Abstract
The biogenesis of eukaryotic COX (cytochrome c oxidase) requires several accessory proteins in addition to structural subunits and prosthetic groups. We have analysed the assembly state of COX and SCO2 protein levels in various tissues of six patients with mutations in SCO2 and SURF1. SCO2 is a copper-binding protein presumably involved in formation of the CuA centre of the COX2 subunit. The function of SURF1 is unknown. Immunoblot analysis of native gels demonstrated that COX holoenzyme is reduced to 10–20% in skeletal muscle and brain of SCO2 and SURF1 patients and to 10–30% in heart of SCO2 patients, whereas liver of SCO2 patients' contained normal holoenzyme levels. The steady-state levels of mutant SCO2 protein ranged from 0 to 20% in different SCO2 patient tissues. In addition, eight distinct COX subcomplexes and unassembled subunits were found, some of them identical with known assembly intermediates of the human enzyme. Heart, brain and skeletal muscle of SCO2 patients contained accumulated levels of the COX1·COX4·COX5A subcomplex, three COX1-containing subcomplexes, a COX4·COX5A subcomplex and two subcomplexes composed of only COX4 or COX5A. The accumulation of COX1·COX4·COX5A subcomplex, along with the virtual absence of free COX2, suggests that the lack of the CuA centre may result in decreased stability of COX2. The appearance of COX4·COX5A subcomplex indicates that association of these nucleus-encoded subunits probably precedes their addition to COX1 during the assembly process. Finally, the consequences of SCO2 and SURF1 mutations suggest the existence of tissue-specific functional differences of these proteins that may serve different tissue-specific requirements for the regulation of COX biogenesis.
Keywords: assembly pathway, CuA centre, cytochrome c oxidase, mitochondria, SCO2, SURF1
Abbreviations: BN-PAGE, Blue Native PAGE; COX, cytochrome c oxidase; CS, citrate synthase; DDM, n-dodecyl-β-D-maltoside; SDH, succinate: ubiquinone oxidoreductase; VDAC, voltage-dependent anion channel
INTRODUCTION
Eukaryotic COX (cytochrome c oxidase), the terminal enzyme of the mitochondrial respiratory chain, is embedded in the inner mitochondrial membrane where it catalyses the transfer of electrons from reduced cytochrome c to molecular oxygen and further couples this reaction with proton translocation across the inner membrane. Mammalian COX is a multisubunit complex of approx. 200 kDa composed of 13 subunits encoded by both the mitochondrial and nuclear genes. The mitochondrially encoded subunits COX1, COX2 and COX3 are evolutionarily conserved and form the catalytic and structural core of the enzyme [1]. The remaining ten evolutionarily younger subunits are encoded by the nuclear genome and are associated with the surface of the complex core. These small polypeptides are required for the stability and assembly of the holoenzyme and are also involved in modulation of its activity in response to various cellular stimuli [2]. Tissue-specific isoforms of subunits COX4, COX6A, COX6B and COX7A were identified in humans [3,4]. In addition to the constituent protein subunits, COX contains several redox-active prosthetic groups directly involved in electron transfer. These are two haem A moieties (a and a3) and two copper centres (CuA and CuB). The CuB centre and both haem A moieties are located within the hydrophobic interior of COX1 subunit composed of 12 transmembrane α-helices. The binuclear CuA centre is located within the ten-stranded β-barrel that forms the polar C-terminal domain of COX2, which protrudes into the intermembrane space. This subunit is anchored to the membrane by two N-terminal transmembrane α-helices that make extensive contacts with COX1 [5]. The assembly pathway of mammalian COX in the inner mitochondrial membrane is a sequential and relatively slow process that is still not fully understood [6,7]. Studies on yeast have identified more than 30 accessory proteins essential for proper biosynthesis or assembly of the enzyme. To date, mutations in six nucleus-encoded factors (SURF1, SCO1, SCO2, COX10, COX15 and LRPPRC) required for the assembly of the COX complex have been identified in humans [6–8].
SCO2 is an inner mitochondrial membrane copper-binding protein presumably involved in copper transfer to the CuA centre of COX2. The molecular mass of its fully processed form is approx. 25 kDa. Mutations in human SCO2 cause fatal infantile COX deficiency with the predominant symptoms being encephalopathy and hypertrophic cardiomyopathy. To date, all patients identified were either compound heterozygotes for 1541G>A (where 1541G>A denotes the guanine>adenine nucleotide transition at the position 1541 of the DNA) mutation, with the other allele carrying either a nonsense or missense mutation, or homozygotes for this common 1541G>A transition, predicting a E140K amino acid substitution near the highly conserved CXXXC putative copper-binding motif [9–11]. The most severe cases (early onset) are compound heterozygotes, while patients homozygous for E140K substitution have a comparatively milder phenotype (delayed onset, less progressive).
Human SURF1 is a 30 kDa transmembrane protein localized in the inner mitochondrial membrane [12,13]. The precise function of this protein is still unknown, but recently it was suggested that human SURF1 promotes the association of COX2 with the COX assembly intermediate composed of COX1, COX4 and COX5A [14], which was originally described as COX1- and COX4-containing assembly intermediate S2 [15]. Mutations in human SURF1 cause Leigh syndrome, a fatal neurological disorder associated with severe isolated COX deficiency and characterized mainly by bilaterally symmetrical necrotic lesions in the basal ganglia and brainstem [16,17]. Nearly all reported SURF1 patients carried loss-of-function mutations that predict either truncated and unstable protein product or unstable mRNA [18].
Histochemical studies and enzyme activity measurements show that SCO2 mutations result in a tissue-specific decrease of COX activity, with heart and skeletal muscle being most severely affected. In contrast, cultured fibroblasts and liver were shown to retain high residual activity [9–11], indicating tissue-specific differences in COX biogenesis or maintenance. The COX activity was reduced to approx. 10% of control values in SURF1 patient fibroblasts [14], and the skeletal muscle of SURF1 patients was repeatedly shown to retain approx. 20% of residual COX activity [19]. In SURF1 fibroblasts, the reduction of COX activity was shown to be accompanied by a similar decrease in holoenzyme levels and also by a marked accumulation of COX subcomplexes, suggesting that the residual enzyme is fully active and that the enzyme deficiency stems from impaired assembly or maintenance of the protein complex [14].
The aim of the present study was to examine and compare the consequences of SCO2 and SURF1 mutations in various human tissues. We have investigated the steady-state levels of COX holoenzyme and the presence and composition of COX subcomplexes in tissues and primary fibroblast cultures from three patients harbouring SCO2 mutations and from three patients carrying mutations in SURF1. We directly demonstrate that mutations in both genes result in tissue-specific decrease of COX holoenzyme levels with a parallel reduction in the COX activity, accompanied by accumulation of specific COX subcomplexes and unassembled subunits. We further show that all investigated SCO2 mutations result in severely decreased levels of mutant SCO2 protein in heart, brain, liver and fibroblasts. The subunit composition of COX subcomplexes identified demonstrates the involvement of human SCO2 in biogenesis or maintenance of COX2 and suggests an addition to the current model of the human COX assembly pathway.
EXPERIMENTAL
Ethics
The present study was carried out in accordance with the Declaration of Helsinki of the World Medical Association, and was approved by the Committees of Medical Ethics at all collaborating institutions. Informed parental consent, in accordance with guidelines of the participating institutions, was obtained for all biopsies and autopsies.
Patients
Patients P1 and P2 were homozygous for 1541G>A transition in SCO2 leading to E140K amino acid substitution. Patient P3 was a compound heterozygote for the 1280C>T transition in SCO2, leading to the formation of a premature termination codon, and a common missense 1541G>A mutation [20]. Patient P4 was a compound heterozygote for the combined frameshift deletion-insertion mutation in SURF1 312_321del 311_312insAT, resulting in the formation of a premature termination codon, and a 821del18 deletion in SURF1 leading to highly unstable mRNA [21]. Patient P5 was homozygous for the frameshift deletion 845_846delCT leading to the formation of a premature termination codon, and patient P6 was homozygous for a 688C>T nonsense substitution in SURF1 leading to the formation of a premature termination codon truncating the protein at Arg230 [22]. Patients P1 and P2 presented with progressive encephalopathy since the third month of age, and they died at <1 year of age [20]. Patient P3 presented with hypertrophic cardiomyopathy since birth and he died at 7 weeks of age [20]. Patients P4–P6 presented with failure to thrive, progressive hypotonia and hypertrichosis at the end of the first year of life, followed by a total arrest of psychomotor development, and all died around the third year of age [21,22].
Cell cultures and tissues
All studied tissues and primary fibroblast cultures were obtained from three patients (P1–P3) harbouring two different combinations of SCO2 mutations, three patients (P4–P6) carrying mutations in SURF1 and age-related controls. Primary skin fibroblast cultures were established from forearm skin biopsies. Open muscle biopsies were obtained from the tibialis anterior muscle and were frozen at −80 °C. Post-mortem heart, liver, brain (basal ganglia) and kidney tissue specimens were removed and frozen less than 2 h after death.
Isolation of mitoplasts and mitochondria
Skeletal muscle, brain and kidney mitochondria were isolated according to standard differential centrifugation procedures [23] in a buffer comprising 150 mM KCl, 10 mM Tris/HCl, 2 mM EDTA and 2 μg/ml aprotinin (pH 7.4) at 4 °C. Heart and liver mitochondria were isolated in a buffer comprising 250 mM sucrose, 20 mM Tris/HCl, 2 mM EDTA and 2 μg/ml aprotinin (pH 7.4) at 4 °C. Mitoplast-enriched fractions were prepared from cultured fibroblasts using digitonin (Sigma–Aldrich) as described in [24], with a final digitonin/protein ratio of 0.6 mg/mg. Protein concentrations were determined with the Bio-Rad Protein Assay kit (Bio-Rad Laboratories). All samples were stored at −80 °C.
Enzyme activity assays
Activities of COX and CS (citrate synthase) were measured spectrophotometrically in fibroblasts and isolated tissue mitochondria essentially as described in [25].
Electrophoresis
BN-PAGE (Blue Native PAGE) [26] was used for separation of mitochondrial membrane protein complexes on polyacrylamide 8–15, 8–16 and 10–18% (w/v) gradient gels using a Mini Protean® 3 System (Bio-Rad Laboratories). Mitoplasts or mitochondria were solubilized with DDM (n-dodecyl β-D-maltoside; Sigma–Aldrich) with a final DDM/protein ratio of 1.0 mg/mg in a buffer containing 1.5 M aminocaproic acid, 2 mM EDTA and 50 mM Bis-Tris (pH 7.0) at 4 °C. Serva Blue G (Serva) was added to solubilized protein at a concentration of 0.1 mg/mg of detergent, and 5–50 μg of protein was loaded for each lane. The electrophoresis was performed at 40 V, 4 °C for 1 h and then at 100 V, 4 °C. Tricine SDS/PAGE was carried out under standard conditions with 12% polyacrylamide, 0.1% (w/v) SDS and 5.5 M urea gels. Mitochondrial fractions were dissociated in 50 mM Tris/HCl (pH 6.8), 12% (v/v) glycerol, 4% SDS, 2% (v/v) 2-mercaptoethanol and 0.01% (w/v) Bromophenol Blue for 30 min at 37 °C, and approx. 10 μg of protein was loaded for each lane. For two-dimensional BN/SDS/PAGE [26], strips of the first-dimension gels were incubated for 40 min in 1% 2-mercaptoethanol and 1% SDS and then for 10 min in 1% SDS, and denatured proteins were then resolved in the second dimension on 13% polyacrylamide, 0.1% SDS and 5.5 M urea gels [14,26].
Preparation of a polyclonal antibody raised against human SCO2
An SCO2-specific antibody was generated by injecting rabbits with a synthetic peptide specific for the C-terminal part of human SCO2 (CGRSRSAEQISDSVRRHMAAF). Testing of the specificity of the SCO2 antiserum revealed that affinity purification was not required, and crude serum was used in all subsequent experiments.
Immunoblot analysis
Proteins were electroblotted from the gels on to Immobilon™-P PVDF membranes (Millipore) using semi-dry transfer for 2 h at a constant current of 0.8 mA/cm2. Membranes were air-dried overnight, rinsed twice with 100% (v/v) methanol and blocked in PBS and 10% (w/v) non-fat dried milk for 1 h. Primary detection was performed with mouse monoclonal antibodies raised against COX subunits COX1 (A-6403; 1 μg/ml), COX2 (A-6404; 1 μg/ml), COX4 (A-21348; 2 μg/ml), COX5A (A-21363; 2 μg/ml) and COX6B (A-21366; 1 μg/ml) (Molecular Probes), with rabbit polyclonal antiserum raised against human SCO2 (1:1000) and with monoclonal antibodies raised against the flavoprotein subunit of SDH (succinate:ubiquinone oxidoreductase) (A-11142; 0.1 μg/ml) (Molecular Probes) and the VDAC (voltage-dependent anion channel) (31HL Ab-1; 1.4 μg/ml) (Calbiochem) at indicated dilutions. Blots were incubated with primary antibodies in PBS, 0.3% (v/v) Tween 20 and 1% non-fat dried milk for 2 h. Secondary detection was carried out with goat anti-mouse IgG–horseradish peroxidase conjugate (A8924; 1:1000) (Sigma–Aldrich) or with goat anti-rabbit IgG–horseradish peroxidase conjugate (A0545; 1:2000) (Sigma–Aldrich) in PBS, 0.1% Tween 20 and 1% non-fat dried milk for 1 h. The blots were developed with West Pico Chemiluminescent substrate (Pierce) and exposed to Kodak BioMax Light films (Kodak). The films were subsequently scanned and digital images were analysed using the Quantity One application (Bio-Rad Laboratories).
RESULTS
Activities of COX in SCO2 and SURF1 patient tissues
Previous respiratory chain enzyme activity measurements of tissues and cell cultures from our patients with SCO2 and SURF1 mutations showed an isolated tissue-specific COX deficiency in all patients [20–22]. To determine the residual COX activity of the mitochondrial preparations used in the present study, we expressed COX activity relative to the activity of the mitochondrial marker enzyme, CS. Severe isolated defects of COX activity were found in the SCO2 patient heart, skeletal muscle and brain, whereas in fibroblasts and liver the activity was only moderately affected (Table 1). In contrast, severe reduction of COX activity was found in all of our SURF1 fibroblast cultures (Table 1).
Table 1. Comparison of relative protein content of COX holoenzyme with relative COX activity in various tissues of SCO2 patients P1–P3 and SURF1 patients P4–P6.
Protein content of COX holoenzyme normalized to protein content of SDH is expressed as a percentage of control values (COX/SDH content). Values of COX activity normalized to CS activity are expressed as a percentage of the mean reference range (COX/CS activity). n.d., not determined.
| P1 | P2 | P3 | P4 | P5 | P6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue | COX/SDH content (%) | COX/CS activity (%) | COX/SDH content (%) | COX/CS activity (%) | COX/SDH content (%) | COX/CS activity (%) | COX/SDH content (%) | COX/CS activity (%) | COX/SDH content (%) | COX/CS activity (%) | COX/SDH content (%) | COX/CS activity (%) |
| Heart | 25 | 8 | 30 | 34 | 10 | 30 | 40 | n.d. | n.d. | n.d. | n.d. | n.d. |
| Muscle | 10 | 19 | 20 | 28 | n.d. | 21 | n.d. | 11 | 15 | 22 | 10 | 9 |
| Brain | 20 | 16 | 20 | 18 | 20 | 13 | 15 | 20 | n.d. | n.d. | n.d. | n.d. |
| Liver | 100 | 76 | 100 | 64 | 100 | 100 | 80 | n.d. | n.d. | n.d. | n.d. | n.d. |
| Fibroblasts | 70 | 62 | 70 | 82 | 60 | 100 | 10 | 12 | 15 | 6 | 15 | 14 |
Steady-state levels of COX holoenzyme in SCO2 and SURF1 patient tissues
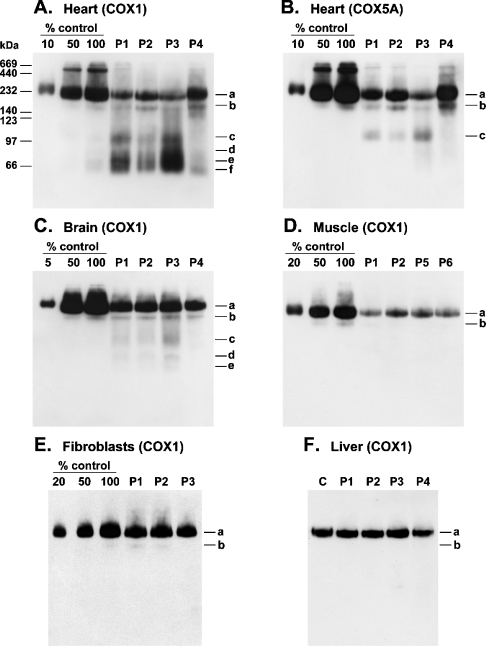
All mitochondrial preparations used in the present study were balanced on the basis of the immunoblot signal of the mitochondrial inner membrane protein complex SDH. To determine the residual steady-state levels of COX holoenzyme in tissues of patients as a percentage of control values, dilutions of control mitochondria were loaded on the same gels. Mitochondrial samples were resolved using BN-PAGE and subsequently probed with an anti-COX1 antibody. In heart mitochondria from patients P1, P2, P3 and P4, the steady-state levels of COX holoenzyme were found to be approx. 25, 30, 10 and 40% of control values respectively (Figures 1A and 1B). Mitochondria from basal ganglia of patients P1–P3 and P4 contained approx. 20 and 15% of residual holoenzyme respectively (Figure 1C). In skeletal muscle from patients P1, P2, P5 and P6, the holoenzyme levels were approx. 10, 20, 15 and 10% of control values respectively (Figure 1D). In primary fibroblasts, the steady-state levels of COX holoenzyme were found to be approx. 70% of control values in the case of patients P1 and P2, approx. 60% in the case of patient P3 (Figure 1E) and approx. 15% in the case of patients P5 and P6 (Table 1). The liver samples of SCO2 patients (P1–P3) were the least affected and contained similar steady-state levels of COX holoenzyme to control samples, whereas in SURF1 patient liver (P4) the holoenzyme was found to be approx. 80% of control values (Figure 1F) (Table 1).
Figure 1. Steady-state levels of COX holoenzyme and subcomplexes in tissues and fibroblast cultures of SCO2 and SURF1 patients.
Mitochondrial fractions from SCO2 (P1–P3) and SURF1 (P4–P6) patient samples were resolved by BN-PAGE (8–15% polyacrylamide), electroblotted on to PVDF membranes and probed with monoclonal antibodies specific for subunits COX1 (A, C–F) or COX5A (B). The amount of protein loaded per lane (∼5 μg) was normalized to SDH. Three aliquots of control mitochondria corresponding to indicated dilutions of control samples were loaded on the same gels. Immunoreactive material was visualized by chemiluminescence. The positions of COX holoenzyme (a), COX subcomplexes (b–f) and molecular-mass standards (kDa) are indicated.
Subcomplexes of COX in SCO2 and SURF1 patient tissues
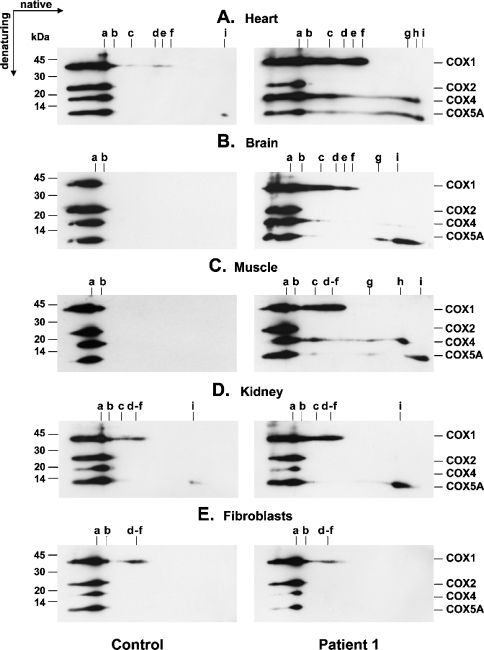
Mitochondrial preparations from various SCO2 and SURF1 patient tissues, primary fibroblast cultures and control samples were resolved using either BN-PAGE or two-dimensional BN/SDS/PAGE and subsequently probed with anti-COX subunit-specific monoclonal antibodies in order to detect the presence and possible accumulation of COX subcomplexes and to uncover their subunit composition. In addition to holoenzyme complex (Figures 1A and 3A, complex a), heart samples of SCO2 patients contained eight distinct COX subcomplexes (b–i). Prolonged exposure of the blots revealed the presence of six of them (b–f and i) also in control heart samples (Figures 2B and 3A). Subcomplexes c–i were found in SCO2 heart samples at highly accumulated levels, whereas only subcomplexes b and f were found slightly increased in the heart of the SURF1 patient (Figure 1A). Mitochondria from SCO2 skeletal muscle contained increased levels of seven distinct subcomplexes (Figure 3C, c–i), apparently identical with that found in SCO2 patient heart. Their steady-state levels were, however, found to be substantially lower in this tissue (Figure 1D), and their full detection thus required higher protein loads (∼50 μg) and longer exposure times (Figure 3C). In line with this, control heart samples contained substantially higher steady-state levels of COX subcomplexes than skeletal muscle controls (Figures 2B and 2C). Also, SCO2 brain samples revealed accumulated COX subcomplexes of approx. 10–120 kDa, detectable mainly with anti-COX1 and anti-COX5A antibodies, but their profile differed from that found in SCO2 heart and skeletal muscle mitochondria (Figures 3A–3C). In particular, the COX4 signal in the brain was very weak in this region. In contrast with SCO2 patient samples, we did not detect any accumulated subcomplexes in SURF1 patient brain (Figure 1C). In skeletal muscle of SURF1 patients, we found substantially decreased levels of subcomplex b and faint accumulation of subcomplexes with similar electrophoretic mobility to subcomplexes c–f from SCO2 samples (Figure 2C). Mitochondria from SCO2 patient kidney contained moderately increased subcomplexes with migration similar to subcomplexes c–f and i from SCO2 patient heart (Figure 3D). We did not detect any conclusive accumulation of COX subcomplexes in SCO2 patient fibroblasts (Figures 1E and 3E).
Figure 3. Subunit composition of COX subcomplexes in various tissues of the SCO2 patient P1.
Mitochondrial fractions (10–50 μg) from various tissues of SCO2 patient P1 and control samples were resolved using two-dimensional BN/SDS/PAGE, electroblotted on to PVDF membranes and probed simultaneously with monoclonal antibodies specific for subunits COX1, COX2, COX4 and COX5A. Sample loads and exposures of films to the blots were chosen such that the signals corresponding to holoenzyme complex a were of similar intensities within both control and patient immunoblots. Immunoreactive material was visualized by chemiluminescence. The polyacrylamide gradient used in the first dimension (BN) was 8–16% for (A, B) and 10–18% for (C–E). The positions of COX holoenzyme (a) and COX subcomplexes (b–i) and the migration of molecular-mass standards (kDa) are indicated.
Figure 2. COX subcomplexes in control heart, heart of SCO2 patients and skeletal muscle of SURF1 patients.
Mitochondrial fractions from SCO2 patient heart (P1–P3) (A, B), SURF1 patient skeletal muscle (P5 and P6) (C) and control samples were resolved by BN-PAGE (8–15% polyacrylamide), electroblotted on to PVDF membranes and probed with monoclonal antibodies specific for subunits COX1 or COX5A. The amount of protein loaded per lane (∼5 μg) was normalized to SDH. Immunoreactive material was visualized by chemiluminescence. The positions of COX holoenzyme (a) and COX subcomplexes (b–f) and the molecular-mass standards (kDa) are indicated.
Alignment of parallel-run immunoblots probed with different antibodies indicated, together with immunoblots of two-dimensional native/denaturing gels, that subcomplex b consists of at least COX1, COX2, COX4, COX5A and COX6B. Subcomplex c comprised at least subunits COX1, COX4 and COX5A, whereas subcomplexes d–f were recognized solely with an anti-COX1 antibody (Figures 1–3). Subcomplex g was detectable with both anti-COX4 and anti-COX5A antibodies, whereas subcomplexes h and i were recognized only with single anti-COX4 and anti-COX5A antibodies respectively (Figures 3A–3C). Low-molecular-mass subcomplexes g–i were not detectable on immunoblots of one-dimensional native gels, since the polyacrylamide gradient used (8–15%) was optimal for fractionation of subcomplexes b–f, while lower-molecular-mass polypeptides were allowed to migrate out of the gel.
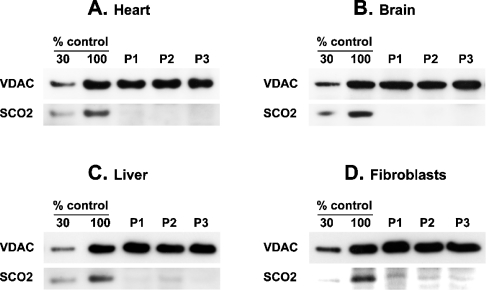
Steady-state levels of SCO2 protein in various tissues of SCO2 patients
Mitochondrial fractions and fibroblast lysates were resolved using SDS/PAGE and subsequently probed with polyclonal antiserum raised against human SCO2. Equal loading was verified with an antibody raised against the mitochondrial outer membrane protein VDAC. The SCO2 protein was undetectable in all SCO2 brain samples and in heart of patients P2 and P3 (Figure 4). In heart of patient P1 and liver of patients P1 and P3, the levels of mutant SCO2 were approx. 5% of control values. In fibroblasts of patients P1–P3, the residual SCO2 was approx. 20% of control values, while in liver of patient P2 it was approx. 10% of control values (Figure 4).
Figure 4. Steady-state levels of SCO2 protein in heart, brain, liver and fibroblasts of SCO2 patients.
Mitochondrial fractions from heart (A), brain (B) and liver (C) samples (∼10 μg) of SCO2 patients (P1–P3) and whole cell lysates (∼10 μg) of SCO2 patient fibroblasts (D) were resolved using SDS/PAGE (12% polyacrylamide), electroblotted on to PVDF membranes and probed with polyclonal antiserum raised against human SCO2 or with monoclonal antibody raised against the mitochondrial outer membrane protein VDAC. Two aliquots of control mitochondria corresponding to indicated dilutions of control samples were loaded on the same gels. Immunoreactive material was visualized by chemiluminescence.
DISCUSSION
The present study represents the first investigation of the assembly state of COX in various clinically affected tissues from patients with SCO2 and SURF1 mutations. Although both SCO2 and SURF1 proteins are thought to act at a similar stage of COX assembly, patients carrying mutations in respective genes present with distinct clinical phenotypes [9–11,16–19]. Quantitative immunoblot analysis of native gels revealed tissue-specific COX assembly defects in all patients studied that corresponded to the enzyme activity measurements. The steady-state levels of mutant SCO2 protein were found severely reduced in all the probed SCO2 patient tissues. The subunit composition of COX subcomplexes identified demonstrates the involvement of human SCO2 protein in biogenesis or maintenance of COX2 and suggests an addition to the current model of the COX assembly pathway.
In our previous work, we have determined the COX activity in several tissues from six patients with mutations in SCO2 [20] that revealed the COX defect to be most pronounced in heart, brain and muscle, moderate or low in liver, and very low or undetectable in fibroblasts. In this study, we present a detailed proteomic analysis of three of these cases in order to reveal how the SCO2 genetic defect manifests at the level of COX biosynthesis and assembly in different tissues.
The COX holoenzyme was repeatedly shown to be reduced to approx. 15% in SURF1 patient fibroblasts, including our patients P4–P6 [12,14,22,27]. Although skeletal-muscle samples of our SURF1 patients revealed a similar decrease in COX holoenzyme to cultured fibroblasts, SURF1-deficient heart and liver contained substantially higher levels of residual holoenzyme. In contrast, the tissue-specific consequences of SCO2 mutations, mainly the profound difference between the residual COX activity in skeletal muscle and fibroblasts, have previously been reported [9,20]. Recently, it was shown that COX holoenzyme is only moderately decreased in immortalized SCO2 patient fibroblasts [28]. In the present study, we show that despite very low levels of mutant protein, the livers of SCO2 patients display practically no reduction of fully assembled COX, corresponding to high residual activity. In our previous study, we have found more pronounced decrease in COX activity (39% of COX/CS) in liver homogenate of an additional SCO2 E140K homozygote patient [20], but due to the lack of material we could not examine the COX assembly state and enzyme content in this case. In contrast, liver and fibroblasts with mutations in SCO1, the paralogous gene of SCO2, were reported with severe reduction of COX activity and holoenzyme content respectively [14,29]. The recombinant human E140K SCO2 protein was shown to have a diminished copper-binding capacity and an altered conformational state [30]. Consistent with the latter finding, our results demonstrate that E140K mutation leads to lowered stability of SCO2 protein, as its levels are drastically decreased in mitochondria of patients. The SCO2 E140K homozygotes present with delayed onset of hypertrophic cardiomyopathy when compared with compound heterozygotes [20,31]. In line with this, heart samples from our E140K homozygotes revealed a substantially milder assembly impairment than that from the E140K/Q53X compound heterozygote. However, we did not detect any substantial difference in the amount of residual SCO2 between the heterozygote and both homozygotes.
In addition to reduced holoenzyme, patient tissues contained varying levels of COX subcomplexes and unassembled subunits. Fibroblasts with mutations in SURF1 were repeatedly shown with increased levels of COX assembly intermediates S1 and S2 [12,14,27], and SCO1 fibroblasts were shown to accumulate subcomplexes comprising COX1 and COX4 [14]. We have identified eight distinct COX subcomplexes (b–i) in various tissues of our SCO2 and SURF1 patients. In addition to monomeric holoenzyme complex (a), all samples contained subcomplex b of approx. 180 kDa that corresponds to the previously identified assembly intermediate S3, which, except for COX6A and either COX7A or COX7B, contains all COX subunits [15]. Heart, skeletal muscle and brain of SCO2 patients contained highly increased levels of a 110 kDa subcomplex c composed of COX1, COX4 and COX5A, very likely identical with the assembly intermediate S2 [15] and with COX1·COX4·COX5A subassembly from SURF1 fibroblasts [14]. In addition, SCO2 heart, skeletal muscle and brain contained increased levels of subcomplexes d–f of 60–90 kDa. They involved solely COX1 subunit and might be identical with subassemblies d–f from SURF1 patient fibroblasts [14]. We assume that subcomplex f represents unassembled COX1 subunit and might thus correspond to assembly intermediate S1 [15]. In SCO2 brain mitochondria, subcomplex c was detectable almost exclusively with anti-COX1 antibody, and the ratio of subcomplexes d–f to subcomplex c was substantially lower here when compared with SCO2 heart and skeletal muscle. This might reflect more efficient clearance of partially assembled COX subunits in brain mitochondria. This is further supported by the fact that, in contrast with heart and skeletal muscle, control brain mitochondria did not reveal, even after very sensitive detection, any COX subcomplexes, except for very low levels of ubiquitous subcomplex b (results not shown). Interestingly, only subcomplexes b and f were found slightly accumulated in the heart of our SURF1 patient, although fibroblasts from this patient contained highly increased levels of four subcomplexes comprising COX1, COX4 and COX5A [14]. Furthermore, despite severe reduction of holoenzyme, the brain sample from the identical patient did not contain any accumulated COX subcomplexes, and also SURF1 skeletal-muscle samples showed rather faint accumulation of subcomplexes, pointing to another tissue-specific aspect of SURF1 deficiency.
In the absence of one or more mitochondrially encoded subunits, the levels of COX4 and COX5A are the least affected [32–39]. Both subunits were also shown to be closely positioned within the X-ray structure of the bovine enzyme [5]. Therefore it was suggested that they might already be associated before assembly with COX1 [40]. Apparently, the COX4·COX5A subcomplex of approx. 40 kDa (g) from SCO2 patient samples represents the proposed heterodimer. According to the current model of the COX assembly pathway, the association of subunits starts with the interaction of COX1 with either COX4 or COX5A [14,15,40]. Instead, our results demonstrate that the mutual association of these nucleus-encoded subunits probably precedes their addition to COX1 during the assembly process (Scheme 1).
Scheme 1. Proposed model of the assembly pathway of human COX.
The Arabic numerals denote subunits of the enzyme. S1–S4 indicate previously identified assembly intermediates. Prosthetic groups are also indicated. Dimerization of the 13-subunit holoenzyme (S4) completes the assembly of the COX complex.
It was previously shown that pools of unassembled COX subunits exist [41]. In fact, subcomplexes h and i with a molecular mass of approx. 10–20 kDa are composed of single COX4 and COX5A subunits respectively and very likely represent unassembled subunits. This indicates that high residual levels of these subunits reported from SCO2 patient tissues [9,11,42] and from cells devoid of one or more mitochondrially encoded subunits [33–40] are attributable to (i) high intrinsic stability of both subunits and (ii) their association with each other and COX1. The lower levels of unassembled COX4, compared with COX5A, observed in most of the probed tissues, very likely reflect the lower intrinsic stability of the COX4 polypeptide. This is further supported by the appearance of detectable levels of free COX5A subunit, but not COX4, under physiological conditions (Figures 3A and 3D).
Most of the subcomplexes that we have identified in probed tissues apparently correspond to known assembly intermediates [15] and/or to subassemblies identified in cultured human cells [14]. Therefore it is unlikely that they represent irrelevant aggregates or parts of labile enzyme with disrupted tertiary interactions. To rule out possible proteolytic breakdown, the correct size of detected subunits was confirmed on two-dimensional native/denaturing immunoblots. Taken together, we assume that the subcomplexes identified in the present study correspond to protected rate-limiting steps relevant to the normal assembly pathway (assembly intermediates) that accumulate in mitochondria of patients due to the impaired biogenesis or maintenance of COX.
The SCO2 mutations carried by our patients are thought to reduce the efficiency with which the CuA centre of COX2 is formed [11,28]. In the present study, we show that, in SCO2-deficient mitochondria, the assembly process is stalled before COX2 associates with the COX1·COX4·COX5A subcomplex. Indeed, all investigated SCO2-deficient tissues with severely reduced holoenzyme contained increased levels of COX1-, COX4- and COX5A-comprising subcomplexes. The fact that the immunoblots did not reveal accumulation of free COX2 or COX2-containing subcomplex(es) suggests that the lack of CuA centre within COX2 results either in (i) decreased stability of the subunit or (ii) diminished efficiency of its interaction with the COX1·COX4·COX5A subcomplex. However, the latter would require that the stability of CuA lacking free COX2 is low enough, albeit not directly responsible for assembly impairment, to prevent its accumulation in mitochondria of patients. Interestingly, the profile of COX assembly defects observed in SCO2 patient tissues closely resembles the situation in cells with inhibited mitochondrial protein synthesis due to doxycycline treatment [15].
The precise molecular basis of tissue-specific consequences of SCO2 and SURF1 mutations remains unresolved. In addition to different levels of COX holoenzyme, variable levels of subcomplexes were found among different tissues, although some of them displayed the same residual level of the holoenzyme (e.g. heart and muscle). This is likely to be attributable to different rates of clearance of partially assembled or unassembled subunits. The tissue-specific pattern of assembly defects only partially overlaps with the expression of particular tissue-specific isoforms of COX subunits, suggesting the involvement of a rather different mechanism. In our patients, the mutant SCO2 protein was almost undetectable in brain and heart with profound COX deficiency, whereas the liver, and particularly fibroblasts, contained small but significant amounts of residual SCO2. However, we find it unlikely that this minor difference could account for the distinct biochemical and clinical involvement of these tissues, unless there is a pronounced difference in ‘spare capacity’ of SCO2 for copper delivery to the CuA centre in these tissues. Therefore we speculate that tissue-specific consequences of SCO2 and SURF1 mutations, in terms of both holoenzyme and subcomplex levels, suggest the existence of tissue-specific functional differences of these proteins that may have evolved to meet different requirements for the regulation of COX biogenesis.
Acknowledgments
This work was supported by grants from the Grant Agency of Charles University (GAUK 17/2004/C to L.S.) and the Grant Agency of Czech Republic (GACR 303/03/D132 to L.C. and GACR 303/03/0749 to J.H.), by the European Union's Sixth Framework Programme for Research, Priority 1 ‘Life sciences, genomics and biotechnology for health’ (LSHM-CT-2004-503116 to P.P. and M.T.), and institutional projects (CAG 1M6837805002 to J.Z. and MSM0021620806 to H.H. and K.V.). We acknowledge S. Knopova for technical assistance.
References
- 1.Capaldi R. A. Structure and assembly of cytochrome c oxidase. Arch. Biochem. Biophys. 1990;280:252–262. doi: 10.1016/0003-9861(90)90327-u. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig B., Bender E., Arnold S., Huttemann M., Lee I., Kadenbach B. Cytochrome c oxidase and the regulation of oxidative phosphorylation. ChemBioChem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.Kadenbach B., Stroh A., Becker A., Eckerskorn C., Lottspeich F. Tissue- and species-specific expression of cytochrome c oxidase isozymes in vertebrates. Biochim. Biophys. Acta. 1990;1015:368–372. doi: 10.1016/0005-2728(90)90042-3. [DOI] [PubMed] [Google Scholar]
- 4.Linder D., Freund R., Kadenbach B. Species-specific expression of cytochrome c oxidase isozymes. Comp. Biochem. Physiol. B. 1995;112:461–469. doi: 10.1016/0305-0491(95)00093-3. [DOI] [PubMed] [Google Scholar]
- 5.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 6.Carr H. S., Winge D. R. Assembly of cytochrome c oxidase within mitochondrion. Acc. Chem. Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- 7.Barrientos A., Barros M. H., Valnot I., Rotig A., Rustin P., Tzagoloff A. Cytochrome oxidase in health and disease. Gene. 2002;286:53–63. doi: 10.1016/s0378-1119(01)00803-4. [DOI] [PubMed] [Google Scholar]
- 8.Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadopoulou L. C., Sue C. M., Davidson M. M., Tanji K., Nishino I., Sadlock J. E., Krishna S., Walker W., Selby J., Glerum D. M., et al. Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 1999;23:333–337. doi: 10.1038/15513. [DOI] [PubMed] [Google Scholar]
- 10.Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H. G., Gerbitz K. D., Shoubridge E. A. Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000;9:795–801. doi: 10.1093/hmg/9.5.795. [DOI] [PubMed] [Google Scholar]
- 11.Jaksch M., Paret C., Stucka R., Horn N., Muller-Hocker J., Horvath R., Trepesch N., Stecker G., Freisinger P., Thirion C., et al. Cytochrome c oxidase deficiency due to mutations in SCO2, encoding a mitochondrial copper-binding protein, is rescued by copper in human myoblasts. Hum. Mol. Genet. 2001;10:3025–3035. doi: 10.1093/hmg/10.26.3025. [DOI] [PubMed] [Google Scholar]
- 12.Tiranti V., Galimberti C., Nijtmans L., Bovolenta S., Perini M. P., Zeviani M. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 1999;8:2533–2540. doi: 10.1093/hmg/8.13.2533. [DOI] [PubMed] [Google Scholar]
- 13.Yao J., Shoubridge E. A. Expression and functional analysis of SURF1 in Leigh syndrome patients with cytochrome c oxidase deficiency. Hum. Mol. Genet. 1999;8:2541–2549. doi: 10.1093/hmg/8.13.2541. [DOI] [PubMed] [Google Scholar]
- 14.Williams S. L., Valnot I., Rustin P., Taanman J. W. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- 15.Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 1998;254:389–394. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A. P., Newbold R. F., Wang J., Chevrette M., et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998;20:337–343. doi: 10.1038/3804. [DOI] [PubMed] [Google Scholar]
- 17.Tiranti V., Hoertnagel K., Carrozzo R., Galimberti C., Munaro M., Granatiero M., Zelante L., Gasparini P., Marzella R., Rocchi M., et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pequignot M. O., Dey R., Zeviani M., Tiranti V., Godinot C., Poyau A., Sue C., Di Mauro S., Abitbol M., Marsac C. Mutations in the SURF1 gene associated with Leigh syndrome and cytochrome c oxidase deficiency. Hum. Mutat. 2001;17:374–381. doi: 10.1002/humu.1112. [DOI] [PubMed] [Google Scholar]
- 19.Sue C. M., Karadimas C., Checcarelli N., Tanji K., Papadopoulou L. C., Pallotti F., Guo F. L., Shanske S., Hirano M., De Vivo D. C., et al. Differential features of patients with mutations in two COX assembly genes, SURF-1 and SCO2. Ann. Neurol. 2000;47:589–595. [PubMed] [Google Scholar]
- 20.Vesela K., Hansikova H., Tesarova M., Martasek P., Elleder M., Houstek J., Zeman J. Clinical, biochemical and molecular analyses of six patients with isolated cytochrome c oxidase deficiency due to mutations in the SCO2 gene. Acta Paediatr. 2004;93:1312–1317. doi: 10.1080/08035250410008761. [DOI] [PubMed] [Google Scholar]
- 21.Williams S. L., Taanman J. W., Hansikova H., Houstkova H., Chowdhury S., Zeman J., Houstek J. A novel mutation in SURF1 causes skipping of exon 8 in a patient with cytochrome c oxidase-deficient Leigh syndrome and hypertrichosis. Mol. Genet. Metab. 2001;73:340–343. doi: 10.1006/mgme.2001.3206. [DOI] [PubMed] [Google Scholar]
- 22.Pecina P., Capkova M., Chowdhury S. K., Drahota Z., Dubot A., Vojtiskova A., Hansikova H., Houstkova H., Zeman J., Godinot C., et al. Functional alteration of cytochrome c oxidase by SURF1 mutations in Leigh syndrome. Biochim. Biophys. Acta. 2003;1639:53–63. doi: 10.1016/s0925-4439(03)00127-3. [DOI] [PubMed] [Google Scholar]
- 23.Rickwood D., Wilson M. T., Darley-Usmar V. M. Isolation and characteristics of intact mitochondria. In: Darley-Usmar V. M., Rickwood D., Wilson M. T., editors. Mitochondria: A Practical Approach. Oxford, U.K.: IRL Press; 1987. pp. 3–5. [Google Scholar]
- 24.Klement P., Nijtmans L. G., Van den Bogert C., Houstek J. Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal. Biochem. 1995;231:218–224. doi: 10.1006/abio.1995.1523. [DOI] [PubMed] [Google Scholar]
- 25.Rustin P., Chretien D., Bourgeron T., Gérard B., Rötig A., Saudubray J. M., Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 26.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 27.Coenen M. J., van den Heuvel L. P., Nijtmans L. G., Morava E., Marquardt I., Girschick H. J., Trijbels F. J., Grivell L. A., Smeitink J. A. SURFEIT-1 gene analysis and two-dimensional blue native gel electrophoresis in cytochrome c oxidase deficiency. Biochem. Biophys. Res. Commun. 1999;265:339–344. doi: 10.1006/bbrc.1999.1662. [DOI] [PubMed] [Google Scholar]
- 28.Leary S. C., Kaufman B. A., Pellecchia G., Guercin G.-H., Mattman A., Jaksch M., Shoubridge E. A. Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet. 2004;13:1839–1848. doi: 10.1093/hmg/ddh197. [DOI] [PubMed] [Google Scholar]
- 29.Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V., Munnich A., Bonnefont J. P., Rustin P., Rotig A. Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 2000;67:1104–1109. doi: 10.1016/s0002-9297(07)62940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foltopoulou P. F., Zachariadis G. A., Politou A. S., Tsiftsoglou A. S., Papadopoulou L. C. Human recombinant mutated forms of the mitochondrial COX assembly Sco2 protein differ from wild-type in physical state and copper binding capacity. Mol. Genet. Metab. 2004;81:225–236. doi: 10.1016/j.ymgme.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Jaksch M., Horvath R., Horn N., Auer D. P., Macmillan C., Peters J., Gerbitz K. D., Kraegeloh-Mann I., Muntau A., Karcagi V., et al. Homozygosity (E140K) in SCO2 causes delayed infantile onset of cardiomyopathy and neuropathy. Neurology. 2001;57:1440–1446. doi: 10.1212/wnl.57.8.1440. [DOI] [PubMed] [Google Scholar]
- 32.Keightley J. A., Hoffbuhr K. C., Burton M. D., Sala V. M., Johnston W. S. W., Penn A. M. W., Buist N. R. M., Kennaway N. G. A microdeletion in cytochrome c oxidase (COX) subunit III associated with COX deficiency and recurrent myoglobinuria. Nat. Genet. 1996;12:410–416. doi: 10.1038/ng0496-410. [DOI] [PubMed] [Google Scholar]
- 33.Comi G. P., Bordoni A., Salani S., Franceschina L., Sciacco M., Prelle M., Fortunato F., Zeviani M., Napoli L., Bresolin L., et al. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann. Neurol. 1998;43:110–116. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- 34.Bruno C., Martinuzzi A., Tang Y., Andreu A. L., Pallotti F., Bonilla E., Shanske S., Fu J., Sue C. M., Angelini C., et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am. J. Hum. Genet. 1999;65:611–620. doi: 10.1086/302546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taanman J.-W., Burton M. D., Marusich M. F., Kennaway N. G., Capaldi R. A. Subunit specific monoclonal antibodies show different steady-state levels of various cytochrome-c oxidase subunits in chronic progressive external ophthalmoplegia. Biochim. Biophys. Acta. 1996;1315:199–207. doi: 10.1016/0925-4439(95)00127-1. [DOI] [PubMed] [Google Scholar]
- 36.Hoffbuhr K. C., Davidson E., Filiano B. A., Davidson M., Kennaway N. G., King M. P. A pathogenic 15-base pair deletion in mitochondrial DNA-encoded cytochrome c oxidase subunit III results in the absence of functional cytochrome c oxidase. J. Biol. Chem. 2000;275:13994–14003. doi: 10.1074/jbc.275.18.13994. [DOI] [PubMed] [Google Scholar]
- 37.Hanson B. J., Carrozzo R., Piemonte F., Tessa A., Robinson B. H., Capaldi R. A. Cytochrome c oxidase-deficient patients have distinct subunit assembly profiles. J. Biol. Chem. 2001;276:16296–16301. doi: 10.1074/jbc.M011162200. [DOI] [PubMed] [Google Scholar]
- 38.Tiranti V., Corona P., Greco M., Taanman J. W., Carrara F., Lamantea E., Nijumans L., Uziel G., Zeviani M. A novel frameshift mutation of the mtDNA COIII gene leads to impaired assembly of cytochrome c oxidase in a patient affected by Leigh-like syndrome. Hum. Mol. Genet. 2000;9:2733–2742. doi: 10.1093/hmg/9.18.2733. [DOI] [PubMed] [Google Scholar]
- 39.Williams S. L., Scholte H. R., Gray R. G., Leonard J. V., Schapira A. H., Taanman J. W. Immunological phenotyping of fibroblast cultures from patients with a mitochondrial respiratory chain deficit. Lab. Invest. 2001;81:1069–1077. doi: 10.1038/labinvest.3780319. [DOI] [PubMed] [Google Scholar]
- 40.Taanman J.-W., Williams S. L. Assembly of cytochrome c oxidase: what can we learn from patients with cytochrome c oxidase deficiency? Biochem. Soc. Trans. 2001;29:446–451. doi: 10.1042/bst0290446. [DOI] [PubMed] [Google Scholar]
- 41.Hundt E., Trapp M., Kadenbach B. Biosynthesis of cytochrome c oxidase in isolated rat hepatocytes. FEBS Lett. 1980;115:95–99. doi: 10.1016/0014-5793(80)80734-4. [DOI] [PubMed] [Google Scholar]
- 42.Tarnopolsky M. A., Bourgeois J. M., Fu M.-H., Kataeva G., Shah J., Simon D. K., Mahoney D., Johns D., MacKay N., Robinson B. H. Novel SCO2 mutation (G1521A) presenting as a spinal muscular atrophy type I phenotype. Am. J. Med. Genet. 2004;125:310–314. doi: 10.1002/ajmg.a.20466. [DOI] [PubMed] [Google Scholar]