Abstract
GSK3 (glycogen synthase kinase-3) regulation is proposed to play a key role in the hormonal control of many cellular processes. Inhibition of GSK3 in animal models of diabetes leads to normalization of blood glucose levels, while high GSK3 activity has been reported in Type II diabetes. Insulin inhibits GSK3 by promoting phosphorylation of a serine residue (Ser-21 in GSK3α, Ser-9 in GSK3β), thereby relieving GSK3 inhibition of glycogen synthesis in muscle. GSK3 inhibition in liver reduces expression of the gluconeogenic genes PEPCK (phosphoenolpyruvate carboxykinase), G6Pase (glucose-6-phosphatase), as well as IGFBP1 (insulin-like growth factor binding protein-1). Overexpression of GSK3 in cells antagonizes insulin regulation of these genes. In the present study we demonstrate that regulation of these three genes by feeding is normal in mice that express insulin-insensitive GSK3. Therefore inactivation of GSK3 is not a prerequisite for insulin repression of these genes, despite the previous finding that GSK3 activity is absolutely required for maintaining their expression. Interestingly, insulin injection of wild-type mice, which activates PKB (protein kinase B) and inhibits GSK3 to a greater degree than feeding (50% versus 25%), does not repress these genes. We suggest for the first time that although pharmacological inhibition of GSK3 reduces hepatic glucose production even in insulin-resistant states, feeding can repress the gluconeogenic genes without inhibiting GSK3.
Keywords: diabetes, glucose, glycogen synthase kinase-3 (GSK3), insulin, phosphoenolpyruvate carboxykinase (PEPCK), sterol regulatory element binding protein 1 (SREBP1)
Abbreviations: CREB, cAMP-response element-binding protein; CBP, CREB-binding protein; DKI, double knockin; FAS, fatty acid synthase; G6Pase, glucose-6-phosphatase; GSK3, glycogen synthase kinase-3; IGFBP1, insulin-like growth factor-binding protein-1; PDK1, 3-phosphoinositide-dependent protein kinase-1; PEPCK, phosphoenolpyruvate carboxykinase; PI3-kinase, phosphoinositol 3-kinase; PKB, protein kinase B; RPA, ribonuclease protection assay; RT, reverse transcription; S6K, S6 kinase; SREBP, sterol regulatory element binding protein; TIRE, thymine rich insulin response element
INTRODUCTION
The molecular defects that are responsible for the large increase in occurrence of Type II diabetes are not yet fully understood but are the subject of intense study. Obesity can promote cellular resistance to insulin, probably by altering the intracellular signalling processes involved in regulation of glucose homoeostasis (for review see [1–3]). GSK3 (glycogen synthase kinase-3), an insulin-inhibited signalling molecule, has been implicated in the hormonal control of many cellular processes [4]. It was originally identified as a protein kinase that phosphorylates and inactivates glycogen synthase in muscle [5]. Two different isoforms of GSK3 are expressed from distinct genes (GSK3α and GSK3β). They share greater than 95% identity in their kinase domains and are ubiquitously expressed in mammals [6]. Inhibition of GSK3 can be achieved through phosphorylation of an N-terminal serine residue (Ser-21 in GSK3α, Ser-9 in GSK3β) [7] and insulin promotes this phosphorylation by activation of PKB (protein kinase B)/c-AKT [8]. This signalling pathway from the insulin receptor, involving PKB activation and GSK3 inhibition results in increased glycogen synthesis and is a major mechanism by which insulin induces glycogen synthesis in muscle [9].
Highly selective inhibitors of GSK3 mimic the action of insulin on glycogen synthesis and hepatic glucose production [10–12]. Insulin controls hepatic glucose production largely through repression of the gluconeogenic genes PEPCK (phosphoenolpyruvate carboxykinase) and G6Pase (glucose-6-phosphatase) [11]. A number of hepatic genes including PEPCK, G6Pase and IGFBP1 (insulin-like growth factor-binding protein-1) share a common TIRE (thymine rich insulin response element) that can be regulated by several transcription factors such as members of the Forkhead (FOXO) family [13–15]. Much data supports a pathway from the insulin receptor to these gene promoters involving activation of PI3-kinase (phosphoinositol-3 kinase), PDK1 (3-phosphoinositide-dependent protein kinase-1) [16] and PKB. Overexpression of active PKB represses the TIRE-containing promoters, potentially through phosphorylation of FOXO [17–19]. However, active PKB will also phosphorylate and inhibit GSK3 [8]. Our laboratory has found that GSK3 overexpression can decrease the insulin sensitivity of the IGFBP1 gene, as well as the isolated IGFBP1 TIRE in hepatoma cells [20]. Other transcription factors such as SREBP1c (sterol regulatory element binding protein 1c) and CBP (CREB-binding protein where CREB stands for cAMP-response element-binding protein)/p300 are capable of affecting insulin regulation of PEPCK [21–23], and SREBP1c is reported to be a substrate for GSK3 [24]. We hypothesized that inhibition of GSK3 activity is part of the mechanism that the hormone uses to repress these insulin response elements.
In an effort to conclusively delineate which of these downstream targets of PKB is important in the regulation of TIRE containing gene promoters we have made use of a mouse model in which GSK3 cannot be regulated by PKB. The genes for GSK3α and GSK3β have been replaced with mutant alleles that contain alanine at the serine residue targeted by PKB (S21A in GSK3α, S9A in GSK3β). Interestingly, these DKI (double knock-in) mice are viable and display no signs of overt diabetes, despite insulin being unable to stimulate glycogen synthase in skeletal muscle of GSK3β knockin mice [9]. In the present study we show that feeding represses PEPCK, G6Pase and IGFBP1 normally in these mice, demonstrating that phosphorylation of GSK3 by PKB is not required for insulin regulation of these genes. Similarly, regulation of SREBP1c expression and a downstream target gene appear relatively normal.
EXPERIMENTAL
Materials
Protein G–Sepharose and [32Pγ]ATP were purchased from Amersham Pharmacia Biotech, protease-inhibitor cocktail tablets and Taq Polymerase were purchased from Roche. Restriction enzymes were from New England Biolabs. Precast SDS polyacrylamide Bis-Tris gels and tissue culture reagents were from Invitrogen. Human insulin from Novo-Nordisk was obtained from Ninewells Pharmacy, Dundee. Phosphocellulose P81 paper and cellulose 31ETCHR paper were from Whatman. Scintillation fluid was ‘Optiscint Hisafe’ from PerkinElmer. All peptides were synthesized by Dr Graham Bloomberg at the University of Bristol.
Animals
The generation of the DKI mouse has been described in detail previously [9]. All animal studies and breeding performed in this study was approved by the University of Dundee ethical committee and performed under a U.K. Home Office project license.
Antibodies
The following antibodies were raised in sheep and affinity purified for the appropriate antigen: total PKB (isolated pleckstrin homology domain of PKBα), total GSK3α (residues 471–483 of rat GSK3 α, QAPDATPTLTNSS) used for immunoblotting, total GSK3β (full-length human protein) used for immunoblotting, S6K (S6 kinase; AGVFDIDLDQPEDAGSEDEL–residues 25–44 of human S6K), S6 protein phospho-S235 (AKRRRLS*SLRASTS – residues 229–242 of human S6). The total PKBα antibody used to immunoprecipitate PKBα was a mouse monoclonal antibody raised against residues 1–149 of human PKB and was purchased from Upstate Inc. The following antibodies were purchased from Cell Signaling Technology: PKB phospho-S473-P, GSK3α/GSK3β-phospho S21-P/S9-P, S6 protein, S6 kinase phospho-T389. Secondary antibodies coupled to horseradish peroxidase were from Pierce.
General buffers
Lysis buffer: 50 mM Tris/HCl (pH 7.5), 1 mM EGTA, 1% (w/v) Triton X-100, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 5 mM sodium pyrophosphate, 0.27 M sucrose, 0.1 μM microcystin-LR, 0.1% (v/v) 2-mercaptoethanol and Complete™ proteinase inhibitor cocktail (Roche) (one tablet per 50 ml). Buffer A was 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA and 0.1% (v/v) 2-mercaptoethanol.
Preparation of tissue extracts
Following an overnight fast±refeeding (for 1 or 6 h), or injection of insulin (150 mg/g), or a combination of insulin and glucose (2 mg/g) for 1 h, the animal was killed by cervical dislocation before removal of the liver. This was immediately frozen in liquid nitrogen. One part was ground and ten volumes of ice-cold lysis buffer added, the solution vortexed, then centrifuged at 4 °C. The supernatant was collected and protein concentration was measured according to the method of Bradford [9a], then snap frozen in aliquots and stored at −80 °C. A second piece of frozen liver was homogenized in Tri Reagent™ (Sigma) and total RNA isolated according to the manufacturer's instructions.
Immunoprecipitation and assay of protein kinases
Liver lysate (500 μg) was used to immunoprecipitate PKBα and S6K. Kinases were immunoprecipitated and assayed as previously described [9].
Immunoblotting
Protein lysate (20 μg) in SDS sample buffer was subjected to SDS/PAGE and transferred to nitrocellulose. For phospho-specific antibody blots the nitrocellulose membranes were immunoblotted at 4 °C for 16 h using the indicated antibodies (1–2 μg/ml for sheep antibodies or 500–1000-fold dilution for commercial antibodies) in the presence of 10 μg/ml of the de-phosphopeptide antigen used to raise the antibody to sheep antibodies. For immunoblotting of total PKBα, GSK3α and GSK3β the nitrocellulose membranes were immunoblotted for 2 h at room temperature using the antibodies described above. Detection was performed using horseradish-peroxidase-conjugated secondary antibodies and the enhanced chemiluminescence reagent.
Measurement of plasma insulin levels
Blood was collected from mice following cervical dislocation and incubated on ice for 30 min. The blood was then centrifuged at 3000 g for 10 min and the plasma supernatant was collected. The plasma insulin level was measured using ultra sensitive Insulin ELISA kit (90060) and mouse insulin standards (90090), both purchased from Crystal Chem Inc. U.S.A. Plasma (15 μl) was used for each assay and mouse insulin standards from 0–6 ng/ml were used in order to generate a standard curve.
RPA (ribonuclease protection assay)
RPA was used to determine the relative expression levels of IGFBP1, G6Pase and β-actin mRNA. Mouse IGFBP1 and G6Pase probes were synthesized by in vitro transcription as described previously [25]. pTRI-β-actin (mouse) linear plasmid (Ambion Inc.) was used as the control linear template. The RPA was carried out using the RPA II Kit (Ambion Inc.). Briefly, 10 μg of total RNA was hybridized to 20000 c.p.m. of each labelled probe. Samples were incubated with RNase to digest single-stranded RNA, and double-stranded products were separated on an 8 M urea/5% polyacrylamide gel. Radioactivity present in the appropriate band was quantified on a PhosphorImager (Fuji) and the data presented as the ratio of IGFBP1 or G6Pase to β-actin mRNA.
Real-time quantitative RT (reverse transcription)-PCR
cDNA was synthesized from total RNA using Superscript™ II Reverse Transcriptase Kit (Invitrogen). PCR analysis was carried out in a model 7700 sequence detector (Applied Biosystems) with primers and probes as follows: PEPCK 5′-ccatcacctcctggaagaaca-3′, sense; 5′-accctcaatgggtactccttctg-3′, antisense and 5′-caggacgcggaaccatgtgcc-3′, probe; SREBP-1 5′-gcggttggcacagagctt-3′, sense; 5′-ggacttgctcctgccatcag-3′, antisense and 5′-cggcctgctatgaggagggtattcctacat-3′, probe; FAS (fatty acid synthase) 5′-ggcatcattgggcactcctt-3′, sense; 5′-gctgcaagcacagcctctct-3′, antisense and 5′-ccatctgcatagccacaggcaacctc-3′, probe. Probes were synthesized with 5′FAM (6-carboxyfluorescein) and 3′-TAMRA (5- and 6-carboxytetramethylrhodamine) modifications. All the mRNA abundancies are presented as ratios relative to 18S rRNA levels. The 18S rRNA Taqman Control Reagent was obtained from Applied Biosystems.
Statistics
Data was analysed by Student's t-test, with 5% significance levels applied.
RESULTS AND DISCUSSION
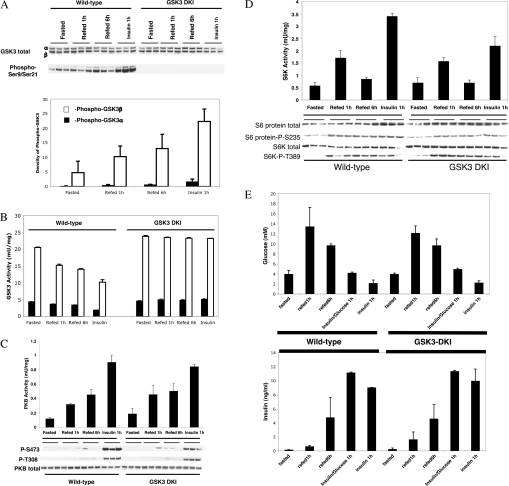
Previous data have suggested several possible mechanisms by which insulin could repress PEPCK, G6Pase and IGFBP1 expression. Overexpression studies, combined with pharmacological inhibition, has implicated GSK3 in the control of these processes by insulin [11,20]. Therefore we examined whether these genes were regulated appropriately in an animal that expressed a mutant form of GSK3 that does not respond to insulin. GSK3 regulation by insulin requires activation of PI3-kinase and PKB. This in turn promotes phosphorylation of GSK3α (at Ser-21) and GSK3β (at Ser-9) significantly decreasing their activity [7,8]. In the GSK3 DKI mouse endogenous GSK3α and GSK3β are replaced with mutant enzymes that are not substrates for PKB (S21A/S9A), and these proteins are expressed from the wild-type gene locus [9]. Expression of the mutant protein in all tissues, including liver, is almost identical to normal expression of wild-type proteins (Figure 1A, upper panel). Feeding or insulin injection promotes phosphorylation of Ser-9/Ser-21 of hepatic GSK3, but only in the wild-type animals (Figure 1A), as these residues are not phosphorylatable in the GSK3 DKI mice. This phosphorylation represents a decrease in GSK3 activity (Figure 1B) of approx. 50% with insulin injection, or 20–30% with feeding. Moreover, activation of PKB (Figure 1C) and S6K (Figure 1D) in liver of wild-type and knockin animals is almost identical. Feeding does not activate the PKB pathway as potently as insulin injection (in either animal) explaining the decreased phosphorylation and inhibition of GSK3 with feeding compared with insulin (Figure 1B). The decreased activation of PKB and inhibition of GSK3 could question the importance of this pathway in the regulation of PEPCK, G6Pase and IGFBP1 following feeding. However, a significant regulation of the pathway is still observed with feeding (Figure 1). Knockin of the mutant GSK3 alleles does not significantly alter the regulation of PKB (Ser-473 or Thr-308) or S6K (Thr-389) phosphorylation, their inherent activity or phosphorylation of the downstream S6 protein (Ser-235) [by either feeding or insulin (Figure 1)]. Therefore GSK3 insensitivity to insulin does not alter regulation of other insulin- sensitive kinases in the liver, or muscle [9]. All of the results presented for insulin injection in Figure 1(A–D) is in the absence of co-injection of glucose.
Figure 1. Intracellular signalling in livers of wild-type and GSK3 DKI mice.
Wild-type (12) and GSK3-DKI mice (12) at 10–12 weeks old were fasted overnight, three were injected with insulin for 1 h, three were refed for 1 h and three refed for 6 h, before being killed and excision of the liver. Three fasted animals were used as a control. (A) GSK3 expression and Ser-21/Ser-9 phosphorylation were assessed by Western blotting. The phospho-GSK3 was quantified by densitometry and is shown in the lower panel. (B) GSK3α and GSK3β activity was measured in each sample. PKB (C) and S6K (D) activity were measured by Western blotting, as well as by immunoprecipitation and assay. Assay data are presented as munit/mg lysate and are the average±S.D. of three separate animals assayed in triplicate. (E) Plasma glucose and insulin levels were determined for each animal following these treatments, as well as an additional treatment where three wild-type and three GSK3 knockin animals were injected with insulin plus glucose for 1 h.
However, identical regulation of these signalling pathways is observed if glucose is co-injected with insulin (results not shown) in order to prevent hypoglycemia (Figure 1E). These mice are perfectly viable and there are no significant differences in the fasted or fed plasma glucose/insulin levels between the GSK3 knockin and wild-type animals (Figure 1E).
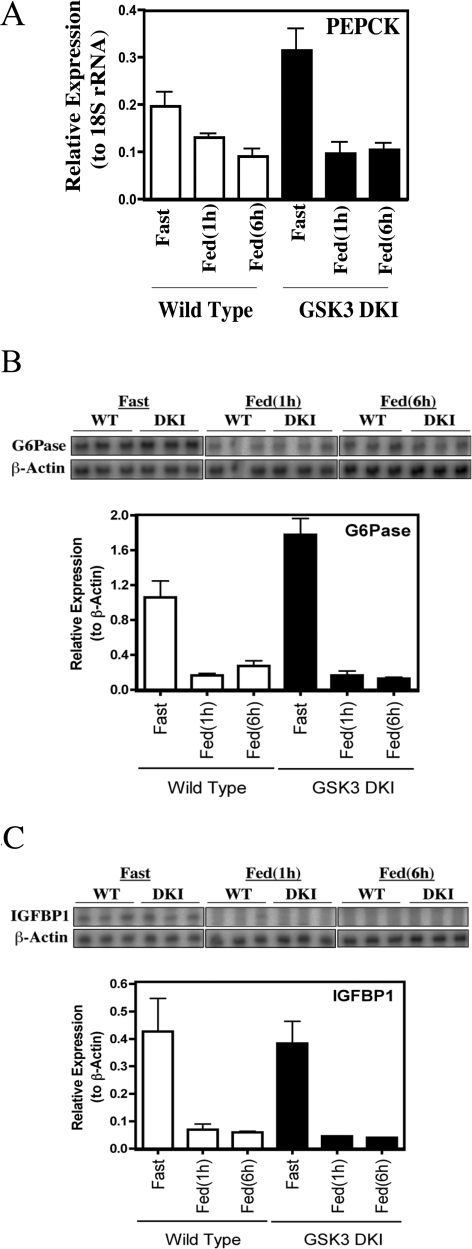
PEPCK and G6Pase are rate-controlling enzymes of gluconeogenesis [26]. Decrease in their expression represents a major mechanism by which feeding turns off hepatic glucose output. Previous work has implicated GSK3 activity as a factor in the regulation of these genes [11,20]. In addition, lithium reduces hepatic gluconeogenesis [27] whereas administration of more selective GSK3 inhibitors to animals decreases hepatic glucose output [12,28]. However, no defects in the repression of PEPCK, G6Pase or IGFBP1 expression following feeding were observed in the DKI animals (Figure 2). GSK3 activity is required for PEPCK, G6Pase and IGFBP1 gene promoter activity in hepatoma cells [11,20], whereas GSK3 overexpression (which is reported in diabetes [29]) will decrease insulin regulation of these promoters [20]. However, under normal GSK3 expression levels, feeding represses these genes without the need to inhibit GSK3 (Figure 2).
Figure 2. Analysis of metabolic gene expression following feeding.
Total RNA was prepared from livers obtained as described in Figure 1. (A) PEPCK gene expression was measured by real-time PCR using 18S ribosomal RNA as an internal control. Data are presented relative to the internal control, as the mean±S.E.M. of three separate animals assayed in triplicate. G6Pase (B) and IGFBP1 (C) gene expression were measured by RPA. Data are presented relative to control (β-actin) as the mean±S.E.M. of three separate animals assayed in triplicate (lower panel) and the autoradiograph of each assay is also presented (upper panel).
There could be several reasons for the apparent difference in the requirement for GSK3 repression in the regulation of these genes in isolated cells and in vivo. First, GSK3 overexpression decreases insulin regulation of IGFBP1 expression in hepatoma cells whereas the GSK3 knockin animals express mutant GSK3 at levels equivalent to normal. Alternatively, the cell work was used to examine insulin regulation of the genes, whereas we have used feeding in the present study. In the wild-type animals the level of inhibition of GSK3 following feeding is less than that obtained following insulin injection (Figure 1).
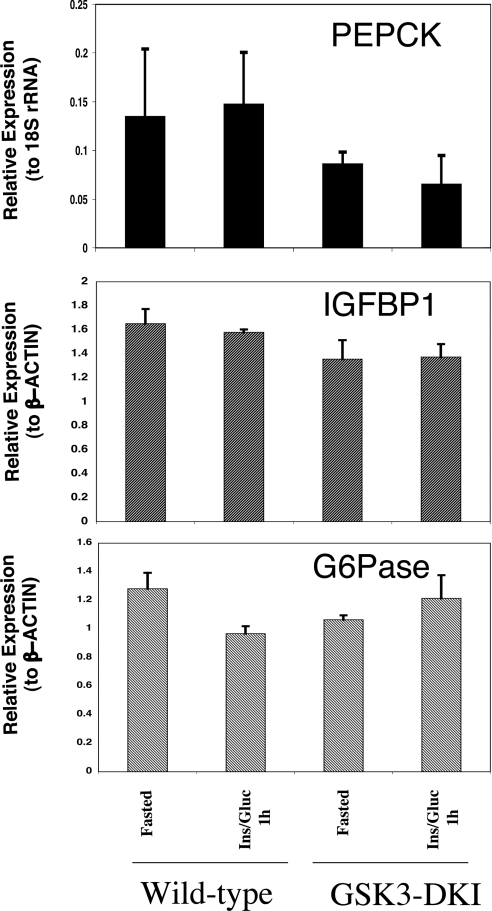
Therefore we examined the effect of insulin injection on the regulation of PEPCK, G6Pase and IGFBP1. Surprisingly, no repression of any of the genes was observed 1 h after injection, despite activation of PKB (Figure 1C), inhibition of GSK3 (Figure 1B) and decrease in blood glucose levels (Figure 1E). Initially we assumed that the hypoglycemia that occurs following insulin injection was affecting insulin regulation of the genes. However, when glucose was co-injected with insulin to maintain blood glucose levels (Figure 1E), no repression of these genes was seen (Figure 3). There are few reports examining regulation of these genes following insulin injection of rodents, but a similar lack of effect on PEPCK [30] or G6Pase [31] expression has been reported. In those studies Granner et al. demonstrated a clear requirement for insulin in PEPCK repression following feeding but proposed the need for an additional intestinal or pancreatic secretion in the response to feeding [30]. Others have reported some repression of PEPCK and G6Pase during hyperinsulinemic euglycemia but at longer incubation times [32]. In addition, the degree of repression in those experiments was not as substantial as seen with feeding.
Figure 3. Insulin injection does not regulate PEPCK, G6Pase or IGFBP1 gene expression.
Total RNA was prepared from the livers of animals fasted overnight, or fasted then injected with insulin plus glucose, as described in the Experimental section. Gene expression was measured as described in Figure 2. Data are presented relative to the internal control, as the mean±S.E.M. of three separate animals assayed in triplicate. Identical results are obtained if the animals are injected with insulin alone.
We investigated the effect of insulin at 1 h, when clear activation of PKB is seen (Figure 1C) and when feeding completely represses all of these genes (Figure 2). Because the repression of the genes by feeding in the GSK3 knockin animals is normal we propose that GSK3 inhibition is not actually required for regulation of the genes by feeding and that the extent of inhibition of GSK3 by insulin injection (50%) is not sufficient in itself to mimic the effects of GSK3 inhibitors on these genes. Indeed, whatever signal is being generated by insulin, it is not sufficient to repress these genes in vivo (Figure 3), but it is sufficient in isolated hepatocytes or hepatoma cells [33].
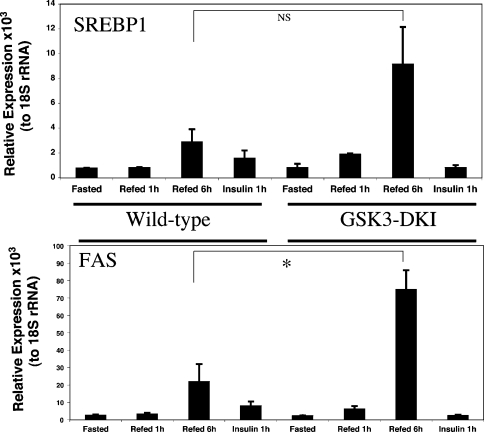
In a previous study we found that deletion of PDK1 activity (upstream of GSK3 regulation) decreases the induction of SREBP1 gene expression in response to feeding [16]. This demonstrates that a PDK1-dependent pathway controls SREBP1 induction by feeding. In addition, GSK3 activity has been reported to directly regulate SREBP1c activity [24], whereas induction of FAS expression following feeding is dependent upon activation of SREBP1 [34–36]. Therefore we examined expression of SREBP1 and FAS in the GSK3 DKI animals. Once more, regulation of SREBP1 expression or activity (FAS expression) by feeding is relatively normal (or possibly even more sensitive to feeding) in the GSK3 knockin animals (Figure 4). Therefore we conclude that inhibition of GSK3 is not a prerequisite for regulation of SREBP1 activity or expression by feeding. Although there is a trend to a more potent induction of SREBP1 expression following feeding, the effect is not significant (Figure 4). There is a significantly greater induction of FAS expression after 6 h refeeding, possibly due to the slightly enhanced induction of SREBP1. Alternatively, direct regulation of SREBP1 by GSK3 [24] may play a role in this effect. There are few metabolic defects in the GSK3 knockin animals suggesting that this increased hepatic FAS expression is compensated for in the intact animal in some way; however, the results still indicate that GSK3 influences the regulation of this gene promoter.
Figure 4. Induction of SREBP1 activity and expression does not require inactivation of GSK3.
Total RNA was prepared from the livers obtained as described in the Experimental section. SREBP1 (A) and FAS (B) gene expression were measured by real-time PCR using 18S ribosomal RNA as control. Data are presented as the mean (n=3) relative to the internal control. * P<0.01.
The fact that the GSK3 knockin animals have little in the way of abnormal phenotype may suggest that GSK3 is not as important a regulator of glucose metabolism as first envisioned. However, there are several possible reasons for the lack of a clear phenotype in this model. For example, most targets of GSK3 require prior phosphorylation (priming) to permit phosphorylation by GSK3 [4]. Therefore if inhibition of the priming kinase occurs then substrate regulation would persist, even in the GSK3 knockin animals. It is possible that the protein(s) that links GSK3 to the TIRE containing genes is also regulated by control of priming following feeding. Hence, the protein would remain sensitive to feeding in the absence of GSK3 inhibition, yet it would be regulated by GSK3 inhibitors. There are at least two examples of GSK3 substrates in which priming is likely to be a regulated event, namely n-myc downstream-regulated gene [37] and CREB [38].
Secondly, phosphorylation is a reversible process, with regulation of phosphatases playing an increasingly acknowledged key role in signalling pathways. It is possible that insulin activation of a phosphatase is sufficient to decrease GSK3 substrate phosphorylation in the knockin animals but not when GSK3 is over-expressed. This may contribute to the discordance between the results obtained in cells after overexpression [20] and these knockin studies (Figure 2).
The target of GSK3 that mediates regulation of the TIRE-containing promoters remains to be identified and much work in our lab is focussed on this question. Its identification will greatly improve our understanding of the connections that allow GSK3 activity to influence hepatic glucose production. Importantly, the results presented in the present study do not rule out GSK3 (or its substrates) as a potential target for intervention in hyperglycemic or insulin-resistant states. Indeed, in subjects with high levels of GSK3 activity this intervention would be predicted to be very effective, as witnessed in animal models of diabetes. Moreover, the GSK3 knockin model is providing important clues as to the complexity of insulin signalling and GSK3 function. The work presented in the present study, using this model, clearly demonstrates that although some GSK3 activity is required for expression of PEPCK, G6Pase and IGFBP1 the physiological repression of these genes by feeding does not require GSK3 inhibition.
Acknowledgments
We thank the protein production and antibody purification teams at DSTT (Division of Signal Transduction Therapy), University of Dundee, co-ordinated by Hilary McLauchlan and James Hastie, for the affinity purification of antibodies, and Professor D. K. Granner (Vanderbilt University, TN, U.S.A.) for helpful discussions. C. L. and E. J. M. are recipients of a BBSRC CASE studentship and an MRC Predoctoral Fellowship respectively and C. S. is the recipient of a Diabetes U.K. Senior Fellowship (02/0002473). This work was supported by Diabetes U.K., the MRC U.K., the Moffat Charitable Trust and the pharmaceutical companies that support the DSTT (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc., Merck KGaA and Pfizer).
References
- 1.Shulman G. I. Cellular mechanisms of insulin resistance. J. Clin. Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puigserver P., Spiegelman B. M. Peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 3.Lazar M. A. How obesity causes diabetes: not a tall tale. Science (Washington, DC) 2005;307:373–375. doi: 10.1126/science.1104342. [DOI] [PubMed] [Google Scholar]
- 4.Frame S., Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Embi N., Rylatt D. B., Cohen P. Glycogen synthase kinase-3 from rabbit skeletal muscle. Eur. J. Biochem. 1980;107:519–527. [PubMed] [Google Scholar]
- 6.Woodgett J. R. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland C., Leighton I. A., Cohen P. Inactivation of glycogen synthase kinase-3β by phosphorylation; new kinase connections in insulin and growth factor signalling. Biochem. J. 1993;296:15–19. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross D. A. E., Alessi D. R., Cohen P., Andjelkovich M., Hemmings B. A. Inhibition of GSK3 by insulin mediated by protein kinase B. Nature (London) 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 9.McManus E. J., Sakamoto K., Armit L. J., Ronaldson L., Shpiro N., Marquez R., Alessi D. R. Role that phosphorylation of GSK3 plays in insulin and Wnt-signalling defined by knockin analysis. EMBO J. 2005;24:1571–1583. doi: 10.1038/sj.emboj.7600633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Coghlan M. P., Culbert A. A., Cross D. A. E., Corcoran S. L., Yates J. W., Pearce N. J., Rausch O. L., Murphy G. J., Carter P. S., Cox L. R., Mills D., Brown M. J., Haigh D., Ward R. W., Smith D. G., Murray K. J., Reith A. D., Holder J. C. Selective small molecule inhibitors of GSK-3 modulate glycogen metabolism and gene transcription. Chem. Biology. 2000;7:793–803. doi: 10.1016/s1074-5521(00)00025-9. [DOI] [PubMed] [Google Scholar]
- 11.Lochhead P. A., Coghlan M. P., Rice S. Q. J., Sutherland C. Inhibition of GSK3 selectively reduces G6Pase and PEPCK gene expression. Diabetes. 2001;50:937–947. doi: 10.2337/diabetes.50.5.937. [DOI] [PubMed] [Google Scholar]
- 12.Cline G. W., Johnson K., Regittnig W., Perret P., Tozzo E., Xiao L., Damico C., Shulman G. I. Effects of a novel GSK3 inhibitor on insulin stimulated glucose metabolism in ZDF (fa/fa) rats. Diabetes. 2002;51:2903–2910. doi: 10.2337/diabetes.51.10.2903. [DOI] [PubMed] [Google Scholar]
- 13.Nakae J., Barr V., Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barthel A., Schmoll D., Kruger K. D., Bahrenberg G., Walther R., Roth R. A., Joost H. G. Differential regulation of endogenous glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression by the forkhead transcription factor FKHR in H4IIE-hepatoma cells. Biochem. Biophys. Res. Commun. 2001;285:897–902. doi: 10.1006/bbrc.2001.5261. [DOI] [PubMed] [Google Scholar]
- 15.Vander Kooi B. T., Streeper R. S., Svitek C. A., Oeser J. K., Powell D. R., O'Brien R. M. The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J. Biol. Chem. 2003;278:11782–11793. doi: 10.1074/jbc.M212570200. [DOI] [PubMed] [Google Scholar]
- 16.Mora A., Lipina C., Tronche F., Sutherland C., Alessi D. R. Deficiency of PDK1 in liver results in glucose intolerance, impairment of insulin regulated gene expression and liver failure. Biochem. J. 2005;385:639–648. doi: 10.1042/BJ20041782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Kops G. J. P. L., Burgering B. M. T. Forkhead transcription factors: new insights into protein kinase B (c-akt) signalling. J. Mol. Med. 1999;77:656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 18.Nakae J., Park B.-C., Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a wortmannin-sensitive pathway. J. Biol. Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 19.Rena G., Prescott A. R., Guo S., Cohen P., Unterman T. G. Roles of the forkhead in rhabdomyosarcoma (FKHR)phosphorylation sites in regulating 14–3–3 binding, transactivation and nuclear targetting. Biochem. J. 2001;354:605–612. doi: 10.1042/0264-6021:3540605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finlay D., Patel S., Dickson L. M., Shpiro N., Marquez R., Rhodes C. J., Sutherland C. Glycogen synthase kinase-3 regulates IGFBP-1 gene transcription through the thymine-rich insulin response element: inhibition is required for full regulation of this promoter element by insulin. BMC Mol. Biol. 2004;5:15. doi: 10.1186/1471-2199-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becard D., Hainault I., Azzout-Marniche D., Bertry-Coussot L., Ferre P., Foufelle F. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]
- 22.Chakravarty K., Wu S. Y., Chiang C. M., Samols D., Hanson R. W. SREBP-1c and Sp1 interact to regulate transcription of the gene for PEPCK in the liver. J. Biol. Chem. 2004;279:15385–15395. doi: 10.1074/jbc.M309905200. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X. Y., Shibusawa N., Naik K., Porras D., Temple K., Ou H., Kaihara K., Roe M. W., Brady M. J., Wondisford F. E. Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nat. Med. 2004;10:633–637. doi: 10.1038/nm1050. [DOI] [PubMed] [Google Scholar]
- 24.Kim K. H., Song M. J., Yoo E. J., Choe S. S., Park S. D., Kim J. B. Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c. J. Biol. Chem. 2004;279:51999–52006. doi: 10.1074/jbc.M405522200. [DOI] [PubMed] [Google Scholar]
- 25.Patel S., Lochhead P. A., Rena G., Fumagalli S., Pende M., Kozma S., Thomas G. M., Sutherland C. Insulin regulation of IGF-binding protein-1 gene expression is dependent on mammalian target of rapamycin (mTOR), but independent of S6K activity. J. Biol. Chem. 2002;277:9889–9895. doi: 10.1074/jbc.M109870200. [DOI] [PubMed] [Google Scholar]
- 26.Granner D. K., Pilkis S. The genes of hepatic glucose metabolism. J. Biol. Chem. 1990;265:10173–10178. [PubMed] [Google Scholar]
- 27.Bosch F., Rodriguez-Gil J. E., Hatzoglou M., Gomez-Foix A. M., Hanson R. W. Lithium inhibits hepatic gluconeogenesis and PEPCK gene expression. J. Biol. Chem. 1992;267:2888–2893. [PubMed] [Google Scholar]
- 28.Henriksen E. J., Kinnick T. R., Teachey M. K., O'Keefe M. P., Ring D., Johnson K. W., Harrison S. D. Modulation of muscle insulin resistance by selective inhibition of glycogen synthase kinase-3 in zucker diabetic fatty rats. Am. J. Physiol. Endocrinol. Metab. 2003;285:E98–E105. doi: 10.1152/ajpendo.00346.2002. [DOI] [PubMed] [Google Scholar]
- 29.Nikoulina S. E., Ciaraldi T. P., Mudaliar S., Mohideen P., Carter L., Henry R. R. Potential role of GSK-3 in skeletal muscle insulin resistance of type 2 diabetes. Diabetes. 2000;49:263–271. doi: 10.2337/diabetes.49.2.263. [DOI] [PubMed] [Google Scholar]
- 30.Granner D. K., Andreone T. L. Insulin modulation of gene expression. Diabetes Metab. Rev. 1985;1:139–170. doi: 10.1002/dmr.5610010108. [DOI] [PubMed] [Google Scholar]
- 31.Massillon D., Chen W., Barzilai N., Prus-Wertheimer D., Hawkins M., Liu R., Taub R., Rossetti L. Carbon flux via the pentose phosphate pathway regulates the hepatic expression of the G-6-Pase and PEPCK genes in conscious rats. J. Biol. Chem. 1998;273:228–234. doi: 10.1074/jbc.273.1.228. [DOI] [PubMed] [Google Scholar]
- 32.Barzilai N., She L., Liu B. Q., Vuguin P., Cohen P., Wang J., Rossetti L. Surgical removal of visceral fat reverses hepatic insulin resistance. Diabetes. 1999;48:94–98. doi: 10.2337/diabetes.48.1.94. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien R. M., Granner D. K. PEPCK gene as a model of inhibitory effects of insulin on gene transcription. Diabetes Care. 1990;13:327–339. doi: 10.2337/diacare.13.3.327. [DOI] [PubMed] [Google Scholar]
- 34.Liang G., Yang J., Horton J. D., Hammer R. E., Goldstein J. L., Brown M. S. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 35.Latasa M. J., Griffin M. J., Moon Y. S., Kang C., Sul H. S. Occupancy and function of the −150 sterol regulatory element and −65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol. Cell Biol. 2003;23:5896–5907. doi: 10.1128/MCB.23.16.5896-5907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosmain Y., Dif N., Berbe V., Loizon E., Rieusset J., Vidal H., Lefai E. Regulation of SREBP-1 expression and transcriptional action on HKII and FAS genes during fasting and refeeding in rat tissues. J. Lipid Res. 2005;46:697–705. doi: 10.1194/jlr.M400261-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Murray J. T., Campbell D. G., Morrice N., Auld G. C., Shpiro N., Marquez R., Peggie M., Bain J., Bloomberg G. B., Grahammer F., Lang F., Wulff P., Kuhl D., P. C. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fiol C. J., Williams J. S., Chou C. H., Wang Q. M., Roach P. J., Andrisani O. M. A secondary phosphorylation of CREB at Ser-129 is required for the cAMP-mediated control of gene expression. A role for GSK3 in the control of gene expression. J. Biol. Chem. 1994;269:32187–32193. [PubMed] [Google Scholar]