Abstract
Previous studies in Trypanosoma cruzi, the etiologic agent of Chagas disease, have resulted in the cloning and sequencing of a pair of tandemly linked genes (TcHA1 and TcHA2) that encode P (phospho-intermediate form)-type H+-ATPases with homology to fungal and plant proton-pumping ATPases. In the present study, we demonstrate that these pumps are present in the plasma membrane and intracellular compartments of three different stages of T. cruzi. The main intracellular compartment containing these ATPases in epimastigotes was identified as the reservosome. This identification was achieved by immunofluorescence assays and immunoelectron microscopy showing their co-localization with cruzipain, and by subcellular fractionation and detection of their activity. ATP-dependent proton transport by isolated reservosomes was sensitive to vanadate and insensitive to bafilomycin A1, which is in agreement with the localization of P-type H+-ATPases in these organelles. Analysis by confocal immunofluorescence microscopy revealed that epitope–tagged TcHA1-Ty1 and TcHA2-Ty1 gene products are localized in the reservosomes, whereas the TcHA1-Ty1 gene product is additionally present in the plasma membrane. Immunogold electron microscopy showed the presence of the H+-ATPases in other compartments of the endocytic pathway such as the cytostome and endosomal vesicles, suggesting that in contrast with most cells investigated until now, the endocytic pathway of T. cruzi is acidified by a P-type H+-ATPase.
Keywords: cytostome, endocytosis, P-type H+-ATPase, proton pump, reservosomes, Trypanosoma cruzi
Abbreviations: ECL, enhanced chemiluminescence; ER, endoplasmic reticulum; FG, fish gelatin; LIT, liver infusion tryptose; P-type, phospho-intermediate-type; PPi, pyrophosphate; TcHA, Trypanosoma cruzi H+-ATPase; V-H+-ATPase, vacuolar-H+-ATPase
INTRODUCTION
Trypanosoma cruzi, the agent of Chagas disease, or American trypanosomiasis has been recognized as a significant cause of morbidity and mortality in Mexico, Central and South America [1]. Chagas' disease remains a problem because of toxicity and lack of effectiveness in the chronic phase by the two drugs available, nifurtimox and benznidazole [1]. Therefore it is important to identify enzymes and metabolic processes in T. cruzi that might be potential targets for drug development. A case in point is that of the P-type (phospho-intermediate-type) H+-ATPases. Previous studies in T. cruzi [2] have resulted in the cloning and sequencing of a pair of tandemly linked genes (TcHA1 and TcHA2) that encode P-type H+-ATPases with homology to fungal and plant proton-pumping ATPases. Full-length TcHA1 and an N-terminal truncated version of TcHA2 were able to complement a Saccharomyces cerevisiae strain deficient in P-type H+-ATPase activity, providing genetic evidence for their function [2]. The absence of electrogenic P-type H+-ATPases in mammalian cells [3,4] and their presence in fungi has led to the proposal that these pumps are promising targets for antifungal therapy [5] and a similar proposal could be made regarding T. cruzi.
The P-type H+-ATPases of fungi and plants are located in the plasma membrane where they act as primary transporters by pumping protons out of the cells, thereby creating pH and electrical potential differences. Transport of many solutes (ions, metabolites etc) both into and out of the cell involves secondary transporters whose ability to function is directly dependent on the proton-motive force created by the H+-ATPases [6]. A similar function has been attributed to the plasma membrane H+-ATPase of T. cruzi [7–9].
Biochemical evidence using permeabilized cells have suggested the presence of an intracellular P-type H+-ATPase in T. cruzi [10]. The presence of an internal P-type H+-ATPase activity is almost unique and has been described elsewhere only in the ER (endoplasmic reticulum) of plant mechanoreceptor organs [11]. In yeasts, the H+-ATPase is made in the rough ER and delivered to the plasma membrane via the secretory pathway [12]. The H+-ATPase travels from the ER to the Golgi into coat protein complex II vesicles and from the Golgi to the plasma membrane via secretory vesicles. The H+-ATPase accumulates in the secretory vesicles of secretion mutants and their isolation has shown that it is able to hydrolyse ATP and pump protons at rates comparable with those seen in the plasma membrane [13]. On the other hand, abnormal H+-ATPases that reach the plasma membrane are retrieved by endocytosis and sent to the vacuole for degradation but no evidence has been presented of their activity in this compartment [14].
On the basis of acidity and K+ content the internal P-type H+-ATPase has been postulated to be located in the reservosomes of T. cruzi [10]. Reservosomes have been described in epimastigote forms as acidic pre-lysosomal compartments [15]. They are large organelles found in the posterior end of the parasite that are rich in the proteinase cruzipain and accumulate macromolecules ingested by the parasite through endocytosis such as albumin, peroxidase, transferrin and low density lipoprotein [15–18]. It has been shown that they also contain lipids [19] and since their number decreases during transformation of epimastigotes into trypomastigotes (metacyclogenesis) they were postulated to have a role in the storage of nutrients necessary for this differentiation step [15]. Interestingly, the vacuolar-type H+-ATPase, which in most eukaryotic cells is involved in acidification of the endocytic pathway, localizes to acidocalcisomes [20] and the plasma membrane [21] of T. cruzi epimastigotes, and is absent from the flagellar pocket and reservosomes [21]. Acidocalcisomes, which do not belong to the endocytic pathway [18], are characterized, in addition to their acidic nature, by their high density (both in weight and by electron microscopy) and high content of PPi (pyrophosphate), polyphosphate, calcium, magnesium and other elements [22], and also contain a vacuolar-type H+-PPase [23].
In the present study we report experiments, using immunofluorescence and immunogold electron microscopy, that provide evidence that both TcHA1 and TcHA2 are located in the reservosomes and other endocytic structures, whereas TcHA1 is the only pump located in the plasma membrane of epimastigotes. Purified reservosomes were shown to have a vanadate-sensitive and bafilomycin A1-insensitive ATP-dependent proton uptake compatible with the presence of a functional P-type H+-ATPase in these organelles. Taken together these results suggest that in contrast with most eukaryotic cells that acidify their endocytic pathway through the action of a vacuolar-type H+-ATPase, in T. cruzi this process occurs through a P-type H+-ATPase.
MATERIALS AND METHODS
Culture methods
Wild-type T. cruzi epimastigotes (Y strain) and transfectants were grown at 28 °C in LIT (liver infusion tryptose) medium [24] supplemented with 10% heat-inactivated newborn calf serum and harvested after 5 days in culture. Trypomastigotes and amastigotes were obtained from the culture medium of infected L6E9 myoblasts as we have described previously [20].
Chemicals
Foetal and newborn calf serum, normal goat serum, BSA, cold fish gelatin, Dulbecco's PBS, EGTA and proteinase inhibitors were purchased from Sigma Chemical Co. (St. Louis, MO, U.S.A.). Alexa Fluor®-labelled secondary antibodies, monoclonal antibody 10D7 against the 100 kDa subunit of the yeast vacuolar H+-ATPase and Prolong Gold® antifade reagent were from Molecular Probes, Inc (Eugene, OR, U.S.A.). The ECL® (enhanced chemiluminescence) detection kit was from Amersham (Arlington Heights, IL, U.S.A.). Probe GT nylon membranes, prestained molecular mass standards and the protein assay were from Bio-Rad (Hercules, CA, U.S.A.). Gold-conjugated secondary antibodies were obtained from Ted Pella, Inc (Reddington, CA, U.S.A.). Affinity purified anti-TcHAf antibody was prepared as described previously [2]. Monoclonal antibody BB2 against the Saccharomyces cerevisiae Ty1 virus-like particle [25] was a gift from Keith Gull (University of Oxford, U.K.). Monoclonal antibody 212-BH6 and polyclonal antibody against cruzipain [26] were a gift from Julio Scharfstein (Federal University of Rio de Janeiro, Brazil). Pfu DNA polymerase was from Stratagene (La Jolla, CA, U.S.A.). All other reagents were of analytical grade.
Isolation of reservosomes
Isolation of reservosomes was carried out as described previously [27] with minor modifications. Briefly, epimastigotes cultured for 5 days were washed twice with ice-cold TMS buffer (20 mM Tris/HCl, pH 7.2, containing 2 mM MgCl2, and 250 mM sucrose) and resuspended in the same buffer containing protease inhibitors cocktail [2 mM PMSF, 100 μg/ml leupeptin, 5 mM ethylenediaminotetraacetic acid, 2 μg/ml aprotinin, 10 μM trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane (E-64), 7 μg/ml pepstatin, and 10 μM antipain] and 1 mM dithiothreitol. Parasites were sonicated on an ice bath using a Branson 250 digital sonifier equipped with a double-step microtip using at least 30 pulses of 2 s on/1 s off, operating at 15% total amplitude. Homogenates were centrifuged at 2400 g for 10 min at 4 °C. The supernatant was collected and mixed 1:1 with 2.3 M sucrose in TM (20 mM Tris/HCl, pH 7.2, containing 2 mM MgCl2) to obtain a final sucrose concentration of 1.27 M. A 12 ml quantity of the mixture was deposited in a Beckman SW-28 centrifuge tube overlaid with 10 ml of 1.2 M, 10 ml of 1.0 M and 5 ml of 0.8 M sucrose and centrifuged at 97000 g for 150 min at 4 °C. All sucrose solutions were prepared in TM buffer containing protease inhibitor cocktail and 1 mM dithiothreitol. The interfaces of 0.8 M/1.0 M (B1), 1.0 M/1.2 M (B2), 1.2/1.27 (B3), the 1.27 layer (B4) and the pellet (P) were collected, diluted in TM buffer and centrifuged at 120000 g for 30 min at 4 °C. Pellets containing the fractions were resuspended in TMS buffer supplemented with the protease inhibitors described above and immediately frozen at −80 °C for further analysis, or fixed for posterior electron microscopy processing.
H+ transport assays
Measurements were carried out immediately after subcellular fractionation. Fractions were diluted in a buffer containing 130 mM KCl, 2 mM MgCl2, 10 mM Hepes and 4 μM Acridine Orange, pH 7.2. Active H+ transport was assayed, as described previously [10], by measuring changes in the absorbance of Acridine Orange (493–530 nm) in an SLM-Aminco DW 2000 dual-wavelength spectrophotometer at 30 °C in the presence, or absence, of ATP, bafilomycin A1 and vanadate.
Enzymatic activity assays
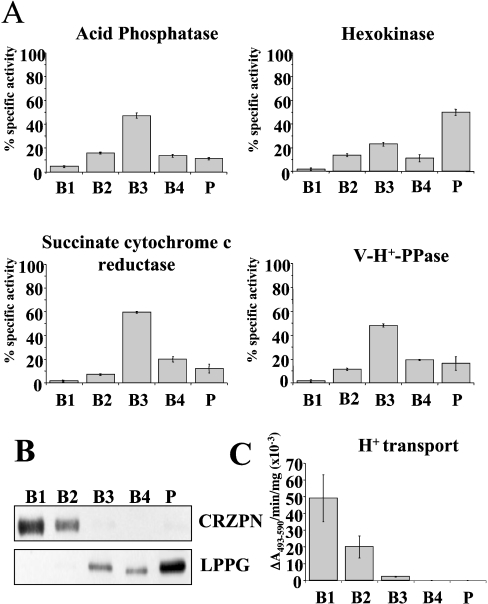
Enzymatic markers for reservosome isolation were: vacuolar H+-pyrophosphatase (acidocalcisomes), hexokinase (glycosomes), succinate-cytochrome c reductase (mitochondria) and acid phosphatase (lysosomes). Assays were modified from previously described methods [23,28–30] in order to fit readings in 96-well microtitre plates. Vacuolar H+-pyrophosphatase activity in terms of phosphate release was assayed by adding samples to 100 μl of a reaction mixture containing 130 mM KCl, 2 mM MgCl2, 10 mM Hepes pH 7.2, 250 μM EGTA, 0.1 mM 2-amino-6-mercapto-7-methylpurine ribonucleoside and 0.4 unit/ml purine nucleoside phosphorylase and the reaction was monitored at 360 nm. Hexo-kinase was assayed by adding samples to 100 μl of a reaction mixture containing 10 mM D-glucose, 0.6 mM ATP, 0.6 mM NADH, 10 mM magnesium chloride, 2.5 units/ml glucose-6-phosphate dehydrogenase, 50 mM K+-Hepes at pH 7.8 and the oxidation of NADH was monitored at 340 nm. Succinate-cytochrome c reductase activity was determined by mixing samples in a reaction buffer containing 6 mM succinate-K+, 0.2 mM cytochrome c, 0.6 mM KCN and 80 mM Hepes at pH 8.0 and monitoring the reaction at 550 nm. Acid phosphatase was assayed by measuring phosphate release from p-nitrophenylphosphate. Samples were added to 50 μl of the reaction mixture containing 0.1 M sodium acetate at pH 5.5, and 10 mM p-nitrophenylphosphate and incubated for 30 min at 28 °C. Reactions were stopped by the addition of 150 mM sodium hydroxide (100 μl) and released p-nitrophenol was detected at 405 nm. Protein amounts of all samples were normalized before each assay. All reactions were performed in a Bio-Tek Instruments PowerWave 340 plate reader at 30 °C.
SDS/PAGE and Western blot analysis
Samples were processed for SDS/PAGE as described by Laemmli [31]. Epimastigotes (5 days old), trypomastigotes (>95% pure) or amastigotes (>90% pure) were collected, washed with PBS and stored at −80 °C in PBS containing 2% of a protease inhibitor cocktail [2 mM PMSF, 100 μg/ml leupeptin, 5 mM EDTA, 2 μg/ml aprotinin, 10 μM trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane (E-64), 7 μg/ml pepstatin and 10 μM antipain]. Samples (10 μg) were applied on to 10% running/4% stacking polyacrylamide gels and electrophoresis was carried out at 100 V in a Bio-Rad Miniprotean II® system. Gels were blotted on to nitrocellulose membranes for 75 min at 100 V and these were processed for antigen immunodetection using the ECL® (Amersham Biosciences) according to the manufacturer instructions.
Immunofluorescence microscopy
For localization of TcHAs using epifluorescence microscopy, parasites were collected, washed 4 times with Dulbecco's PBS and fixed with 4% formaldehyde in PBS for 15 min. The fixed cells were adhered to poly-L-lysine-coated coverslips, permeabilized using 0.3% Triton X-100 for 5 min and blocked using 3% BSA and 50 mM NH4Cl in PBS for 30 min. A 1:100 dilution of affinity purified antibody against the 35 kDa expressed protein in PBS was applied at room temperature for 30 min (Figure 1B) or 3 h (Figure 2) and a fluorescein isothiocyanate-coupled goat anti-rabbit immunoglobulin G (IgG) secondary antibody (1:150) was then applied at room temperature for 30 min. Control preparations were incubated with pre-immune serum. Slides were observed using an Olympus BX-60 microscope, and digital images obtained using the system previously described [28]. For co-localization studies using confocal microscopy, epimastigotes were washed with PHEM (60 mM Pipes, 25 mM HEPES, 10 mM EGTA and 2 mM MgCl2), pH 7.2, and fixed with 4% freshly prepared formaldehyde and 0.05% grade I glutaraldehyde in PHEM for 10 min at room temperature and 50 min at 4 °C. The fixed cells were adhered to poly-L-lysine-coated coverslips, allowed to attach for 60 min and permeabilized with 0.1% Triton X-100 in PBS for 2–5 min at room temperature. Samples were blocked in 1% BSA, 0.5% fish gelatin and 2% normal goat serum for 30 min at room temperature. Primary antibodies, diluted in PBS-1% fish gelatin, were applied at room temperature for 60 min. After washing with PBS-1% fish gelatin, samples were incubated with Alexa 488-conjugated goat-anti mouse or Alexa 568-conjugated goat anti-rabbit antibodies diluted 1:500–1:1000 in PBS-1% fish gelatin at room temperature for 60 min. Cover-slips were washed in PBS and mounted on slides using Prolong Gold® antifade reagent. Primary antibody titres were: polyclonal anti-TcHAf, 1:5–1:50; polyclonal anti-cruzipain, 1:200; monoclonal 212-BH6 anti-cruzipain, 1:500; monoclonal BB2 anti-Ty1, 1:20 and monoclonal 10D7 anti-V (vacuolar)-H+-ATPase, 1:500. Images were obtained using an Olympus Fluoview FV 300 confocal laser scanning microscope.
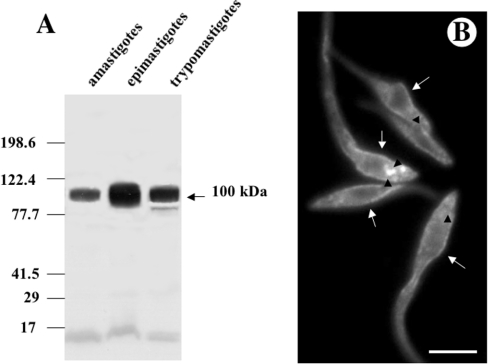
Figure 1. Western blot analysis of T. cruzi H+-ATPase (A) and immunofluorescence microscopy showing the localization of TcHAs in epimastigotes of T. cruzi (B).
(A) Homogenates containing 10 μg of protein from amastigotes, epimastigotes and trypomastigotes were subjected to SDS/PAGE on 10% polyacrylamide gels and transferred to nitrocellulose membranes. Lanes were probed with affinity purified anti-TcHAf antibody. Migration positions of prestained molecular mass standards are shown to the left of the gel. (B) Epimastigotes were treated with the affinity-purified antibody against TcHAs. Labelling of the plasma membrane (arrows) and large vacuoles (arrowheads) is evident. Scale bar, 5 μm.
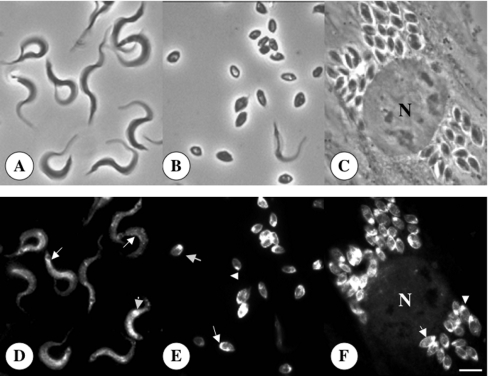
Figure 2. Immunofluorescence microscopy showing the localization of TcHAs in trypomastigote (D) and amastigote (E, F) forms of T. cruzi.
Upper panels show the same cells as in lower panels by bright field microscopy. N, host cell nucleus. Scale bar, 4 μm (A–F). Labelling of the plasma membrane (arrows) and intracellular structures (arrowheads) is evident in both trypomastigotes and amastigotes.
Electron microscopy
Subcellular fractions were immediately fixed with 2.5% grade II glutaraldehyde, washed with 0.1 M cacodylate buffer at pH 7.2 and post fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide at room temperature in the same buffer. In addition, epimastigotes cultured for 5 days were washed in PBS and fixed as above. After washes in 10% ethanol samples were dehydrated by incubation in increasing concentrations of ethanol (25, 50, 75 and 100%) at room temperature. Infiltration and embedding were performed in eponate resin and blocks were polymerized at 60 °C. Ultrathin sections were collected on copper grids and stained with uranyl acetate and lead citrate.
For immunocytochemistry epimastigotes, amastigotes or trypomastigotes were harvested, washed with PBS and fixed for 60 min at room temperature in a solution containing 4% freshly prepared formaldehyde, 0.1% grade I glutaraldehyde and 0.8% picric acid in 0.1 M cacodylate buffer at pH 7.2. Fixed cells were washed in PBS, dehydrated in increasing concentrations (25, 50, 75, 95, 100 and 100%) of methanol, embedded in Unicryl at −20 °C and polymerized at the same temperature under UV irradiation. Ultrathin sections were collected on formvar-coated nickel grids. Grids were blocked in PBS containing 1% FG (fish gelatin), 0.1% (v/v) Tween 20 (PBS-FG-Tw) and 50 mM ammonium chloride for 30 min, washed in PBS-FG-Tw and incubated with a 1:50 dilution of affinity-purified anti-TcHAf in PBS-FG-Tw for 120 min at room temperature. After washing in PBS-FG-Tw, grids were incubated with goat anti-(rabbit IgG) labelled with 15 nm gold (1:50) in PBS-FG-Tw for 60 min at room temperature. For double-labelling experiments in epimastigotes, grids were blocked with PBS-FG-Tw at 37 °C for 60 min and incubated with affinity-purified anti-TcHAf (1:10) and monoclonal 212-BH6 anti-cruzipain (1:500) for 30 min at 37 °C. After washing in PBS-FG-Tw, samples were incubated with a 20 nm gold-conjugated goat anti-rabbit antibody (1:20) and a 10 nm gold-conjugated goat anti-mouse antibody (1:75) in PBS-FG-Tw for 30 min at 37 °C. After antibody incubations grids were washed first in PBS-FG-Tw and then with distilled water, stained with uranyl acetate and lead citrate. All micrographs were obtained using a Hitachi H600 transmission electron microscope at 80 or 100 kV. Controls were carried out using an unrelated antibody or incubation in the presence of the secondary antibody only.
Cellular localization of TcHA1 and TcHA2
The Ty1 epitope-tag [25] was inserted into the central portion of the TcHA1 and TcHA2 genes and between codons in such a way that there was no disruption of the open reading frames. For TcHA1 the tag was inserted between the E480 and M481 and for TcHA2 the insertion was between the E530 and M531. Tag addition was performed by PCR amplification of 1440 bp and 1590 bp fragments, corresponding to the N-terminal ends of TcHA1 and TcHA2 respectively, using the λGEM 5–1 clone as template [2]. For TcHA1 the primers 5′-GAATTCATGCTACCGCCGTCCAAGGG-3′ and 5′-ACTAGTGTCAAGTGGATCCTGGTTAGTATGGACCTCCTCCTTCGCAATCAGCAC-3′ were used. For TcHA2 the primers used were 5′-GAATTCATGAACCAGAAGAACGATAA-3′ and 5′-ACTAGTGTCAAGTGGATCCTGGTTAGTATGGACCTCCTCCTTCGCAATCAGCAC-3′. The PCR products having EcoRI and SpeI linkers (underlined) and the Ty1 epitope-tag were subcloned into the EcoRI and SpeI sites of the pTEX vector [32] in order to generate the plasmids pTEX/TcHA1 and pTEX/TcHA2. Subsequently, fragments of 1185 bp and 1161 belonging to the C-terminal end of TcHA1 and TcHA2 respectively, were amplified from the λGEM 5–1 clone DNA [2] using the primers 5′-ACTAGTATGTGCCGCATGCTTAACCT-3′ and 5′-CTCGAGTTACACCGTGGGTTCCTTTG-3′ for TcHA1, and 5′-ACTAGTATGTGCCGCATGCTTAACCT-3′ and 5′-CTCGAGTTAATTGGCAGGCTCAGTGATC-3′ for TcHA2. The PCR products with SpeI and XhoI linkers (underlined) were subcloned into the SpeI and XhoI sites of plasmids pTEX/TcHA1 and pTEX/TcHA2 generating the plasmids pTEX/TcHA1-Ty1 and pTEX/TcHA2-Ty1 respectively. Alternatively TcHA1 or TcHA2 were amplified and subcloned without the epitope-tag into the pTEX vector generating the plasmids pTEX/TcHA1Φ and pTEX/TcHA2Φ. For TcHA1 the primers 5′-GAATTCATGCTACCGCCGTCCAAGGG-3′ and 5′-CTCGAGTTACACCGTGGGTTCCTTTG-3′ were used. For TcHA2 the primers used were 5′-GAATTCATGAACCAGAAGAACGATAA-3′ and 5′-CTCGAGTTAATTGGCAGGCTCAGTGATC-3′. The PCR products with EcoRI and XhoI linkers (double underlined) were inserted in the EcoRI and XhoI sites of the pTEX vector. All PCR amplifications were carried out using the Pfu DNA polymerase to assure a high accuracy for the amplified DNAs. The PCRs were performed for 25 cycles of 96 °C for 1 min, 55–60 °C for 1 min, and 72 °C for 1.5 min using a thermal cycler. All constructs were sequenced to confirm their identity. Epimastigotes were transfected by electroporation with a single discharge of 1.5 kV, 50 μF, with pTEX vector alone, or with the plasmids pTEX/TcHA1Φ, pTEX/TcHA2Φ, pTEX/TcHA1-Ty1 or pTEX/TcHA2-Ty1. Cells were suspended in LIT medium with 10% newborn calf serum and incubated at 28 °C for 24 h, followed by a 1:5 dilution in the same medium containing 500 μg/ml of G418. After 48 h, another 1:5 dilution was carried out under the same conditions. Selection was applied by growing the parasites in the presence of 500 μg/ml G418 for 3 or 4 weeks to allow the establishment of stable lines.
RESULTS
Localization of TcHAs in T. cruzi
Total homogenates prepared from different stages of T. cruzi were subjected to Western blot analysis with affinity-purified antibodies obtained against the protein product of TcHAf [2]. This polypeptide encodes a fragment of about 35 kDa, from amino acid 345–604 of the TcHA2 sequence, that is almost identical to the corresponding sequence of TcHA1 (amino acid 295–554), except for three conservative substitutions (Gln to Pro, Val to Leu, and Leu to Met). These antibodies detected a band of approximately 100 kDa, close to the predicted molecular mass of the gene products, in amastigote, epimastigote and trypomastigote homogenates (Figure 1A), in agreement with a previous report [2]. An additional band of 85 kDa was occasionally detected in trypomastigotes (Figure 1A, lane trypomastigotes). This band could be a proteolysis product of TcHAs since its presence was not reproducible [2]. No detectable bands were observed using pre-immune serum (results not shown). The localization of the TcHAs in different forms of T. cruzi was determined by indirect immunofluorescence assays using the anti-TcHAf antibody. Different forms of T. cruzi were fixed and permeabilized before antibody binding (Figures 1B and 2). As Figure 1(B) shows, TcHAs were detected in epimastigotes. In agreement with the Western blots (Figure 1A), weaker signals were detected in amastigotes and trypomastigotes when the same incubation time was used for all stages (Figure 1B and results not shown). No detectable signal was observed when the pre-immune serum was used (results not shown). In all stages fluorescence was detected in association with the plasma membrane (Figure 1B and 2, arrows). Reaction was also detected in large intracellular vacuoles of epimastigotes (Figure 1B, arrowheads). When the antibody was used in longer incubation periods, a punctuate intracellular staining was observed in amastigotes (Figure 2E, arrowhead) and trypomastigotes (Figure 2D, arrowhead). In amastigotes, there was always an intracellular labelling at the anterior end of the cells (arrowhead). The expression of TcHAs was also observed in intracellular amastigotes in infected myoblasts with no staining of the host cells (Figure 2F). Here, as occurred in isolated amastigotes, TcHAs were mainly localized to the plasma membrane (Figure 2 F, arrow) and to intracellular structures in the anterior region (Figure 2F, arrowhead).
Immunoelectron microscopy
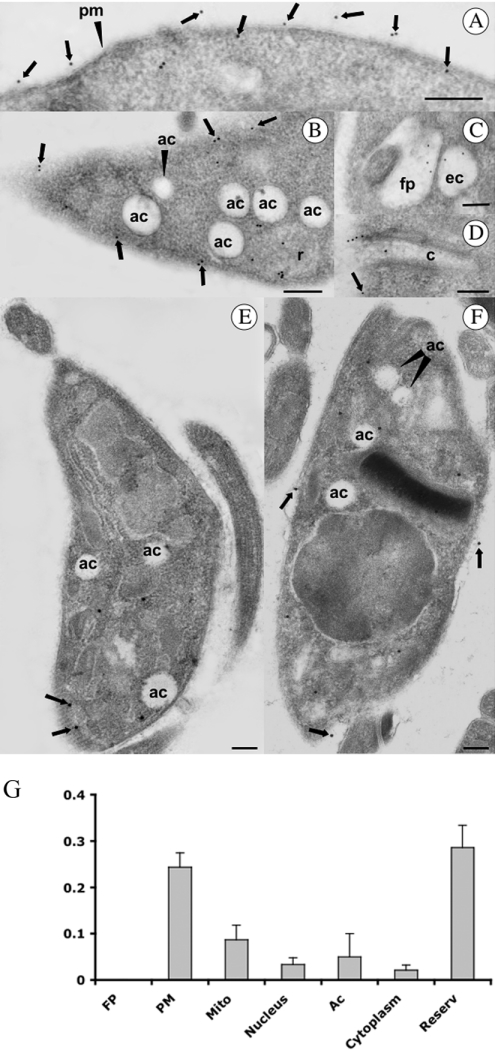
In order to analyse in more detail the structures labelled with the antibodies, immunoelectron microscopy was performed on thin-sections of parasites embedded in the hydrophilic resin Unicryl. The results obtained confirmed that in epimastigotes, gold particles were seen on the cell surface (arrows in Figures 3A, 3B and 3D), the flagellar pocket (Figure 3C), endocytic vacuoles (Figure 3C), the membrane lining the cytostome, (Figure 3D) and in large cytoplasmic vacuoles resembling reservosomes (Figure 3B). Labelling was weaker in the plasma membrane of amastigotes (Figure 3F) and trypomastigotes (Figure 3E), which is in agreement with the results from immunofluorescence (Figures 1B and 2). No labelling of acidocalcisomes, vacuoles that appear empty using conventional electron microscopy [22], was detected in the different stages of T. cruzi (Figure 3B, 3E and 3F). A TcHAf immunogold density histogram was constructed for the following subcellular compartments: flagellar pocket, plasma membrane, mitochondria, nucleus, acidocalcisomes, cytosol (cytoplasm) and reservosomes (Figure 3G). The density of TcHAf gold particles in the plasma membrane and reservosomes was significantly higher than in other compartments. The flagellar pocket was not significantly labelled.
Figure 3. Immunocytochemical localization of H+-ATPase in epimastigotes (A–C), trypomastigote (E) and amastigote (F) stages of T. cruzi.
Particles (15 nm) were used to localize the H+-ATPase. Ac, acidocalcisome; C, cytostome; EV, endocytic vacuole; FP, flagellar pocket; PM, plasma membrane; R, reservosome. Arrowheads show the plasma membrane localization. Scale bars, 0.5 μm. (G) The density of gold particles was determined in the FP, mitochondria (Mito), nucleus, acidocalcisomes (ac), cytosol (cytoplasm) and reservosomes (reserv). Higher densities were found in the plasma membrane and reservosomes. Results are expressed as mean±S.E.M.
Co-localization of the H+-ATPases with cruzipain and lack of co-localization with the V-H+-ATPase
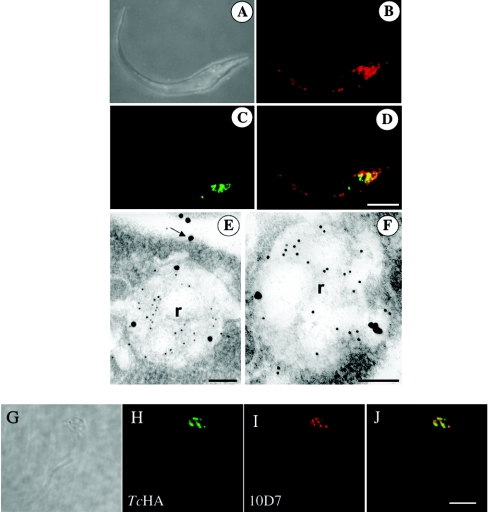
To investigate whether the intracellular labelling obtained using antibodies against TcHAf corresponded to reservosomes, we performed co-localization studies using cruzipain, a known marker of these organelles [33–35]. Immunofluorescence microscopy of epimastigotes showed co-localization in reservosomes of TcHAs (Figure 4B) and cruzipain (Figure 4C), as detected with the polyclonal antibody against TcHAf and monoclonal antibody 212-BH6 against cruzipain respectively. TcHAs were also localized to the plasma membrane (Figure 4B). Figure 4(D) shows the merged image indicating co-localization in yellow. Similar results were obtained by immunoelectron microscopy. Antibodies against cruzipain gave a strong reaction in reservosomes whereas a weaker reaction was observed using antibodies against TcHAf (Figures 4E and 4F), which also labelled the plasma membrane (Figure 4E, arrow). By contrast, monoclonal antibodies against the V-H+-ATPase produced a strongly punctate reaction (Figure 4I; probably corresponding to acidocalcisomes [20]), which did not co-localize with the TcHAs as detected using antibodies against TcHAf (Figures 4H and 4J).
Figure 4. Localization of TcHAs in epimastigotes by confocal laser scanning microscopy (B–D, H–J) and immunoelectron microscopy (E–F).
(B–D) Fluorescence images of epimastigote forms probed with antibody against TcHAf in red (B and D), and antibody against cruzipain in green (C and D). (D) Overlay of (B) and (C) showing co-localization in yellow. (A) Differential interference contrast image of the same cell. Scale bar, 5 μm (A–D). (E, F) Epimastigote forms probed with antibody against TcHAf (20 nm particles) and antibody against cruzipain (10 nm particles) labelling the reservosomes (R). Labelling of the plasma membrane with antibodies against TcHAf was also evident (arrow). Scale bars, 125 nm. (H–J) Fluorescence images of epimastigote forms probed with antibody against TcHAf (at a 1:50 dilution to detect only the reservosomes) in green (H and J) and antibody against the V-H+-ATPase in red (I and J). (J) Overlay of (H) and (I) showing lack of co-localization. Scale bar, 5 μm.
Isolation of reservosomes
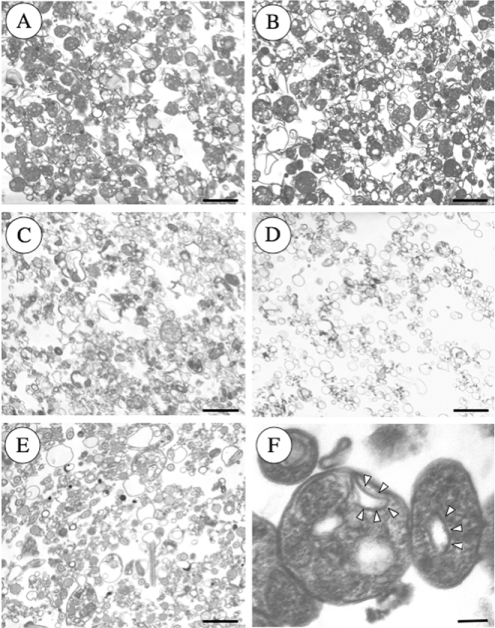
Reservosomes were isolated by the method of Cunha-e-Silva et al. [27] with the modifications indicated under the Materials and methods section. The utility of the method was assessed by assaying marker enzymes (Figure 5). The reservosome fraction (B1) was well resolved from organelle markers for mitochondria (succinate cytochrome c reductase) [29], glycosomes (hexokinase) [36], lysosomes (acid phosphatase) [30], and acidocalcisomes (V-H+-PPase) [23] (Figure 5A), as well as plasma membrane (LPPG, Figure 5B) [37], but was highly enriched in cruzipain (Figure 5B). Examination of the different fractions by electron microscopy (Figure 6) showed great enrichment of reservosomes in fractions B1 (Figure 6A) and B2 (Figure 6B), as previously reported [27]. Fractions B3 (Figure 6C) and B4 (Figure 6D) contained organelles resembling mitochondria, glycosomes and acidocalcisomes, whereas the pellet fraction (Figure 6E) also contained flagella and plasma membrane vesicles. When fraction B1 was examined at higher magnification (Figure 6F) reservosomes were shown to contain internal membranes (arrowheads).
Figure 5. Distribution of H+-ATPase activity from epimastigotes on sucrose density gradients.
ATP-dependent bafilomycin A1 (0.5 μM)-insensitive proton transport activity (C) is concentrated in fractions B1 and B2. Its distribution was compared with that of established organelle markers: acid phosphatase (lysosomes); hexokinase (glycosomes); succinate cytochrome c reductase (mitochondria), and vacuolar proton pyrophosphatase (V-H+-PPase, acidocalcisomes). Scale bars show mean±S.E.M. as percentage of total recovered activity (or total activity for H+-transport (C) in absorbance/min per mg protein from 4 experiments. (B) Western blot analysis showing that cruzipain (CRZPN), which concentrates in reservosomes and lipopeptidephosphoglycan (LPPG, a plasma membrane marker) are in the different fractions of the gradient. Anti-cruzipain reactivity is present in fractions B1 and B2 whereas LPPG reactivity is absent in these fractions.
Figure 6. Representative electron micrographs of the subcellular fractions of epimastigotes.
Note that reservosomes are enriched in B1 (A) and B2 (B) fractions. B3 (C) and B4 (D) contain other vacuoles that resemble mitochondrial fragments and microsomes, whereas the pellet (E) fraction also contains flagella and acidocalcisomes. (F) Shows reservosomes of fraction B1 at higher magnification showing the presence of internal membrane-bound vesicles (arrowheads). Scale bars, 0.5 μm (A–E) and 0.1 μm (F).
H+-ATPase activity of reservosomes
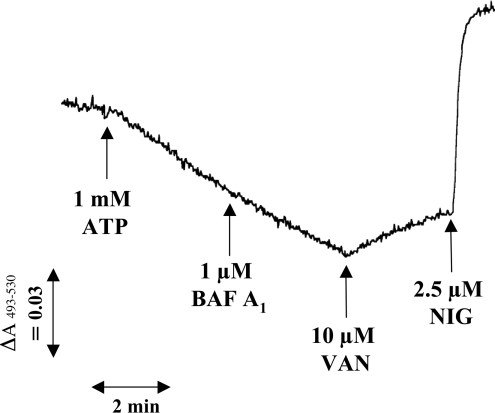
H+-ATPase activity was detectable in the purified reservosome fraction using Acridine Orange (Figures 5C and 7). In this assay, vesicle acidification induced by ATP may be measured as the decrease in absorbance at 493–530 nm [38]. Activity was 49±14 absorbance units/min per μg of protein in the standard assay (3 independent preparations) (Figure 5C). ATP-induced acidification was inhibited by concentrations of sodium o-vanadate (10 μM) previously used to inhibit the recombinant enzyme [2], as shown by the release of Acridine Orange indicating deacidification of the vesicles. This effect demonstrates the alteration of the pH gradient maintained by the P-type H+-ATPase. By contrast H+-uptake was not affected by bafilomycin A1, a specific inhibitor of vacuolar-type H+-ATPases when used at low concentrations [39]. In previous work we demonstrated that bafilomycin A1 [21], as well as concanamycin [10] are effective inhibitors of the V-H+-ATPase of T. cruzi. Addition of nigericin collapsed the pH gradient (Figure 7). No change in absorbance was observed when PPi was used instead of ATP (results not shown). Taken together, these results are compatible with the presence of a P-type H+-ATPase activity in reservosomes.
Figure 7. ATP-induced acidification of reservosomes measured by Acridine Orange uptake.
In the experiment shown, 0.13 mg of reservosome protein (fraction B1) was added per ml of assay medium. ATP (1 mM), bafilomycin A1 (BAF A1, 1 μM), sodium o-vanadate (VAN, 10 μM) and nigericin (NIG, 2.5 μM) were added where indicated.
Different localization of TcHA1 and TcHA2
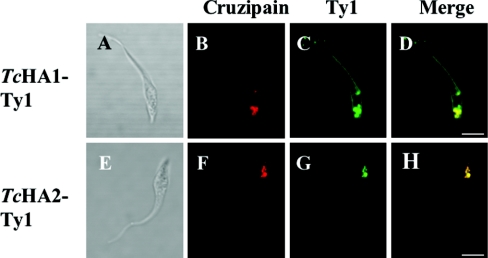
To investigate whether TcHA1 and TcHA2 have different subcellular localizations we inserted a 10 amino acid epitope-tag to the central region of TcHA1 and TcHA2 and expressed these modified proteins in epimastigotes. This 10 amino acid sequence (Glu-Val-His-Thr-Asn-Gln-Asp-Pro-Leu-Asp) is derived from the immunologically well characterized major structural protein of S. cerevisiae Ty1 virus-like protein and has been used previously to study protein targeting and organelle biogenesis [25], as well as the localization of different Ca2+-ATPase isoforms in T. brucei [40]. Polyclonal antibodies against cruzipain (Figures 8B and 8F) were used as controls. We observed strong labelling of intracellular vacuoles with monoclonal antibody BB2 against the Ty1 epitope in epimastigotes transfected with both TcHA1-Ty1 (Figure 8C) and TcHA2-Ty1 (Figure 8G), whereas labelling was also present in the plasma membrane when the antibodies were used against epimastigotes transfected with TcHA1-Ty1 (Figure 8C). Co-localization was evident in reservosomes (Figures 8D and 8H). No immunofluorescence was observed in control wild-type parasites incubated with polyclonal antibody against Ty1 (results not shown).
Figure 8. Epimastigote forms expressing TcHA1-Ty1 and TcHA2-Ty1 fusion proteins.
Epimastigotes were transfected with expression constructs pTEX-TcHA1-Ty1 (A–D) and pTEX-TcHA2-Ty1 (E–H) respectively. Stable transformants were labelled with monoclonal antibody BB2 anti-Ty1 in green (C and G) and polyclonal antibody against cruzipain in red (B and F). TcHA1-Ty1 localized to the reservosomes and the plasma membrane whereas TcHA2-Ty1 localized only to the reservosomes. (D) and (H) are overlays of (B) and (C), and (F) and (G) respectively, showing co-localization in yellow. (A) and (E) are differential interference contrast images of the same cells. Scale bars, 5 μm.
DISCUSSION
In the present study, we have demonstrated that P-type ATPases are present in the plasma membrane and intracellular compartments of the three different stages of T. cruzi (Figures 1–3). The main intracellular compartment containing these ATPases in epimastigotes was identified as the reservosome. This was accomplished by immunofluorescence assays (Figures 1, 2 and 4) and by immunoelectron microscopy (Figure 4) showing their co-localization with cruzipain, and by subcellular fractionation (Figures 5–7). ATP-dependent proton transport by isolated reservosomes was sensitive to vanadate and insensitive to bafilomycin A1, in agreement with the localization of P-type H+-ATPases in these organelles (Figures 5C and 7). Our results also indicate that both P-type H+-ATPases TcHA1 and TcHA2 [2] are located in the reservosomes whereas one of them (TcHA1) is additionally present in the plasma membrane (Figure 8). Immunogold electron microscopy allowed the detection of the H+-ATPases in other components of the endocytic pathway of T. cruzi such as the cytostome and endosomal vesicles (Figure 3) suggesting that these H+-ATPases are involved in acidification of the endocytic pathway of T. cruzi. This is the first report of the presence of P-type H+-ATPases in the endocytic pathway of any eukaryotic cell. An internal staining was also observed in Dictyostelium discoideum using antibodies against a P-type H+-ATPase although this was attributed to cross-reaction of the antiserum with other proteins [11].
Acidification of the endocytic pathway in all eukaryotic cells investigated until now, occurs through the activity of a vacuolar-type H+-ATPase [41] but this does not appear to be the case with T. cruzi. The V-H+-ATPase was not found in the flagellar pocket of T. cruzi [21], which is a region of endocytosis and exocytosis in trypanosomatids, and was found instead localized to the plasma membrane and acidocalcisomes [20]. In addition, the V-H+-ATPase did not co-localize with the TcHAs (Figures 4G–4J).
The endocytic pathway of T. cruzi is highly polarized. The cytostome and the flagellar pocket are the only sites of uptake [16]. The cytostome opens near the flagellar pocket and invaginates deeply forming the cytopharynx [42]. Endocytic smooth vesicles bud off from either the flagellar pocket [17] or the cytopharynx [16] and endocytic cargo is then transferred to an endosomal network, composed of many tubules and vesicles extending along the cell body, until reaching the posterior end of the parasite [43]. Finally, the nutrients are delivered to reservosomes. The distinction between reservosomes and lysosomes is not clear since identification of the latter organelles has been difficult, due in part to the absence of typical lysosomal markers such as Lamp 1 in trypanosomatids.
Interestingly, the cytostome and cytopharynx are already known to be acidic [43], which is in agreement with the presence of proton pumps in these structures (Figure 3D). Since acidification of the endocytic pathway in T. cruzi is carried out through a P-type H+-ATPase that is absent in mammalian cells, our results suggest that this enzyme could represent an attractive target for novel anti-trypanosomatid chemotherapy.
Acknowledgments
We thank Keith Gull (University of Oxford, U.K.) for the antibody BB2 against Ty1, Julio Scharfstein (Federal University of Rio de Janeiro, Brazil) for antibodies against cruzipain, Wen Yan for her initial help in this project and Linda Brown for technical assistance. This work was supported by U.S. (N.I.H.) National Institutes of Health Grant AI-23259 to R.D. This investigation was conducted in part in a facility constructed with support from Research Facility Improvement Grant Number C06 RR16515–01 from the National Center for Research Resources, N.I.H.
References
- 1.Urbina J. A., Docampo R. Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol. 2003;19:495–501. doi: 10.1016/j.pt.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Luo S., Scott D. A., Docampo R. Trypanosoma cruzi H+-ATPase 1 (TcHA1) and 2 (TcHA2) genes complement yeast mutants defective in H+ pumps and encode plasma membrane P-type H+-ATPases with different enzymatic properties. J. Biol. Chem. 2002;277:44497–44506. doi: 10.1074/jbc.M202267200. [DOI] [PubMed] [Google Scholar]
- 3.Axelsen K. B., Palmgren M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 4.Møller J. V., Juul B., le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim. Biophys. Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 5.Monk B. C., Perlin D. S. Fungal plasma membrane proton pumps as promising new antifungal targets. Crit. Rev. Microbiol. 1994;20:209–223. doi: 10.3109/10408419409114555. [DOI] [PubMed] [Google Scholar]
- 6.Michelet B., Boutry M. The plasma membrane H+-ATPase (a highly regulated enzyme with multiple physiological functions) Plant Physiol. 1995;108:1–6. doi: 10.1104/pp.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanderHeyden N., Benaim G., Docampo R. The role of a H(+)-ATPase in the regulation of cytoplasmic pH in Trypanosoma cruzi epimastigotes. Biochem. J. 1996;318:103–109. doi: 10.1042/bj3180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.VanderHeyden N., Docampo R. Intracellular pH in mammalian stages of Trypanosoma cruzi is K+-dependent and regulated by H+-ATPases. Mol. Biochem. Parasitol. 2000;105:237–251. doi: 10.1016/s0166-6851(99)00184-x. [DOI] [PubMed] [Google Scholar]
- 9.VanderHeyden N., Docampo R. Proton and sodium pumps regulate the plasma membrane potential of different stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002;120:127–139. doi: 10.1016/s0166-6851(01)00444-3. [DOI] [PubMed] [Google Scholar]
- 10.Scott D. A., Docampo R. Characterization of isolated acidocalcisomes of Trypanosoma cruzi. J. Biol. Chem. 1998;275:24215–24221. doi: 10.1074/jbc.M002454200. [DOI] [PubMed] [Google Scholar]
- 11.Bockelmann C., Liss H., Weiler E. W. Evidence for a functional P-type H+-ATPase at the rough endoplasmic reticulum of Bronia dioica Jacq. tendrils. J. Plant Physiol. 1998;152:194–198. [Google Scholar]
- 12.Ferreira T., Mason A. B., Slayman C. W. The yeast Pma1 proton pump: a model for understanding the biogenesis of plasma membrane proteins. J. Biol. Chem. 2001;276:29613–29616. doi: 10.1074/jbc.R100022200. [DOI] [PubMed] [Google Scholar]
- 13.Nakamoto R. K., Rao R., Slayman C. W. Expression of the yeast plasma membrane [H+]ATPase in secretory vesicles. A new strategy for directed mutagenesis. J. Biol. Chem. 1991;266:7940–7949. [PubMed] [Google Scholar]
- 14.Ferreira T., Mason A. B., Pypaert M., Allen K. E., Slayman C. W. Quality control in the yeast secretory pathway: a misfolded PMA1 H+-ATPase reveals two checkpoints. J. Biol. Chem. 2002;277:21027–21040. doi: 10.1074/jbc.M112281200. [DOI] [PubMed] [Google Scholar]
- 15.de Souza W. Basic cell biology of Trypanosoma cruzi. Current Pharmac. Design. 2002;8:269–285. doi: 10.2174/1381612023396276. [DOI] [PubMed] [Google Scholar]
- 16.Soares M. J., de Souza W. Endocytosis of gold-labeled proteins and LDL by Trypanosoma cruzi. Parasitol. Res. 1991;77:461–468. doi: 10.1007/BF00928410. [DOI] [PubMed] [Google Scholar]
- 17.Soares M. J., Souto-Padrón T., de Souza W. Identification of a large pre-lysosomal compartment in the pathogenic protozoon Trypanosoma cruzi. J. Cell Sci. 1992;102:157–167. doi: 10.1242/jcs.102.1.157. [DOI] [PubMed] [Google Scholar]
- 18.Scott D. A., Docampo R., Dvorak J. A., Shi S., Leapman R. D. In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J. Biol. Chem. 1997;272:28020–28029. doi: 10.1074/jbc.272.44.28020. [DOI] [PubMed] [Google Scholar]
- 19.Soares M. J., de Souza M. F., de Souza W. Ultrastructural visualization of lipids in trypanosomatids. J. Protozool. 1987;34:199–203. doi: 10.1111/j.1550-7408.1987.tb03160.x. [DOI] [PubMed] [Google Scholar]
- 20.Lu H.-G., Zhong L., de Souza W., Benchimol M., Moreno S. N. J., Docampo R. Ca2+ content and expression of an acidocalcisomal calcium pump are elevated in intracellular forms of Trypanosoma cruzi. Mol. Cell. Biol. 1998;18:2309–2323. doi: 10.1128/mcb.18.4.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benchimol M., de Souza W., VanderHeyden N., Zhong L., Lu H.-G., Moreno S. N. J., Docampo R. Functional expression of a vacuolar-type H+-ATPase in the plasma membrane and intracellular vacuoles of Trypanosoma cruzi. Biochem. J. 1998;332:695–672. doi: 10.1042/bj3320695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Docampo R., de Souza W., Miranda K., Rohloff P., Moreno S. N. J. Acidocalcisomes-conserved from bacteria to man. Nat. Rev. Microbiol. 2005;3:251–261. doi: 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- 23.Scott D. A., de Souza W., Benchimol M., Zhong L., Lu H. G., Moreno S. N., Docampo R. Presence of a plant-like proton-pumping pyrophosphatase in acidocalcisomes of Trypanosoma cruzi. J. Biol. Chem. 1998;273:22151–22158. doi: 10.1074/jbc.273.34.22151. [DOI] [PubMed] [Google Scholar]
- 24.Bone G. J., Steinert M. Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature (London) 1956;178:308–309. doi: 10.1038/178308a0. [DOI] [PubMed] [Google Scholar]
- 25.Bastin P., Bagherzadeh Z., Matthews K. R., Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- 26.Murta A. C., Persechini P. M., Padron T. de S., de Souza W., Guimaraes J. A., Scharfstein J. Structural and functional identification of GP57/51 antigen of Trypanosoma cruzi as a cysteine proteinase. Mol. Biochem. Parasitol. 1990;43:27–38. doi: 10.1016/0166-6851(90)90127-8. [DOI] [PubMed] [Google Scholar]
- 27.Cunha-e-Silva N. L., Atella G. C., Porto-Carreiro I. A., Morgado-Diaz J. A., Pereira M. G., de Souza W. Isolation and characterization of a reservosome fraction from Trypanosoma cruzi. FEMS Microbiol. Lett. 2002;214:7–12. doi: 10.1111/j.1574-6968.2002.tb11317.x. [DOI] [PubMed] [Google Scholar]
- 28.Cannata J. J. B., Valle E., Docampo R., Cazzulo J. J. Subcellular localization of phosphoenolpyruvate carboxykinase in the trypanosomatids Trypanosoma cruzi and Crithidia fasciculata. Mol. Biochem. Parasitol. 1982;6:151–160. doi: 10.1016/0166-6851(82)90074-3. [DOI] [PubMed] [Google Scholar]
- 29.Sottocasa G. L., Kuylenstierna B., Ernster L., Bergstrand A. An electron-transport system associated with the outer membrane of liver mitochondria. A biochemical and morphological study. J. Cell Biol. 1967;32:415–438. doi: 10.1083/jcb.32.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett A. J., Heath M. F. In: Lysosomes: A Laboratory Handbook. Dingle J. T., editor. Amsterdam: Elsevier/North-Holland Biomedical Press; 1977. pp. 36–40. [Google Scholar]
- 31.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Kelly J. M., Ward H. M., Miles M. A., Kendall G. A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids. Res. 1992;20:3963–3969. doi: 10.1093/nar/20.15.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Souto-Padrón T., Campetella O. E., Cazzulo J. J., de Souza W. Cysteine proteinase in Trypanosoma cruzi: immunocytochemical localization and involvement in parasite-host cell interaction. J. Cell Sci. 1990;96:485–490. doi: 10.1242/jcs.96.3.485. [DOI] [PubMed] [Google Scholar]
- 34.Engel J. C., Doyle P. S., Palmer J., Hsieh I., Bainton D. F., McKerrow J. H. Cysteine protease inhibitors alter Golgi complex ultrastructure and function in Trypanosoma cruzi. J. Cell Sci. 1998;111:596–606. doi: 10.1242/jcs.111.5.597. [DOI] [PubMed] [Google Scholar]
- 35.Engel J. C., Torres C., Hsieh I., Doyle P. S., McKerrow J. H. Upregulation of the secretory pathway in cysteine protease inhibitor-resistant Trypanosoma cruzi. J. Cell Sci. 2000;113:1345–1354. doi: 10.1242/jcs.113.8.1345. [DOI] [PubMed] [Google Scholar]
- 36.Taylor M. B., Gutteridge W. E. Trypanosoma cruzi: subcellular distribution of glycolytic and some related enzymes of epimastigotes. Exp. Parasitol. 1987;63:84–97. doi: 10.1016/0014-4894(87)90081-6. [DOI] [PubMed] [Google Scholar]
- 37.de Lederkremer R. M., Lima C., Ramirez M. I., Casal O. L. Structural features of the lipopeptidophosphoglycan from Trypanosoma cruzi common with the glycophosphatidylinositol anchors. Eur. J. Biochem. 1990;192:337–345. doi: 10.1111/j.1432-1033.1990.tb19232.x. [DOI] [PubMed] [Google Scholar]
- 38.Palmgren M. G. Acridine orange as a probe for measuring pH gradients across membranes: mechanism and limitations. Anal. Biochem. 1991;192:316–321. doi: 10.1016/0003-2697(91)90542-2. [DOI] [PubMed] [Google Scholar]
- 39.Bowman E. J., Siebers A., Altendorf K. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl Acad. Sci. U.S.A. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo S., Rohloff P., Cox J., Uyemura S. A., Docampo R. Trypanosoma brucei plasma membrane-type Ca2+-ATPase 1 (TbPMC1) and 2 (TbPMC2) genes encode functional Ca2+-ATPases localized to the acidocalcisomes and plasma membrane, and essential for Ca2+ homeostasis and growth. J. Biol. Chem. 2004;279:14427–14439. doi: 10.1074/jbc.M309978200. [DOI] [PubMed] [Google Scholar]
- 41.Forgac M. Structure and properties of the vacuolar (H+)-ATPases. J. Biol. Chem. 1999;274:12951–12954. doi: 10.1074/jbc.274.19.12951. [DOI] [PubMed] [Google Scholar]
- 42.Milder R., Deane M. P. The cytostome of Trypanosoma cruzi and T conorhini. J. Protozool. 1969;16:730–737. doi: 10.1111/j.1550-7408.1969.tb02335.x. [DOI] [PubMed] [Google Scholar]
- 43.Porto-Carreiro I., Attias M., Miranda K., de Souza W., Cunha-e-Silva N. Trypanosoma cruzi epimastigote endocytic pathway: cargo enters the cytostome and passes through an early endosomal network before storage in reservosomes. Eur. J. Cell Biol. 2000;79:858–869. doi: 10.1078/0171-9335-00112. [DOI] [PubMed] [Google Scholar]