D (Figures 4B and 4C). Indeed, in the presence of cytochalasin D, the responses in cells transfected with full-length IP3KB were not significantly different from those transfected with IP3KB262–934 (Figure 4C), supporting the likelihood that the different localizations rather than activities of the two constructs may be the major factor in their different properties in this context.
Similar results were obtained for full-length IP3KB with the drug latrunculin A (5 μM), which disrupts F-actin by a different mechanism (Figure 4D, right panel) [27]. Conversely, we if we blocked actin dynamics with jasplakinolide (1 μM), cables of F-actin formed throughout the cell (results not shown), and Ca2+-imaging experiments showed that, in jasplakinolide-treated cells, the ability of IP3KB to attenuate histamine-induced Ca2+ increases was unaltered (Figure 4D). Overall, these results suggest that localization of IP3KB by its binding to F-actin has a significant effect on its efficacy, as judged by its ability to attenuate a Ca2+ response in histamine-activated HeLa cells. An analogous influence on the Ca2+ response caused by localization has been previously demonstrated for the type I Ins(1,4,5)P3 5-phosphatase, which is anchored to the plasma membrane by a lipid modification [28].
Proteolysis of IP3KB
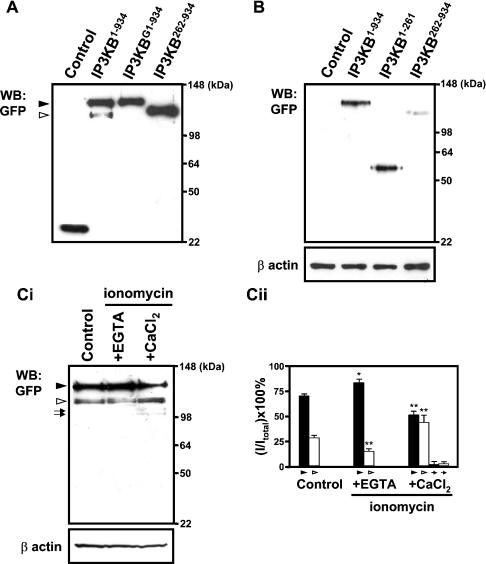
One puzzling aspect of our data in Figure 2 was that the full-length IP3KB showed a partly heterogeneous localization, being in part of the plasma membrane and the cytoskeleton and also in the ER (Figure 2). This is particularly so in COS-7 cells, and, as we have argued above that the localization and truncation of IP3KB may have significant effects on its ability to remove Ins(1,4,5)P3 in vitro and in intact cells (Figures 3 and 4), this heterogeneity may have functional implications. One possibility is that the enzyme is being proteolysed. Previous studies on rat IP3KB have demonstrated the presence of several PEST sequences, a common feature of CaM-binding proteins [29]. The PEST sequences are thought to be targets for rapid proteolysis, particularly by calpain, a Ca2+-dependent protease. Indeed, Pattni et al. have shown that calpain can hydrolyse IP3KB in vitro [15]. We therefore sought evidence that proteolysis of IP3KB occurred in living COS-7 cells.
Firstly, we carried out immunoblots of total lysate protein of COS-7 cells transfected with IP3KB tagged with eGFP. As shown in Figure 5(A), the anti-GFP antibody detected a major protein of ≈135 kDa (IP3KB1–934; ▸), but there was also clearly a minor counterpart of ∼110 kDa (⊳) which we suggest represents a proteolytic product (see also below). Interestingly, when the eGFP protein was fused to the N-terminus of IP3KB, no proteolytic products were seen on the immunoblot (IP3KBG1–934). These data imply that the position of eGFP at either N- or C-terminus has an influence on the proteolysis of IP3KB, such that eGFP fused to the N-terminus of the enzyme protects it from proteolysis. A smaller tag, such as FLAG, has no such protective effect (see below).
Figure 5. Western-blot (WB:) analysis of IP3KB after transfection of different GFP-tagged IP3KB expression constructs.
(A) COS-7 cells were transfected with empty vector (Control) or individual IP3K expression plasmids (IP3KB1–934, IP3KBG1–934, IP3KB262–934). At 48 h after transfection, cells were harvested and the cell lysates were then immunoblotted with an antibody recognizing GFP. (B) At 48 h after transfection, living cells were extracted in 1% Triton X-100 as described in th Experimental section. Lysates were then prepared from the unextractable (cytoskeleton-associated) fractions and then immunoblotted with polyclonal anti-GFP antibody or with monoclonal anti-β-actin antibody to indicate the amount of protein in each lane. (Ci) At 16 h after transfection, cells were treated with EGTA (500 μM) or CaCl2 (10 mM) in the presence of ionomycin (1 μM). Transfected cells incubated with normal medium are used as negative control (Control). After 32 h of incubation, cells were harvested and analysed by Western blotting. ▸, un-cleaved major protein; ⊳, minors (n=7). (Cii) Quantitative analysis of GFP expression was performed as described in the Experimental section. The quantitative data were normalized relative to F-actin. The percentage of the density of each fraction relative to the total was calculated. Results from seven separate experiments are expressed as mean±S.E.M. *P<0.01, **P<0.001 versus control (one-way ANOVA as described in the Experimental section).
We hypothesized that the proteolytic product shown in the immunoblot (Figure 5A) might be responsible for the heterogeneous localization shown above, given that, for example, removing the first 261 amino acids would lead to a translocation from F-actin to the ER (Figure 2I). To confirm that the proteolytic product detected in Figure 5(A) (lane 3) does not bind to F-actin, we specifically examined actin-bound proteins by Western-blotting cell lysates enriched in F-actin (see the Experimental section). As shown in Figure 5(B), the presence of a band at about 135 kDa indicated that, as expected, uncleaved IP3KB is bound to the cytoskeleton of the cells. However, there was no counterpart to the proteolytic cleavage band in the actin skeleton (Figure 5B, lane 2), suggesting that the proteolytic product of IP3KB does not contain an F-actin binding domain. Moreover, when cells were transfected (to an equal degree, as assessed by whole cell lysates; results not shown) with the C-terminally eGFP-tagged individual peptide fragments IP3KB1–261 and IP3KB262–934, and the Triton-resistant fraction examined in the same way, while IP3KB1–261 was bound to the actin cytoskeleton (Figure 5B, lane 3), IP3KB262–934 showed almost no binding (Figure 5B, lane 4).
Finally, given that the Ca2+-regulated protease calpain has been implicated in the proteolysis in vitro of IP3Ks (e.g. [15,29]), we sought indirect evidence that it might be responsible for the data shown in Figures 5(A) and 5(B). We therefore treated cells with ionomycin, with either extracellular Ca2+ or EGTA present; that is, we adjusted the intracellular Ca2+ level 16 h post transfection by adding EGTA (500 μM) or CaCl2 (10 mM) to the medium in the presence of ionomycin (1 μM). Examining the cells did not reveal any obvious changes in cell number or morphology, implying that, at least superficially, the cells were not being damaged or killed by this protocol. We then examined cell lysates by Western blotting, and found that, at least as far as this simple protocol suggests, the generation of the lower-molecular-mass fragment (⊳ in Figure 5Ci) is increased by Ca2+ (Figure 5Cii; 46.5±5.2%; control; 28.4±2.3%, P<0.001), and decreased by EDTA (19.4±2.1%, P<0.001 versus control). There also appears to be some further proteolysis in the ionomycin/Ca2+-treated cells (compared with controls), judged by faint lower-molecular-mass bands visible in Figure 5(C) (lane 3, arrows). Our results suggested that IP3KB is cleaved to generate a major fragment seen in Figure 5. We cannot say from our results whether this cleavage has taken place at any putative calpain cleavage sites [15,29], and the assignation of possible protease(s) to such cleavage must await purification and sequencing of this fragment.
Regulation of IP3KB localization by proteolysis in intact cells
The above data imply that IP3KB is subject to proteolytic cleavage, which generates protein species that would have different localizations and activities. However, as with all such studies so far, this is based on in vitro evidence [15] and on Western blotting of cell lysates (above). We cannot eliminate the possibility that some of the proteolysis might be occurring during the processing of lysates for Western blotting. Moreover, although we infer that proteolysis is responsible for the heterogeneous localization, this could also be due to unrelated mechanisms, such as protein phosphorylation. In order to strengthen the case for the physiological relevance of the proteolysis, we explored the proteolytic cleavage of IP3KB in intact cells by double-tagging full-length IP3KB at the N- and C-terminal ends with FLAG and mRFP1 respectively (Figure 6A). When COS-7 cells were transfected with full-length of IP3KB in normal growth medium before fixation, the FLAG signal was observed mostly in the cytoskeleton and plasma membrane, whereas the mRFP1 signal was clearly more heterogeneous (Figure 6B, upper panels). This separation of the two ends of the IP3KB molecule was analysed quantitatively (see the Experimental section), and the number obtained by analysis of randomly selected cells (R2=0.37±0.04, n=41) is direct evidence for its proteolysis in intact cells and its differential localization as a result.
We also subjected the cells to the same Ca2+ or EGTA protocol as that described above for Figure 5(C) and examined the localization of the two tags. In the presence of Ca2+ (Figure 6B, lower panels), cells expressing IP3KB had the FLAG signal either mostly localized to the plasma membrane and cytoskeleton, or in some cells there was also some in the ER. The mRFP1 signal (Figure 6B, lower panels), however, was very much more localized to the ER, with very few cells showing any in the cytoskeleton, and this striking increase in separation of the two ends of the IP3KB molecule is confirmed by quantitative analysis (R2=0.06±0.02, n=37). These data confirm our implication from the data in Figure 5(C) that a Ca2+-activated protease is indeed hydrolysing the enzyme in an intact cell.
Although this prolonged Ca2+ treatment could be argued to be unphysiological, some evidence for physiological significance comes from Figure 6(B) (middle panels). Thus, in the presence of EGTA, all cells expressing IP3KB exhibited a closer co-localization of the FLAG and mRFP1 signal, both being associated with the cytoskeleton and plasma membrane. This is significantly different from the control cells (Figure 6B middle panels, merge; and R2=0.61±0.09, n=43) and confirms the data in Figure 5(C), suggesting less proteolysis of IP3KB if EGTA is present. Crucially, though subject to the caveat that all these data are obtained with the high expression levels resulting from transfection, the comparison of EGTA-treated cells with control cells in both Figures 5 and 6 show that, in the controls, that is, in intact COS-7 cells grown under standard cell culture conditions and with no other stimuli or treatments present, IP3KB is subject to significant constitutive proteolysis, leading to a heterogeneity in its subcellular localization.
Conclusion
Since the first report of the partial sequence of IP3KB was published in 1992 [12], many groups have investigated its localization and functional expression in various cell types. In this paper we have shown that N- and C-termini of IP3KB are differentially and predominantly localized to the actin cytoskeleton and ER respectively and that this localization has a direct impact on the ability of the enzyme to attenuate an Ins(1,4,5)P3-induced Ca2+ response. Moreover, we found that transfected IP3KB undergoes proteolysis constitutively in intact cells, leading to relocation and accumulation of a stable C-terminal domain of ≈83 kDa to the ER, while the uncleaved full-length enzyme remains on the actin cytoskeleton and plasma membrane. Thus this proteolysis may have a physiologically important role in the complex spatiotemporal regulation of Ins(1,4,5)P3 signalling.
Acknowledgments
This study is supported by a Programme Grant from The Wellcome Trust. J.C.H.Y. is supported by the Croucher Foundation, S.M.L.-B. by Merck Sharp and Dohme, and R.F.I and M.J.S. by The Royal Society.
References
- 1.Berridge M. J., Irvine R. F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature (London) 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 2.Berridge M. J. Inositol trisphosphate and calcium signalling. Nature (London) 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 3.Drayer A. L., Pesesse X., De Smedt F., Communi D., Moreau C., Erneux C. The family of inositol and phosphatidylinositol polyphosphate 5-phosphatases. Biochem. Soc. Trans. 1996;24:1001–1005. doi: 10.1042/bst0241001. [DOI] [PubMed] [Google Scholar]
- 4.Irvine R. F., Schell M. J. Back in the water: the return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001;2:327–328. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- 5.Communi D., Vanweyenberg V., Erneux C. Molecular study and regulation of D-myo-inositol 1,4,5-trisphosphate 3-kinase. Cell Signalling. 1995;7:643–650. doi: 10.1016/0898-6568(95)00035-n. [DOI] [PubMed] [Google Scholar]
- 6.Nalaskowski M. M., Mayr G. W. The families of kinases removing the Ca2+ releasing second messenger Ins(1,4,5)P3. Curr. Mol. Med. 2004;4:277–290. doi: 10.2174/1566524043360726. [DOI] [PubMed] [Google Scholar]
- 7.Pattni K., Banting G. Ins(1,4,5)P3 metabolism and the family of IP3-3kinases. Cell Signalling. 2004;16:643–654. doi: 10.1016/j.cellsig.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Schell M. J., Erneux C., Irvine R. F. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 2001;276:37537–37546. doi: 10.1074/jbc.M104101200. [DOI] [PubMed] [Google Scholar]
- 9.Jun K., Choi G., Yang S. G., Choi K. Y., Kim H., Chan G. C., Storm D. R., Albert C., Mayr G. W., Lee C. J., Shin H. S. Enhanced hippocampal CA1 LTP but normal spatial learning in inositol 1,4,5-trisphosphate 3-kinase(A)-deficient mice. Learn. Mem. 1998;5:317–330. [PMC free article] [PubMed] [Google Scholar]
- 10.Pouillon V., Hascakova-Bartova R., Pajak B., Adam E., Bex F., Dewaste V., Van Lint C., Leo O., Erneux C., Schurmans S. Inositol 1,3,4,5-tetrakisphosphate is essential for T lymphocyte development. Nat. Immunol. 2003;4:1136–1143. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- 11.Wen B. G., Pletcher M. T., Warashina M., Choe S. H., Ziaee N., Wiltshire T., Sauer K., Cooke M. P. Inositol 1,4,5-trisphosphate 3 kinase B controls positive selection of T cells and modulates Erk activity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5604–5609. doi: 10.1073/pnas.0306907101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mailleux P., Takazawa K., Albala N., Erneux C., Vanderhaeghen J. J. Astrocytic localization of the messenger RNA encoding the isoenzyme B of inositol 1,4,5-trisphophate 3-kinase in the human brain. Neurosci. Lett. 1992;148:177–180. doi: 10.1016/0304-3940(92)90833-s. [DOI] [PubMed] [Google Scholar]
- 13.Hascakova-Bartova R., Pouillon V., Dewaste V., Moreau C., Jacques C., Banting G., Schurmans S., Erneux C. Identification and subcellular distribution of endogenous Ins(1,4,5)P3 3-kinase B in mouse tissues. Biochem. Biophys. Res. Commun. 2004;323:920–925. doi: 10.1016/j.bbrc.2004.08.152. [DOI] [PubMed] [Google Scholar]
- 14.Brehm M. A., Schreiber I., Bertsch U., Wegner A., Mayr G. W. Identification of the actin-binding domain of inositol 1,4,5-trisphosphate 3-kinase isoform B [Ins(1,4,5)P3 3-kinase-B] Biochem. J. 2004;382:353–362. doi: 10.1042/BJ20031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattni K., Millard T. H., Banting G. Calpain cleavage of the B isoform of Ins(1,4,5)P3 3-kinase separates the catalytic domain from the membrane anchoring domain. Biochem. J. 2003;375:643–651. doi: 10.1042/BJ20030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soriano S., Thomas S., High S., Griffiths G., D'Santos C., Cullen P., Banting G. Membrane association, localization and topology of rat inositol 1,4,5-trisphosphate 3-kinase B: implications for membrane traffic and Ca2+ homoeostasis. Biochem. J. 1997;324:579–589. doi: 10.1042/bj3240579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewaste V., Moreau C., De Smedt F., Bex F., De Smedt H., Wuytack F., Missiaen L., Erneux C. The three isoenzymes of human inositol 1,4,5-trisphosphate 3-kinase show specific intracellular localization but comparable Ca2+ responses on transfection in COS-7 cells. Biochem. J. 2003;374:41–49. doi: 10.1042/BJ20021963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irvine R. F., Letcher A. J., Heslop J. P., Berridge M. J. The inositol tris/tetrakisphosphate pathway – demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues. Nature (London) 1986;320:631–634. doi: 10.1038/320631a0. [DOI] [PubMed] [Google Scholar]
- 20.Thorn P. Calcium influx during agonist and Ins(2,4,5)P3-evoked Ca2+ oscillations in HeLa epithelial cells. J. Physiol. (London) 1995;499:307–314. doi: 10.1113/jphysiol.1995.sp020516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rintoul G. L., Baimbridge K. G. Effects of calcium buffers and calbindin–D28k upon histamine-induced calcium oscillations and calcium waves in HeLa cells. Cell Calcium. 2003;34:131–144. doi: 10.1016/s0143-4160(03)00041-1. [DOI] [PubMed] [Google Scholar]
- 22.Bootman M. D. Hormone-evoked subcellular Ca2+ signals in HeLa cells. Cell Calcium. 1996;20:97–104. doi: 10.1016/s0143-4160(96)90099-8. [DOI] [PubMed] [Google Scholar]
- 23.Bootman M. D., Berridge M. J. Subcellular Ca2+ signals underlying waves and graded responses in HeLa cells. Curr. Biol. 1996;6:855–865. doi: 10.1016/s0960-9822(02)00609-7. [DOI] [PubMed] [Google Scholar]
- 24.Millard T. H., Cullen P. J., Banting G. Effects of elevated expression of inositol 1,4,5-trisphosphate 3-kinase B on Ca2+ homoeostasis in HeLa cells. Biochem. J. 2000;352:709–715. [PMC free article] [PubMed] [Google Scholar]
- 25.Bubb M. R., Spector I., Beyer B. B., Fosen K. M. Effect of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- 26.Cooper J. A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spector I., Shochet N. R., Blasberger D., Kashman Y. Latrunculins – novel marine marcolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil. Cytoskeleton. 1989;13:127–144. doi: 10.1002/cm.970130302. [DOI] [PubMed] [Google Scholar]
- 28.De Smedt F., Missiaen L., Parys J. B., Vanweyenberg V., De Smedt H., Erneux C. Isoprenylated human brain type I inositol 1,4,5-trisphosphate 5-phosphatase controls Ca2+ oscillations induced by ATP in Chinese hamster ovary cells. J. Biol. Chem. 1997;272:17367–17375. doi: 10.1074/jbc.272.28.17367. [DOI] [PubMed] [Google Scholar]
- 29.Dewaste V., Roymans D., Moreau C., Erneux C. Cloning and expression of a full-length cDNA encoding human inositol 1,4,5-trisphosphate 3-kinase B. Biochem. Biophys. Res. Commun. 2002;291:400–405. doi: 10.1006/bbrc.2002.6456. [DOI] [PubMed] [Google Scholar]