Abstract
In eukaryotic cells, several mRNAs including those of c-myc and c-fos are localized to the perinuclear cytoplasm and associated with the cytoskeleton. The localization element of c-myc mRNA is present within its 3′UTR (3′-untranslated region) but the precise nature of this signal has remained unidentified. Chemical/enzymatic cleavage with RNases (ribonucleases) and lead have identified single-stranded and double-stranded regions in RNA transcripts of nucleotides 194–280 of the c-myc 3′UTR. Combined with computer predicted structure these results indicate that this region folds so that part of it forms a stem-loop structure. A mutation, that has been previously shown to prevent localization, leads to a different secondary RNA structure in this region as indicated by altered cleavage patterns. Competitive gel-retardation assays, using labelled transcripts corresponding to nucleotides 205–280 of c-myc 3′UTR, and fibroblast extracts revealed that the stem-loop region was sufficient for RNA–protein complex formation. In situ hybridization studies in cells transfected with reporter constructs, in which all or parts of the region corresponding to this stem-loop were linked to β-globin, indicated that this region was sufficient for localization and that deletion of the nucleotides corresponding to the proposed upper-stem or terminal loop prevented localization. Our hypothesis is that an AU-rich stem-loop structure within nt 222–267 in the c-myc 3′UTR forms the perinuclear localization signal. Bioinformatic analysis suggests that this signal shares features with 3′UTRs of other localized mRNAs and that these features may represent a conserved form of signal in mRNA localization mechanisms.
Keywords: c-myc, mRNA localization signal, 3′untranslated region (3′UTR), RNA secondary structure, perinuclear localization, trafficking
Abbreviations: CMCT, 1-cyclohexyl-3-(2-morpholino-ethyl)carbodi-imide metho-p-toluene sulphonate; DMEM, Dulbecco's minimal Eagle's medium; EMSA, electrophoretic mobility-shift assay; FBS, foetal bovine serum; Ltk, leukocyte tyrosine kinase; MT-1, metallothionein-1; RNase, ribonuclease; TBE, Tris/borate/EDTA; 3′UTR, 3′-untranslated region
INTRODUCTION
In eukaryotic cells, mRNA localization to different regions in the cytoplasm provides a mechanism for synthesis of proteins close to where they are required [1–3]. For example, in fibroblasts some mRNAs, such as those encoding β-actin and creatine kinase isoform M, are transported to the cell periphery [4,5], whereas c-myc, c-fos, MT-1 (metallothionein-1) and vimentin mRNAs remain localized to the perinuclear cytoplasm [6–9]. The mRNAs localized around the nucleus, including c-myc, are associated with cytoskeletal-bound polysomes or the cytoskeleton itself [10]. Such perinuclear localization has been shown to be required for the nuclear localization of MT-1 during the G1/S phase transition in the cell cycle [11]. c-myc and c-fos are proto-oncogenes that encode nuclear transcription factors critical in the control of cell growth; therefore localization of their mRNAs around the nucleus could play an important role in allowing efficient nuclear import of the newly synthesized proteins.
In the majority of cases, localization of mRNAs is due to cis-acting signals, or ‘zipcodes’, within the 3′UTR (3′-untranslated region) [2–9]. Deletion analysis and mutagenesis suggest that the entire 3′UTR is not necessary for localization but rather that one or more restricted regions or localization elements are sufficient. However, the nature of such cis-acting signals has in most cases proved elusive [3] and it has been difficult to define exactly what features make up localization signals. This is because the signals appear to be variable in length, structure and complexity with some appearing to contain short single or repetitive motifs and others multi-component units spanning hundreds of nucleotides. For example, a relatively short region containing a CAC repeat appears sufficient to localize β-actin mRNA [12] and a short 21 nt region is adequate for myelin basic protein mRNA transport [13] whereas a complex signal is required in the case of bicoid [14].
In the case of mRNAs that demonstrate a perinuclear localization, the precise nature of the cis-acting signal within the 3′UTR has not been defined. Perinuclear localization of c-myc mRNA has been shown to require a signal that resides between nt 194–280 in the 3′UTR [15]. However, the sequences, motifs or structure that form the cis-acting localization signal within this section are not known. Since the signal resides in such a relatively small region, further analysis of the c-myc 3′UTR provides an opportunity to investigate the nature of a cis-acting perinuclear localization signal in detail. In the present study RNA structure and deletion analysis together with in situ hybridization and gel retardation assays were used to define the nature of the cis-acting RNA localization signal present in the c-myc 3′UTR.
MATERIALS AND METHODS
Gene constructs
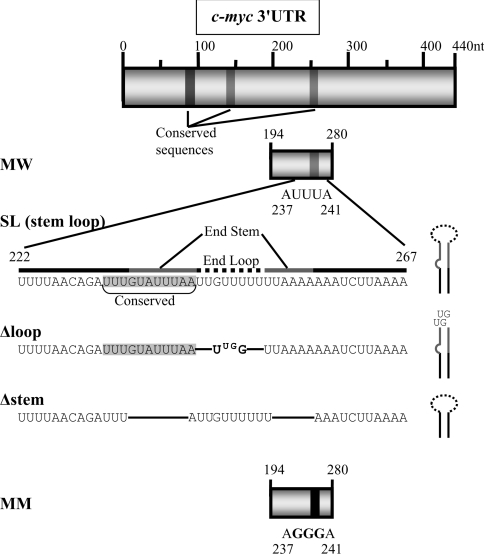
A series of gene constructs were made containing sections of the wild-type c-myc 3′UTR and various deletions (Figure 1). pcglobinSL contains the region corresponding to the proposed stem-loop in the c-myc 3′UTR (nt 222–267) linked to the β-globin coding sequence. The entire rabbit β-globin cDNA coding sequence was amplified by PCR from pcKGG [8] using oligonucleotides 1 and 2 (sequences shown in supplementary Table http://www.BiochemJ.org/bj/392/bj3920475add.htm) as forward and reverse primers respectively. The reverse primer included bases corresponding to the sequence of nt 222–267 of the mouse c-myc 3′UTR so that amplification resulted in this c-myc sequence being attached to the 3′ end of the β-globin coding sequence. In order to introduce specific KpnI and XbaI restriction sites, this product was then used as the template in a second PCR amplification with oligonucleotides 3 and 4 as forward and reverse primers. The product of this second PCR was digested with KpnI and XbaI restriction enzymes and ligated into pcDNA3 (Invitrogen).
Figure 1. Details of regions in c-myc 3′UTR used in gene constructs.
Diagram of the c-myc 3′UTR, highlighting regions of sequence conservation and regions within nucleotides 194–280, that were used or deleted in various gene constructs. MW refers to the wild-type sequence 194–280 containing the conserved AUUUA at nt 237–241; MM is a mutated sequence in which AUUUA is replaced by AGGGA [15]. SL is the sequence 222–267 that corresponds to the predicted stem-loop region. Δstem and Δloop contain deletions within this SL region and their predicted effects on secondary structure are illustrated schematically.
Mutated c-myc 3′UTR stem-loop sequences were attached to the β-globin coding region using a strategy that involved annealing of synthetic complementary oligonucleotides corresponding to different fragments of the c-myc 3′UTR, followed by ligation into pcKGG and removal of the β-globin 3′UTR by site-directed mutatgenesis. The complementary oligonucleotides (5 and 6 to generate pcglobinΔloop; 7 and 8 to generate pcglobinΔstem) were annealed by mixing, heating to 95 °C and then allowed to cool slowly. This produced double-stranded oligonucleotides encoding the mutant c-myc 3′UTR sequences with added XbaI-cohesive ends and these were inserted into the XbaI site of pcKGG [8] by ligation. Removal of the β-globin 3′UTR and vector sequence was achieved by a modification of the site-directed mutagenesis method (Stratagene) using the Pfu Turbo enzyme with the relevant oligonucleotide 5–8 as forward primer and oligonucleotide 9 as reverse primer, both of which were phosphorylated at the 5′ end using the polynucleotide kinase. PCR amplification produced a product which was then circularized in a ligation reaction and the original plasmid was removed by digestion with DpnI enzyme. Correct orientation of the oligonucleotide sequences was confirmed by sequencing.
In vitro transcription of labelled and unlabelled RNAs
Sequences from the mouse c-myc 3′UTR corresponding to bases 194–280 and containing a conserved AUUUA (MW), and to bases 194–280 with a 3 base change within the AUUUA sequence (MM), were transferred from vectors PM13Δ3, pSVc-myc1 and pSVc-mycSK/Cl [15] into the polylinker of pBluescriptII SK (Stratagene) so as to maintain the RNA polymerase sites. Templates containing the T7 promoter sequence for transcription of the mouse c-myc 3′UTR nucleotides 194–280 (MW or MM) were generated by PCR from the latter plasmid using primers 10 (forward) and 11 (reverse). Vector sequences 3′ of the T7 promoter (including a tract of 7 cysteine residues) and bases 194–205 of the 3′UTR sequence were also removed from the MW construct by digestion with XhoI and KpnI to generate Δ205 which contains bases 205–280 of the 3′UTR.
Templates for transcription of nucleotides corresponding to the stem-loop of the mouse c-myc 3′UTR (nt 222–267) or the mutant sequences (Δloop and Δstem) linked to the last 100 nt of the β-globin coding region were generated by PCR from pcglobinSL, pcglobinΔloop and pcglobinΔstem using the T7 promoter-containing oligonucleotide 12 as forward primer and the relevant reverse primer (oligos 2, 6 and 8). The template to generate control transcripts corresponding to the last 100 nt of the β-globin coding region were made by PCR using primers 12 (forward) and 13 (reverse). Finally, templates for transcription of the mouse c-fos 3′UTR 144 nt region which includes a perinuclear localization signal were generated by PCR from the plasmid pIREglo-fos [7] using primers 14 (forward) and 15 (reverse). All PCR products were purified using QIAquick columns (Qiagen). Unlabelled transcripts were produced using the Megashortscript kit (Ambion). For EMSA (electrophoretic mobility-shift assay), Δ205 transcripts were labelled with [α-32P]CTP (800 Ci/mmol) using the MAXIscript kit (Ambion). Unlabelled and labelled RNAs were then extracted with phenol/chloroform and precipitated in ethanol. Incorporation of the radionucleotide was assessed by scintillation counting and unlabelled RNA quantified spectrophotometrically. RNA integrity was controlled by denaturing gel electrophoresis with comparison against known standards.
EMSA
Ltk− (leukocyte tyrosine kinase null)-fibroblasts were grown in DMEM (Dulbecco's minimal Eagle's medium) supplemented with 10% FBS (foetal bovine serum) and S100 protein extracts were prepared as described previously [16,19]. Gel-retardation reactions were carried out using 2 μg of S100 extract and 12 fmol of 32P-labelled RNA (heated to 70 °C and allowed to cool and refold slowly) in binding buffer (30 mM Tris/HCl, pH 7.6, 5 mM MgCl2, 40 mM NaCl, 2 mM dithiothreitol and EDTA-free protease inhibitor cocktail) in a total volume of 8 μl, at 22 °C for 15 min. For competition experiments, labelled and unlabelled RNA were added simultaneously. Products were digested with RNase T1 by adding 40 units of enzyme to the binding-reaction and incubation continued for 5 min before the addition of 2 μl of 20% (w/v) Ficoll and separation on 5% (w/v) nondenaturing polyacrylamide gels [79:1, 0.5×TBE (1×TBE=45 mM Tris/borate/1 mM EDTA) at 20 V/cm for 3 h at 4 °C]. Gels were dried and analysed by autoradiography.
In situ hybridization
Studies of RNA distribution were carried out in cells grown in multi-well chamber slides so that the different cell lines could be studied under identical conditions and the quantification of staining be directly comparable. In situ hybridization was carried out using digoxigenin-labelled riboprobes as described previously [7,17]. Cells were washed in PBS, fixed in 4% (w/v) paraformaldehyde in PBS for 10 min at 4 °C, dehydrated in 70% (v/v) ethanol and permeabilized using 4% paraformaldehyde/0.2% Triton X-100 in PBS for 10 min. After pre-hybridization in 2×SSC containing 50% (v/v) formamide for 10 min at room temperature, hybridization was carried out overnight at 55 °C using 200 ng of digoxigenin-labelled antisense riboprobe. The antisense globin probe was a 511 nt ApaI/BamHI fragment generated using SP6 RNA polymerase as described previously [17]. In addition control sense probes were generated from the same fragment using T7 RNA polymerase and other controls were carried out using either no probe in the hybridization mixture or using RNase A treatment of the samples. After hybridization, cells were washed in 5× SSC for 30 min at room temperature, and then for 30 min in 2× SSC/50% formamide at 55 °C, before treatment with 20 μg/ml RNase A in wash buffer (10 mM Tris/HCl, pH 7.5, containing 0.4 M NaCl and 5 mM EDTA) for 30 min at 37 °C to remove non-specifically bound probe. After a brief wash in buffer and two further washes in 2× SSC, the bound probe was detected by incubation with alkaline phosphate-linked anti-digoxigenin IgG (Roche) and then with 4-Nitro Blue Tetrazolium for 16 h (Roche). Staining was quantified by examining fields of view at random and assigning localization characteristics (presence or absence of a distinct ring of perinuclear staining) to approximately 100 cells in each of at least 3 separate experiments. Cells were classified as exhibiting perinuclear localization of mRNA or no localization.
RNA secondary structure analysis
RNA containing the c-myc 3′UTR nt 194–280 (MW or MM version) or the c-fos 3′UTR 144 nt region implicated in localization [7] was purified on an 8% polyacrylamide gel in 7 M urea. Full-size RNA was eluted from the gel in 0.5 M ammonium acetate, 1 M EDTA, 0.1% (w/v) SDS in a diethyl pyrocarbonate-treated Eppendorf tube overnight at room temperature and precipitated using ethanol. RNA was labelled at the 5′-end with [γ-32P]ATP using the KinaseMax kit (Ambion) according to the manufacturer's instructions. After purification using spin-column chromatography (Chromaspin-30 columns; B.D. Biosciences), 32P-labelled RNA (approx. 105 c.p.m.) was supplemented with 2–5 μg of yeast tRNA and incubated separately with RNase T1 (1 unit/μl, Ambion), V1 (0.1 unit/μl, Ambion), T2 (30 units/μl, Sigma) or A (1 μg/μl, Ambion) at room temperature in 20 mM Tris/HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl according to the conditions indicated in the legend of the Figures. For lead-mediated cleavage, RNA was supplemented with 4 μg of yeast tRNA and incubated for 5 min at room temperature with 2 or 5 mM Pb2+ in 20 mM Hepes/NaOH (pH 7.5), 7 mM magnesium acetate, 50 mM potassium acetate. Enzymatic and lead cleavage reactions were stopped by chilling the samples on ice and addition of 20 μl of inactivation/precipitation solution from Ambion (plus 2 μl of 100 mM EDTA for the lead samples). RNA was ethanol precipitated in the presence of 1 μg of Glycoblue (Ambion), air-dried and resuspended in loading buffer. RNA cleavage products were separated on a 10% polyacrylamide gel (19:1, 7 M urea and 1×TBE). Gels were analysed by autoradiography after drying. Alkaline and G-specific RNase T1 ladders were generated in parallel to identify the cleavage positions. Limited alkaline hydrolysis of 32P-labelled RNA was performed by incubation in sodium carbonate (pH 9) at 90 °C for 4 min. Ladders of G were produced by pre-incubation of the labelled RNA at 55 °C for 10 min in 20 mM sodium citrate (pH 5), 1 mM EDTA, 7 M urea and a further 10 min incubation at 55 °C in the presence of 0.5 unit of RNase T1. For primer extension analysis, unlabelled RNA was subjected to modification by CMCT [1-cyclohexyl-3-(2-morpholino-ethyl)carbodi-imide metho-p-toluene sulphonate] using the standard procedure described by Ehresmann et al. [18]. Briefly, 1 pmol of RNA was supplemented with 1 μg of yeast tRNA and incubated in the presence of CMCT (10 mg/ml) for 20 min at 30 °C in 50 mM borate/NaOH (pH 8), 20 mM Mg acetate and 300 mM KCl. The reaction was stopped by ethanol precipitation of RNA. Detection of the modified nucleotides was accomplished by hybridization and extension of the 5′-end-labelled 5′-GGAGCTCGCCCTATTTAC-3′ primer (approx. 105 c.p.m.) that annealed to the RNA downstream of c-myc 86 nt region. Positions of the modifications were identified by running in parallel a didesoxysequencing reaction. The reverse transcripts were separated on a 10% sequencing gel.
RESULTS
Sequence and structural analysis of the region in c-myc 3′UTR responsible for perinuclear localization
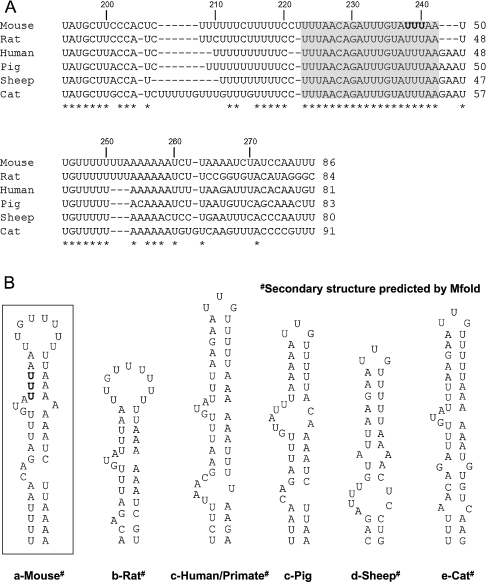
Previous work has shown that the nucleotides between nt 194 and 280 within the c-myc 3′UTR are sufficient for both localization of β-globin reporter transcripts to the perinuclear cytoplasm [15] and binding of annexin A2 [19]. Sequence comparison of this section of c-myc 3′UTR between various mammalian species (Figure 2A) shows an interspecies identity of 58%. In particular, a block of 20 consecutive nucleotides (shaded in grey) appears to be totally conserved between the different species, suggesting that this region of the 3′UTR is functionally important. Interestingly, this block contains the conserved AUUUA motif, mutation of which to AGGGA has been previously shown to abolish perinuclear localization [15]. The region of the c-myc 3′UTR from nt 194–280 was further analysed by applying the Mfold program [20] in order to search for putative secondary structure(s). A well-defined stem–loop structure rich in AU base pairs and containing two bulges was predicted to form in the mouse sequence (Figure 2B, within the square). A comparable structure displaying similar features (AU base pair content and presence of bulges) could also be obtained in the sequences of the other species either using Zuker's algorithm [20] (rat, primate, sheep and cat) or by visual inspection (human and pig).
Figure 2. Sequence comparison and folding prediction for the c-myc 3′UTR nt region 194–280 in various mammalian species.
(A) Mouse (reference), rat, human, pig, sheep and cat sequences from the National Center for Biotechnology Information were aligned and insertions introduced (−) so as to maximize identity. Nucleotides are numbered based on the mouse sequence starting at position 194 of the 3′UTR. 100% interspecies identity is indicated by an asterisk under the bottom line. (B) Secondary structure predicted for the different species sequences.
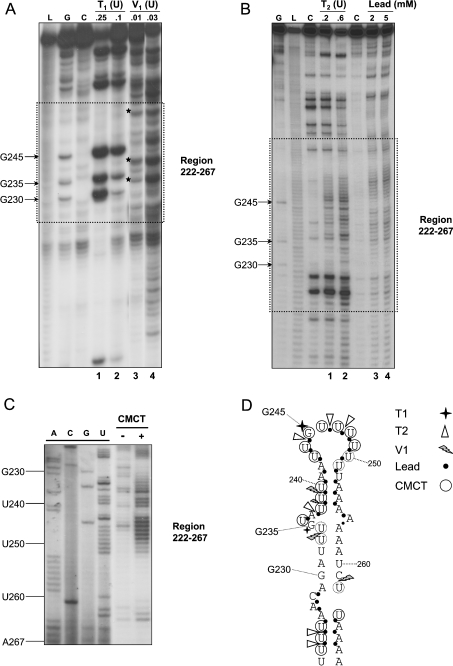
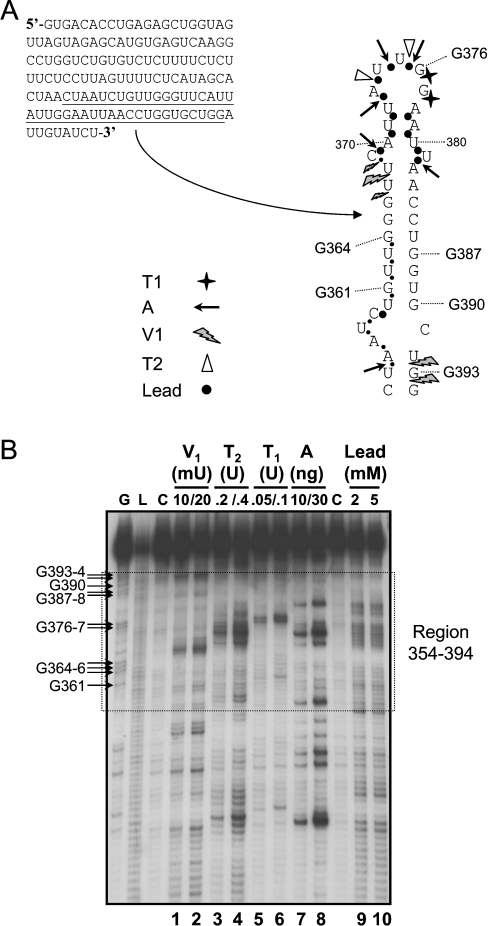
In order to determine if this region of the c-myc 3′UTR can form such a stem–loop structure, enzymatic and chemical probing was carried out on RNA containing nt 194–280 of the mouse c-myc 3′UTR (Figure 3). This region corresponds to the MW section shown previously to have a localization function [15]. The 5′-end-labelled RNA was analysed for the presence of single- and double-stranded regions using RNases (T1, T2 and V1) and Pb2+ while participation of U (and to a lesser extent G) residues in base pairing was further investigated using CMCT modification. Several separate experiments were conducted for each probe and representative gels are presented in Figures 3(A), 3(B) and 3(C). We observed that within the region nt 222–267 of c-myc 3′UTR, the guanine G245 was the major cleavage site of the nuclease T1 with a less sensitive cleavage at G235 (Figure 3A, lanes 1 and 2), suggesting that these two Gs are in single-stranded regions. By contrast, three preferential positions, namely U233, U239 and C261 (each indicated by an asterisk in Figure 3A, lane 3), were consistently cleaved by the double-strand-specific nuclease V1, suggesting the presence of these nucleotides in a double-stranded, helical configuration. The presence of single-stranded regions was probed by using lead and the nuclease T2 in parallel (Figure 3B). Three regions between nt 222–267 were clearly sensitive to Pb2+ cleavage (nt 223–228, 236–255 and 263–265; Figure 3B, lanes 3 and 4). Cleavage by nuclease T2 within these regions (at nt U223–U224, A237, A241, U244 and U246–U247; Figure 3B, lanes 1 and 2) is consistent with the sensitivity to Pb2+. In addition, only nucleotides U232, U252 and U260 showed some protection against modification by CMCT, suggesting they are base-paired, while all other U residues present in the nt 222–267 region demonstrated some reactivity towards the chemical (Figure 3C). As illustrated in Figure 3(D), these data are compatible with several features of the folding predicted by Mfold, in which there is a terminal loop, an upper bulge and a stem region. For example, the presence of the terminal loop and the upper bulge are largely corroborated by the T1, T2 and Pb2+ cleavage results, as well as CMCT modifications. Furthermore, the major stem structure involving nt 229–234 and complementary sequences 257–262 is consistent with the observed protection against these single-strand-specific probes and the presence of two major cleavage sites (U233 and C261) by the nuclease V1. The apparently contradictory results concerning the existence of the upper stem of the structure (the presence of V1 cuts alongside Pb2+ cleavage and CMCT modifications) suggest a significant degree of ‘looseness’ in the upper part of this structure starting from the upper bulge. Overall, the probing results and computer predictions indicate that this region of the c-myc 3′UTR, particularly nt 229–262, forms an AU-rich stem–loop.
Figure 3. Enzymatic and chemical probing of nt 194–280 in c-myc 3′UTR.
(A) 5′-End-labelled RNA containing nt 194–280 of c-myc 3′UTR was cleaved using 0.25 and 0.1 unit of the RNase T1 (cleaves 3′ to Gs in single-stranded regions of RNA; lanes 1 and 2) for 5 min at 20 °C or 0.01 and 0.03 unit of the double-strand-specific RNase V1 (lanes 3 and 4) for 5 min at 20 °C. Uncleaved 32P-labelled RNA is shown in lane C. For reference, an alkaline hydrolysis ladder and a G-specific ladder generated by T1 under denaturing conditions are shown in lanes L and G, respectively. (B) Cleavage patterns were obtained by incubation of the labelled RNA with 0.2 and 0.6 unit of the RNase T2 (cuts in single-stranded regions of RNA without pronounced base specificity; lanes 1 and 2) for 3 min or with 2 and 5 mM lead acetate (cleaves single-stranded regions of RNA; lanes 3 and 4) for 5 min at room temperature. (C) Unlabelled RNA containing nt 194–280 of c-myc 3′UTR was also modified (+) by CMCT (10 mg/ml) for 20 min at 30 °C. In such experiments, protection of the chemical groups in the bases against chemical modification is indicative of their participation in base pairing. The lane noted (−) corresponds to the control reaction (unmodified RNA). A, C, G and U are sequencing lanes. (D) Proposed secondary structure for nt 222–267 of c-myc 3′UTR as compiled from several experiments using each probe. Cleavage sites produced by the different nucleases, and lead, are indicated by the symbols noted in the Figure. The size of the symbols is indicative of the relative strength of cleavage. Residues modified by CMCT are circled.
Mutation of a conserved AUUUA sequence induces an alteration in the secondary structure
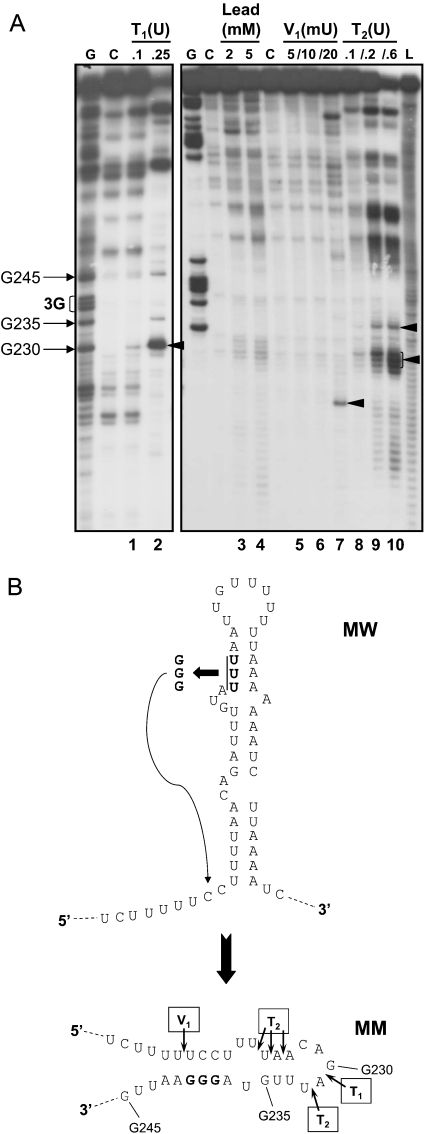
Substitution of a conserved AUUUA motif (nt 237–241) within the 194–280 region of c-myc 3′UTR by AGGGA has been previously shown to abrogate both perinuclear localization of β-globin reporter transcripts and protein binding [15,19]. The role of the stem–loop structure within nt 194–280 was investigated by comparing the secondary structure of the mutated version of this region (MM) with that obtained for the wild-type RNA (MW, Figure 3) using enzymatic and chemical probing. Probing of both MM and MW were conducted in parallel in several experiments and representative gels for MM are presented in Figure 4(A). Compared with MW, significant changes (indicated by arrows) were noticeable in the cleavage patterns obtained with the different RNases, indicating an alteration to the secondary structure. This analysis showed a clear change in the degree of sensitivity of the G nucleotides present in the nt 222–267 region towards the RNase T1 (G230 becoming the preferential cleavage site of this enzyme; Figure 4A, lanes 1 and 2). The cleavage patterns obtained with Pb2+ (Figure 4A, lanes 3 and 4) and the nucleases V1 (Figure 4A, lanes 5–7) and T2 (Figure 4A, lanes 8–10) were also distinctly altered. In particular, clear cuts by the RNase T2 were evident at positions U224-A226 and A231 whereas a new band corresponding to U218 (Figure 4A, arrow, lane 7) was observed using the RNase V1. These probing results obtained with the mutant MM version of the c-myc 86 nt region indicate a change in the structure of this region and are compatible with a hypothetical model in which the three inserted Gs in MM form new base pairs with nt 219–221 (UCC) located just upstream of the stem–loop (Figure 4B), so disrupting the stem–loop proposed for the wild-type sequence. Since the MM mutation abolishes localization [15] and protein binding [19], this observation suggests that the stem–loop structure is important for both mRNA localization and binding of annexin A2.
Figure 4. Secondary structure analysis of c-myc mutant 3′UTR 86 nt region (MM).
(A) 5′-End-labelled-RNA containing nt 194–280 of c-myc 3′UTR in which the AGGGA substitution (MM) was cleaved using 0.1 and 0.25 unit of the single-strand-specific RNase T1 (lanes 1 and 2) for 5 min at 20 °C; 2 and 5 mM lead acetate for 5 min at 20 °C (lanes 3 and 4); 0.005, 0.01 and 0.02 unit of the double strand-specific RNase V1 (lanes 5–7) for 5 min at 20 °C or 0.1, 0.2 and 0.6 unit of the single-strand-specific RNase T2 (lanes 8–10) for 3 min at 20 °C. C, control lanes; G, ladders of Gs; L, alkaline ladders. (B) Hypothetical secondary structure model proposed for the change in structure to the 86 nt region of c-myc 3′UTR when the AUUUA motif (nt 237–241; MW) was mutated to AGGGA (MM).
Protein binding to the nt 222–267 region
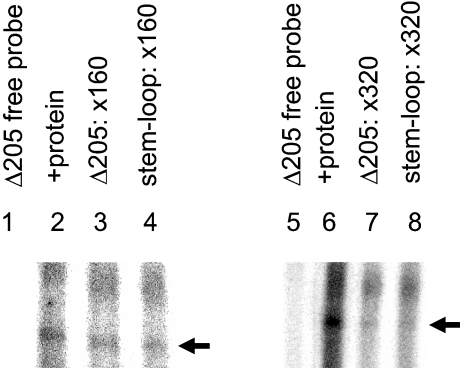
Recent work has shown annexin A2 to bind nt 205–280 of the c-myc 3′UTR and gel retardation assays followed by RNase T1 digestion reveal formation of RNA–protein complexes [19]. To investigate the role of the proposed stem–loop in binding of annexin A2, competitive-gel-retardation experiments were carried out using labelled transcripts corresponding to nt 205–280 of the c-myc 3′UTR and unlabelled competitor transcripts corresponding to the proposed stem–loop region (nt 222–267). As shown in Figure 5 and Mickleburgh et al. [19], transcripts corresponding to nt 205–280 (Δ205) formed a complex with S100 extracts from Ltk−-fibroblasts. Unlabelled transcripts of this region showed self-competition at ×160 and ×320 molar excess, and greater competition at the higher concentration. Transcripts corresponding to the stem–loop alone also showed competition at both these concentrations and the extent of competition was similar to that found using Δ205 transcripts. Control β-globin transcripts did not compete for binding (results not shown). These results suggest that nt 222–267 are sufficient for protein-binding.
Figure 5. RNA–protein complex formation monitored by competitive gel-retardation assay.
Complex formation was studied using [α-32P]CTP-labelled c-myc Δ205 RNA (12 fmol) and 2 μg of S100 extract protein from Ltk−-fibroblasts and prepared as described in the Materials and Methods section. RNase T1 digestion was performed after the binding-reaction and then heparin was added. Ficoll was added to binding-reactions before 5% (79:1) native PAGE. Lanes 1 and 5 show free probe, lanes 2 and 6 show complex-formation with the cell extract, and lanes 3, 4, 7 and 8 show competitive assays using labelled Δ205 transcripts and unlabelled Δ205 (lanes 3 and 7) or stem-loop (lanes 4 and 8) transcripts at 160- or 320-fold molar excess. Complex-formation is indicated by the black arrowhead and this corresponds to the complex formed with annexin A2 [19].
The nt 222–267 region is sufficient to localize β-globin reporter transcripts
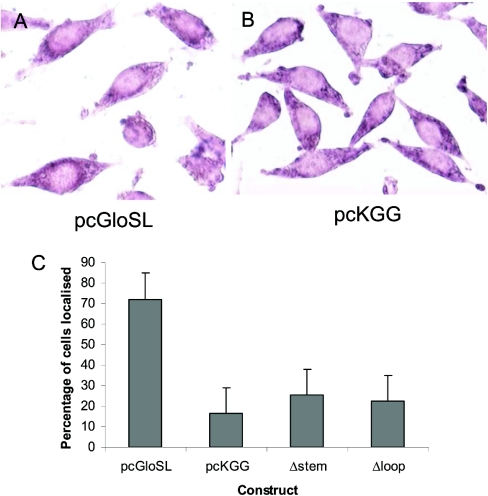
In order to determine the role of this stem-loop structure in localization, a gene construct was made, in which the region corresponding to the proposed stem-loop in the c-myc 3′UTR (nt 222–267) was linked to the β-globin coding region and inserted into pcDNA3. This plasmid (pcglobinSL, Figure 1) was then introduced into Ltk−-fibroblasts by transfection and localization of the globin transcripts assessed by in situ hybridization of stable transfectants. The pcglobinSL cells showed a marked ring of staining around the nucleus indicating a perinuclear localization of globin transcripts (Figure 6A). By contrast, cells expressing globin transcripts linked to the native globin 3′UTR (pcKGG, [8]) showed no ring of perinuclear staining (Figure 6B), as observed previously [7,8,21]. Using the presence of a ring of perinuclear staining as being indicative of perinuclear localization, the staining pattern was assessed in at least 100 cells at random so as to quantify the extent of localization (Figure 6C). Such quantification confirmed visual inspection of the staining pattern by indicating that in pcglobinSL cells at least 70% of cells exhibited localized transcripts whereas in pcKGG-transfected cells less than 20% of cells showed any evidence of a perinuclear ring of transcripts. These results suggest that the region of the c-myc 3′UTR corresponding to the proposed stem-loop (nt 222–267) is sufficient for localization. To further investigate the importance of the stem-loop in localization, the effect of two deletions in this region on perinuclear localization of reporter β-globin transcripts was determined in cells stably transfected using constructs in which the mutant sequences (Figure 1, Δstem, Δloop) were linked to the β-globin coding region. As shown in Figure 6(C), quantification of the staining pattern showed that deletion of either the predicted terminal loop or the upper stem caused a large decrease in localization of globin transcripts as measured by the number of cells exhibiting localization.
Figure 6. The AU-rich stem-loop region from the c-myc 3′UTR is sufficient for localization of globin reporter transcripts.
In situ hybridization of cells after incubation with a digoxigenin-labelled riboprobe that recognizes transcripts containing globin coding sequences. Examples are shown of cells expressing pcgloSL (globin coding region linked to the stem-loop region) exhibiting a distinct perinuclear localization of globin transcripts (A) and cells expressing β-globin coding region linked to its own 3′UTR showing a diffuse, non-localized distribution (B). The staining pattern was quantified in cells transfected using a range of constructs by examining 50– 100 cells for perinuclear localization of globin transcripts and scored as showing localization or not. The percentage of cells exhibiting localization is shown in a histogram (C) as mean±S.E.M. for 3 separate experiments.
The region of c-fos 3′UTR required for perinuclear localization can also form a stem-loop structure
As well as in c-myc, the 3′UTR of the c-fos proto-oncogene mRNA has been shown to contain a perinuclear localization signal and this has been mapped to between nt 260 and 403 [7]. This 144 nt region (see Figure 7A) is highly conserved between species (results not shown), implying an important functional role for this portion of c-fos 3′UTR. In order to determine if this particular region of the c-fos 3′UTR could fold into a structure comparable to that found in the c-myc 3′UTR, enzymatic and chemical probing in solution was performed on RNA corresponding to nt 260–403 of c-fos 3′UTR (Figure 7B). Between nt 354–394, a clear single-stranded region encompassing nt 369–381 was revealed by Pb2+ cleavage (Figure 7B, lanes 9 and 10). This region also included preferential sites for cleavage by the single-strand-specific RNases T1 (G376 and G377; Figure 7B, lanes 5 and 6), T2 (A373 and U375; Figure 7B, lanes 3 and 4) and A (C369, U372, U374–U375 and U381; Figure 7B, lanes 7 and 8). By contrast, cleavage at positions G366, U367 and U368 by the double-stranded RNase V1 suggested that these nucleotides were in a helical configuration. Overall, the cleavage results obtained from these probing experiments were consistent with a structure formed by a major stem region and a single-stranded region including a terminal loop, as proposed in the model shown in Figure 7(A). Thus the probing results indicate that a stem-loop structure similar to the one proposed for c-myc exists in the region of c-fos 3′UTR containing a perinuclear localization signal.
Figure 7. Secondary structure analysis of c-fos 3′UTR 144 nt region.
(A) Sequence in region 260–403 of c-fos 3′UTR containing a perinuclear localization signal and proposed secondary structure for nt 354–394 of this region. (B) 5′-End-labelled RNA containing nt 260–403 of c-fos 3′UTR was cleaved using 0.01 and 0.02 unit of the double-strand-specific RNase V1 (lanes 1 and 2) for 5 min at 20 °C; 0.2 and 0.4 unit of the single-strand-specific RNase T2 (lanes 3 and 4) for 3 min at 20 °C; 0.05 and 0.1 unit of the single-strand-specific RNase T1 (lanes 5 and 6) for 5 min at 20 °C; 0.01 and 0.03 μg of the single-strand-specific RNase A (cleaves 3′ to Us and Cs in single-stranded regions of RNA; lanes 7 and 8) for 3 min at 20 °C; 2 and 5 mM lead acetate for 5 min at 20 °C (lanes 9 and 10). C, control lanes; G, ladder of G nucleotides; L, alkaline ladder. The probing results obtained from several separate experiments are combined in the model shown in (A). Cleavage sites produced by the different ribonucleases, and lead, are indicated by the symbols noted in the Figure. The size of the symbols is indicative of the relative cleavage strength.
DISCUSSION
A perinuclear localization signal was previously mapped to the region of the c-myc 3′UTR between nt 194 and 280 [15]. The results presented here extend analysis of this region and provide the first evidence for the detailed nature of this localization signal, indicating that nt 222–267 are sufficient for localization and that the structure of this region is important for the localization signal.
In silico analysis using bioinformatic protocols such as the Mfold program predicted that the central region of the c-myc 3′UTR previously implicated in localization [15] forms a stem-loop. Enzymatic and chemical probing analysis (Figure 3) was largely consistent with this model by indicating that nt 229–262 form an upper stem region containing a bulge and a terminal loop. However, cleavage using RNase T2 and Pb2+ suggested that the lower region (nt 222–226 and 263–267) was single-stranded rather than base-paired as suggested by computer predictions of structure. Using the mouse sequence as a reference, the bioinformatics approach suggested that the stem-loop in this part of the c-myc 3′UTR was conserved among various mammalian species. In situ hybridization experiments using chimaeric transcripts, in which the β-globin coding region was linked to the sequence corresponding to the predicted stem-loop, showed that this small 46 nt region of c-myc 3′UTR alone was sufficient to direct localization of reporter transcripts to the perinuclear cytoplasm (Figure 6), suggesting that a stem-loop structure was a feature of the localization signal.
This view was supported by the effects of mutations. Firstly, the loss of perinuclear localization caused by mutation of a conserved AUUUA motif, present in this region of c-myc 3′UTR, to AGGGA [15] was correlated with a structural change to this region and the disruption of the proposed stem-loop (Figure 4). Secondly, mutations that were predicted to affect the stem-loop structure either by shortening the terminal loop (Δloop) or deleting the upper stem (Δstem) resulted in the loss of transcript localization around the nucleus. All of the above results are consistent with the view that a stem-loop formed within region 222–267 is a critical part of the perinuclear localization signal. However, further detailed mutagenesis studies are required to determine the precise features of the stem-loop required for localization and whether sequence motifs play a role in localization in addition to structural features. Interestingly, perinuclear localization of MT-1 and vimentin mRNAs has also been found to be dependent on their 3′UTRs [8,21,22] and the regions responsible for localization are predicted to fold into a stem with an internal loop, or Y-shaped region containing a stem-loop respectively [21,23], structures which although apparently different, nevertheless demonstrate common features. Furthermore, in the case of MT-1, the localization signal appears to require both sequence and structural features [21]. It remains to be seen if there are consensus features in these signals.
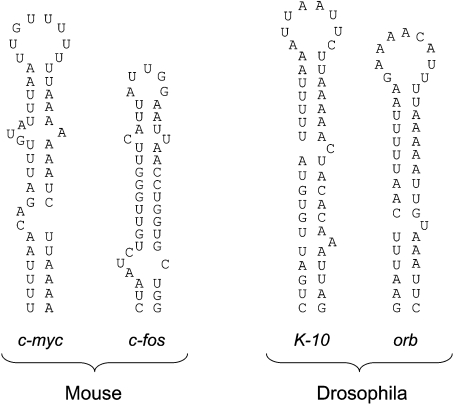
The stem-loop structure proposed here as the determinant of c-myc mRNA perinuclear localization is reminiscent of localization elements previously described in the 3′UTR of two mRNAs in Drosophila [24]. A single AU-rich stem-loop of 44 nt is responsible for the transport and the localization of the K-10 transcripts to the oocyte's anterior cortex while a very similar structure was proposed to transport and localize orb mRNA to the same region. However, during the course of our study, we also observed that part of the 144 nt region of c-fos 3′UTR that contains a peri-nuclear localization signal [7] could fold into a well defined stem-loop structure of 41 nt comparable to the one described for c-myc, although the role of this structure in c-fos mRNA peri-nuclear localization remains to be experimentally confirmed. The structures present in the 3′UTR of these four different mRNAs are shown in Figure 8. The four stem-loop structures are very similar and all are rich in AU base pairs (GU base pairs in the case of c-fos) and contain one or two bulges, features that would give a significant degree of ‘looseness’ to the structures so that they can easily form alternative open structures. The stem-loop has not been observed in other localized transcripts, either perinuclear or non-perinuclear.
Figure 8. Comparison of secondary structure of the localization elements in K-10, orb, c-myc and c-fos mRNAs.
Structures shown are those proposed for the Drosophila K-10 and orb mRNAs [24] and those for mouse c-myc and c-fos determined in the present study by a combination of chemical cleavage and computer prediction using Mfold.
In all of the above cases, the specific determinant(s) that form the recognition site(s) for trans-acting protein(s) involved in localization still remain to be defined. In the case of the c-myc 3′UTR the region from nt 205–280 has been shown to be important for binding of the protein annexin A2 [19] and in the present work gel-retardation assays provided evidence that the 46 nt sequence corresponding to the stem-loop (nt 222–267) is sufficient for protein binding. Although the proteins binding to the related signals need to be identified, it is conceivable that localization of these mRNAs to specific subcellular sites involves evolutionarily conserved or closely related trans-acting factor(s).
Both c-myc and c-fos mRNAs are unstable, containing instability elements within their 3′UTRs [25]. The regions of 3′UTR responsible for instability are distinct from those implicated in localization [7,15] and thus the cis-acting signals described here form part of a mechanism that is separate from that involved in the instability of these mRNAs.
In conclusion, the results presented here indicate that part of the nt 194–280 region of c-myc 3′UTR responsible for the mRNA targeting folds into a stem-loop structure, which is able to direct reporter transcripts to the perinuclear cytoplasm. Our hypothesis is that the AU-rich stem-loop structure formed by nt 222–267 of c-myc 3′UTR represents a perinuclear mRNA localization signal. Both structural and computer prediction analyses suggest that this signal shares features with 3′UTRs in other localized mRNAs (c-fos, orb and K-10) and therefore such features may represent a conserved signal in mRNA localization mechanisms.
Online data
Acknowledgments
This work was supported by grants C13737 and C14236 from the BBSRC (Biotechnology and Biological Sciences Research Council), U.K. We are grateful to Zofia Chrzanowska-Lightowlers for useful discussions.
References
- 1.Jansen R. P. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 2.Oleynikov Y., Singer R. H. RNA localization: different zipcodes, same postman? Trends Cell. Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- 3.Chabanon H., Mickleburgh I., Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomics Proteomics. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 4.Kislauskis E. H., Li Z., Taneja K. L., Singer R. H. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson I. A., Brindle K. M., Fulton A. M. Differential localization of the mRNA of the M and B forms of creatine kinase in myoblasts. Biochem. J. 1995;308:599–605. doi: 10.1042/bj3080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesketh J., Campbell G., Piechaczyk M., Blanchard J. M. Targeting of c-myc and β-globin coding sequences to cytoskeletal-bound polysomes by c-myc 3′ untranslated region. Biochem. J. 1994;298:143–148. doi: 10.1042/bj2980143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgleish G., Veyrune J. L., Blanchard J. M., Hesketh J. mRNA localisation by a 145-nucleotide region of the c-fos 3′untranslated region. J. Biol. Chem. 2001;276:13593–13599. doi: 10.1074/jbc.M001141200. [DOI] [PubMed] [Google Scholar]
- 8.Mahon P., Partridge K., Beattie J. H., Glover L. A., Hesketh J. E. The 3′untranslated region plays a role in the targeting of metallothionein-1 mRNA to the perinuclear cytoplasm and cytoskeletal-bound polysomes. Biochim. Biophys. Acta. 1997;1358:153–162. doi: 10.1016/s0167-4889(97)00058-x. [DOI] [PubMed] [Google Scholar]
- 9.Hesketh J. E., Campbell G. P., Whitelaw P. F. c-myc mRNA in cytoskeletal-bound polysomes in fibroblasts. Biochem. J. 1991;274:607–609. doi: 10.1042/bj2740607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesketh J. E. Sorting of messenger RNAs in the cytoplasm: mRNA localization and the cytoskeleton. Exp. Cell Res. 1996;225:219–236. doi: 10.1006/excr.1996.0172. [DOI] [PubMed] [Google Scholar]
- 11.Levadoux M., Mahon C., Beattie J. H., Wallace H. M., Hesketh J. E. Nuclear import of metallothionein requires its mRNA to be associated with the perinuclear cytoskeleton. J. Biol. Chem. 1999;274:34961–34966. doi: 10.1074/jbc.274.49.34961. [DOI] [PubMed] [Google Scholar]
- 12.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ainger K., Avossa D., Diana A. S., Barry C., Barbarese E., Carson J. H. Transport and localization elements in myelin basic protein mRNA. J. Cell. Biol. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald P. M., Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell Biol. 1998;18:3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veyrune J. L., Campbell G. P., Wiseman J., Blanchard J. M., Hesketh J. E. A localisation signal in the 3′untranslated region of c-myc mRNA targets c-myc and β-globin reporter sequences to the perinuclear cytoplasm and cytoskeletal-bound polysomes. J. Cell Sci. 1996;109:1185–1194. doi: 10.1242/jcs.109.6.1185. [DOI] [PubMed] [Google Scholar]
- 16.Behar L., Marx R., Sadot E., Barg J., Ginzburg I. cis-acting signals and trans-acting proteins are involved in tau mRNA targeting in neurites of differentiating neuronal cells. Int. J. Dev. Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- 17.Bermano G., Hesketh J. E. The study of mRNA-cytoskeleton interactions and mRNA sorting in mammalian cells. In: Carraway K. L., Carraway C. A. C., editors. Cytoskeleton Signalling and Cell Regulation; A Practical Approach. Oxford University Press; 1999. pp. 209–244. [Google Scholar]
- 18.Huntzinger E., Possedko M., Winter F., Moine H., Ehresmann C., Romby P. Probing RNA structures with enzymes and chemicals in vitro and in vivo. In: Hartmann R. K., Bindereif A., Schon A., Westhof E., editors. Handbook of RNA Biochemistry. Weinheim: Wiley–VCH; 2005. pp. 151–171. [Google Scholar]
- 19.Mickleburgh I., Burtle B., Hollas H., Campbell G., Chrzanowska-Lightowlers Z., Vedeler A., Hesketh J. Annexin A2 binds to the localization signal in the 3′ untranslated region of c-myc mRNA. FEBS J. 2005;272:413–421. doi: 10.1111/j.1742-4658.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 20.Zuker M. Mfold webserver for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nury D., Chabanon H., Levadoux-Martin M., Hesketh J. A 11-nucleotide section of the 3′untranslated region is required for perinuclear localization of rat metallothionein-1 mRNA. Biochem. J. 2005;387:419–428. doi: 10.1042/BJ20040630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bermano G., Shepherd R. K., Zehner Z. E., Hesketh J. E. Perinuclear mRNA localisation by vimentin 3′untranslated region requires a 100 nucleotide sequence and intermediate filaments. FEBS Lett. 2001;497:77–81. doi: 10.1016/s0014-5793(01)02438-3. [DOI] [PubMed] [Google Scholar]
- 23.Zehner Z. E., Shepherd R. K., Gabryszuk J., Fu T. F., Al-Ali M., Holmes W. M. RNA-protein interactions within the 3′ untranslated region of vimentin mRNA. Nucleic Acids Res. 1997;25:3362–3370. doi: 10.1093/nar/25.16.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serano T. L., Cohen R. S. A small predicted stem-loop structure mediates oocyte localization of drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 25.Chen C. Y., You Y., Shyu A. B. Two cellular proteins bind specifically to a purine-rich sequence necessary for the destabilization function of a c-fos protein-coding region determinant of mRNA instability. Mol. Cell. Biol. 1992;12:5748–5757. doi: 10.1128/mcb.12.12.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.